The TREMs (triggering receptors expressed on myeloid cells) represent a family of 5 receptors clustered on murine chromosome 17. TREMs 1 and 2 affect various aspects of myeloid cell activation and development, including responsiveness to lipopolysaccharide and regulation of dendritic cell maturation, yet no inhibitory receptor has been demonstrated within this cluster. Here we characterize TLT-1 (TREM-like transcript-1), a putative inhibitory receptor within the TREM cluster that contains an extracellular V-set Ig domain, a proline-rich region, and an immune receptor tyrosine-based inhibitory motif (ITIM) in its cytoplasmic tail. To our knowledge, TLT-1 is the first ITIM-containing receptor carrying a potential Src homology 3 domain ligand. TLT-1 transcripts are abundant in bone marrow cells, but not in lymphocytes, and phosphorylated TLT-1 associates with SHP-1, suggesting that it is indeed an inhibitory receptor. Based on these characteristics, it is likely that TLT-1 regulates the signaling of the TREM family receptors.
Introduction
The triggering receptors expressed on myeloid cells (TREMs) are an emerging family of activating receptors expressed on various cells of the myeloid lineage.1-4 The TREMs represent a loose cluster (150 kb) on mouse chromosome 17, and the cluster's genomic organization is highly conserved on human chromosome 6 (Figure 1A). Although the family members possess only 30% amino acid identity, each member consists of a leader sequence, single V-set Ig domain, short cytoplasmic tail, and transmembrane domain containing a positively charged residue, suggesting interaction with a signaling polypeptide.3,4Biochemical analysis has demonstrated that of the 4 TREM sequences described to date, TREMs 1, 2, and 3 associate with the activating signaling chain DAP 12, and TREM 4 is predicted to as well.1-5
Genomic organization and expression of TREM-like genes in the human and mouse, and comparison of the predicted amino acid sequences of mouse and human TLT-1 sequences.
(A) Schematic showing the relationship of the human TREM cluster (top) and the mouse TREM cluster (bottom). (B) The exon structure of the TLT-1 gene is shown. The blackened area of exon 2 represents the deletion caused by the alternative splice event detected by RT-PCR in RAW264.7 cells and dendritic cell cultures. The asterisk denotes the premature stop in this smaller mRNA species. (C) The predicted amino acid sequences of murine TLT-1 (mTLT-1) and human TLT-1 (hTLT-1). The leader sequence is bolded, the cystines forming disulfide bonds within the Ig V-type domain are boxed with dotted lines, potential O-glycosolation sites are marked by “Ŝ” for serines or “Ť” for theonines, the transmembrane domain is underlined, polyproline-rich region is boxed, and the ITIM sequence is boxed in gray. Asterisk indicates stop codon. (D) Northern expression of TREM-like genes in bone marrow (lane 1), lung (lane 2), lymph node (lane 3), testis (lane 4), and thymus (lane 5). (E) Northern expression of TREM-like genes in RAW264.7 (lane 1) and P815 (lane 2).
Genomic organization and expression of TREM-like genes in the human and mouse, and comparison of the predicted amino acid sequences of mouse and human TLT-1 sequences.
(A) Schematic showing the relationship of the human TREM cluster (top) and the mouse TREM cluster (bottom). (B) The exon structure of the TLT-1 gene is shown. The blackened area of exon 2 represents the deletion caused by the alternative splice event detected by RT-PCR in RAW264.7 cells and dendritic cell cultures. The asterisk denotes the premature stop in this smaller mRNA species. (C) The predicted amino acid sequences of murine TLT-1 (mTLT-1) and human TLT-1 (hTLT-1). The leader sequence is bolded, the cystines forming disulfide bonds within the Ig V-type domain are boxed with dotted lines, potential O-glycosolation sites are marked by “Ŝ” for serines or “Ť” for theonines, the transmembrane domain is underlined, polyproline-rich region is boxed, and the ITIM sequence is boxed in gray. Asterisk indicates stop codon. (D) Northern expression of TREM-like genes in bone marrow (lane 1), lung (lane 2), lymph node (lane 3), testis (lane 4), and thymus (lane 5). (E) Northern expression of TREM-like genes in RAW264.7 (lane 1) and P815 (lane 2).
Recently, Bouchon et al uncovered the importance of this family in the regulation of multiple facets of the immune response.1,2,5These studies defined TREM 1 as an important mediator of septic shock1,5-7 and TREM 2 as playing a unique role in dendritic cell maturation and, therefore, T-cell priming.2 8 Taken together, these data demonstrate the intriguing potential for receptors of the TREM family to be key regulators of both the innate and adaptive immune response. Despite the recent advances in TREM immunobiology, TREM ligands and mode of regulation remain ill-defined.
Here we report the initial characterization of a putative inhibitory receptor within the TREM locus. This gene represents the only potential TREM regulator identified thus far, suggesting it may play a critical role in the regulation of both innate and adaptive immunity.
Animal care was provided in accordance with the procedures outlined in “A Guide for the Care and Use of Laboratory Animals” (National Institutes of Health Publication No. 86-23, 1985).
Study design
Sequence information
Sequence information and genomic structural characteristics were determined using the Celera Discovery System. In some cases public information was used and the sequences confirmed by the Celera Discovery System. The validity of both the TREM-like and non–TREM-like murine sequences of the cluster was confirmed using reverse transcription–polymerase chain reaction (RT-PCR) and the following primer pairs: TREM 1: 5′-gagcttgaaggatgaggaag-3′, 5′-gctcctcctgtgaaatagac-3′; TREM 2: 5′-cccaagcttaacaccacggtgctgcagg-3′, 5′-cgcggatcctgactggacttaagctgta-3′; TREM 3: 5′-gttagcacaccaggaaggag-3′, 5′-ctgtttctcagagactccctg-3′; FJL13693: 5′-atggatggatttgtcacgac-3′, 5′-aaatccagccatcatcacag-3′.
Epitope tagging
Full-length murine TLT-1 (TREM-like transcript-1; GenBank accession number AY078502) was generated by RT-PCR using Platinum HiFi-Supermix (Gibco-BRL, Grand Island, NY) with bone marrow cDNA as a template. The resulting cDNA was epitope tagged using the TOPO V5 epitope tag system from Invitrogen (Carlsbad, CA). The primers were 5′-agaacctactactgcccag-3′, 5′-gccaatatgtaatgacggtag-3.
Tissue and cell line expression
The expression of TLT-1 in normal tissue and cell lines was determined using RT-PCR. Total RNA was made using Trizol (Gibco-BRL) according to the manufacturer's specification. First-strand synthesis was achieved using the Superscript cDNA synthesis kit (Gibco-BRL). PCR cycles were as follows: 95°C for 2 minutes and 30 cycles, 94°C, 30 sec; 55°C, 30 sec; and 72°C, 1 minute. For Northern analysis, 30 μg RNA was used per lane according to the methods as decribed.9
Transfections and immunoprecipitation
Phosphorylation and protein-protein interactions were analyzed using HEK293T cells as described.10
Results and discussion
In light of our understanding of the killer immunoglobulinlike receptor (KIR), leukocyte Ig-like receptor (LIR), and Ly49 gene families, the existence of a family of activating receptors suggested the presence of negative regulatory receptors within the TREM locus.11 Analysis of the murine TREM cluster revealed a putative regulatory receptor we have termed TLT-1, just telomeric to TREM 2. This cDNA encodes a single open reading frame predicting a 322–amino acid protein containing a leader sequence and a single V-set Ig domain (Figure 1). In stark contrast to the TREMs, the TLT-1 functional domain appears to be in the carboxy-terminus (Figure1C).5 The cytoplasmic region of TLT-1 contains an immunoreceptor tyrosine-based inhibitory motif (ITIM), implying the ability to mediate inhibition through the recruitment of Src homology (SH) 2 domain-containing protein tyrosine phosphatases. In addition, TLT-1 contains a polyproline-rich segment, suggesting that it has the ability to interact with SH3 domain-containing targets. The apparent human ortholog shows 70% identity at the amino acid level with murine TLT-1 and is similarly located within the TREM cluster (Figure 1A).
Given the similarities between TLT-1 and the TREMs, we evaluated TLT-1 expression in relation to TREM 1 and 2. Preliminary screening by RT-PCR revealed TLT-1 message in murine bone marrow, brain, liver, peritoneal monocytes, P815 mastocytoma cells, and RAW264.7 macrophages. TLT-1 transcript was not seen in spleen, lung, or thymus (data not shown). In RAW and dendritic cells, RT-PCR demonstrated a minor mRNA species lacking 235 bp (Figure 1B). This mRNA predicts a truncated polypeptide with no apparent homology to the TREMs. In contrast to RT-PCR, Northern analysis demonstrated significant 1.2-kb TLT-1 mRNA only in bone marrow (Figure 1D). The 1.2-kb mRNA confirmed that the nucleotide sequence of TLT-1 represents a full-length transcript. Similar to TLT-1, TREM 1 and 2 mRNA also was found predominantly in bone marrow, suggesting significant coexpression of TLT-1 and the activating TREM. TREM 2, but not TLT-1 or TREM 1, also is highly expressed in the RAW cells (Figure 1E).
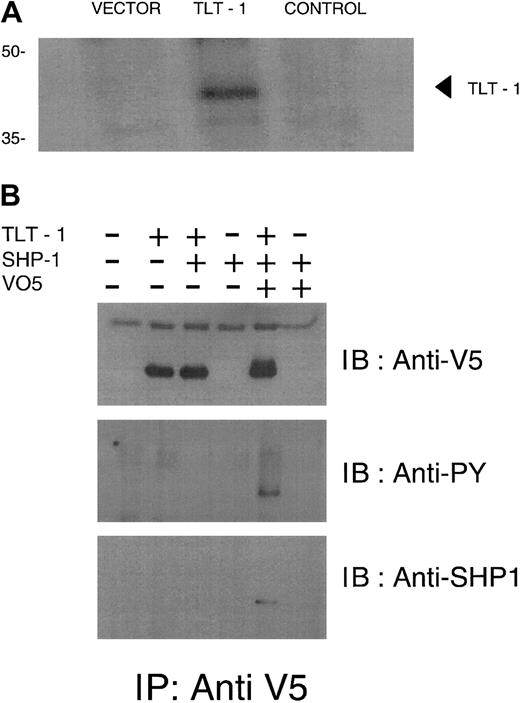
Immunoprecipitation and Western blot analysis of surface biotinylated cells expressing epitope-tagged TLT-1 confirmed TLT-1 surface expression by revealing biotin-labeled receptor of the expected molecular weight (Figure 2A). Using nonreducing buffer, some TLT-1 protein migrates at 75 kDa, suggesting it can exist as a homodimer on the cell surface (data not shown).
TLT-1 is a receptor expressed in myeloid cells and binds SHP-1 when phosphorylated.
(A) Biotinylation of surface proteins in TLT-1–transfected cells. Vector, vector alone; TLT-1, V5 epitope-tagged TLT-1; control, full-length TLT without epitope tag. Lysates were immunoprecipitated (IP) with anti-V5 antibody and immunoblotted (IB) with streptavidin. (B) HEK293T cells were transfected as indicated. Lysates were immunoprecipitated with anti-V5 then immunoblotted with anti-V5 (top), antiphosphotyrosine (middle), or anti–SHP-1 (bottom).
TLT-1 is a receptor expressed in myeloid cells and binds SHP-1 when phosphorylated.
(A) Biotinylation of surface proteins in TLT-1–transfected cells. Vector, vector alone; TLT-1, V5 epitope-tagged TLT-1; control, full-length TLT without epitope tag. Lysates were immunoprecipitated (IP) with anti-V5 antibody and immunoblotted (IB) with streptavidin. (B) HEK293T cells were transfected as indicated. Lysates were immunoprecipitated with anti-V5 then immunoblotted with anti-V5 (top), antiphosphotyrosine (middle), or anti–SHP-1 (bottom).
TLT-1 contains an ITIM and, therefore, might recruit a protein phosphatase such as SHP-1 when phosphorylated. To test this possibility, HEK293T cells were transfected with TLT-1 alone or together with SHP-1, treated with pervanadate, then immunoprecipitated with anti-V5. The resulting immunoblots were then serially probed with antiphosphotyrosine, anti–SHP-1, then anti-V5. These experiments demonstrated that once phosphorylated, TLT-1 interacts with SHP-1 (Figure 2B). Based on these results, TLT-1 clearly has the potential for inhibition.
Taken together, our findings provide several lines of evidence suggesting that TLT-1 is a regulatory component of the TREM cluster. First, the homology of TLT-1 with the TREM proteins indicates a common ancestor and possibly even similar ligands. Second, in our analysis, the pattern of TLT-1 expression overlaps with TREM 2 and is identical to TREM 1, making them potential targets for TLT-1–mediated inhibition. Third, TLT-1 possesses the physical characteristics necessary for inhibitory signaling, most importantly, the ability to recruit SHP-1.
Regardless of the ultimate role of TLT-1, the identification of an inhibitor within the TREM family adds the TREM to the growing list of paired immune receptor systems.11 Although a member of the ever-growing superfamily of inhibitory receptors, TLT-1 appears to be unique in that it contains cytoplasmic motifs for the recruitment of both SH2 and SH3 domain-containing proteins. The existence of a proline-rich segment in the cytoplasmic domain of a receptor is rare. Although we have not yet identified TLT-1 binding partners other than SHP-1, it is tempting to speculate that this proline-rich domain may be involved in recruiting the kinases necessary to mediate phosphorylation of TLT-1. Alternatively, the proline-rich domain may be involved in bringing SH3 domain-containing phosphoproteins into proximity so they can be dephosphorylated by TLT-1–bound SHP-1. Considering that TLT-1 is one of the 3 TREM-like transcripts that is conserved between humans and mice and is the sole inhibitory receptor within the TREM cluster, it is expected to play a prominent role in the regulation of myeloid cell function.
By acceptance of this article, the publisher or recipient acknowledges the right of the United States government to retain a nonexclusive, royalty-free license in and to any copyright of the article. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States government.
Prepublished online as Blood First Edition Paper, July 12, 2002; DOI 10.1182/blood-2002-02-0523.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Daniel W. McVicar, NCI-FCRDC Building 560/Room 31-93, Frederick, MD 21702; e-mail:mcvicar@nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal