The αIIb/β3-integrin receptor is present at high levels only in megakaryocytes and platelets. Its presence on platelets is critical for hemostasis. The tissue-specific nature of this receptor's expression is secondary to the restricted expression of αIIb, and studies of the αIIb proximal promoter have served as a model of a megakaryocyte-specific promoter. We have examined the αIIb gene locus for distal regulatory elements. Sequence comparison between the human (h) and murine (m) αIIb loci revealed high levels of conservation at intergenic regions both 5′ and 3′ to the αIIb gene. Additionally, deoxyribonuclease (DNase) I sensitivity mapping defined tissue-specific hypersensitive (HS) sites that coincide, in part, with these conserved regions. Transgenic mice containing various lengths of the hαIIb gene locus, which included or excluded the various conserved/HS regions, demonstrated that the proximal promoter was sufficient for tissue specificity, but that a region 2.5 to 7.1 kb upstream of the hαIIb gene was necessary for consistent expression. Another region 2.2 to 7.4 kb downstream of the gene enhanced expression 1000-fold and led to levels of hαIIb mRNA that were about 30% of the native mαIIb mRNA level. These constructs also resulted in detectable hαIIb/mβ3 on the platelet surface. This work not only confirms the importance of the proximal promoter of the αIIb gene for tissue specificity, but also characterizes the distal organization of the αIIb gene locus and provides an initial localization of 2 important regulatory regions needed for the expression of the αIIb gene at high levels during megakaryopoiesis.
Introduction
It is known that αIIb/β3 is a prototypic member of the integrin superfamily of cell surface receptors that include the receptors for such ligands as von Willebrand factor, fibrinogen, vitronectin, fibronectin, and collagen.1 αIIb/β3 is found at high levels only on the surface of megakaryocytes and platelets.2 The approximately 80 000 copies of αIIb/β3 found on the platelet surface make it the most abundant receptor present, representing half of the total molar number of receptors.3 Normal activity of this receptor is critical for platelet function during thrombus development.4
The tissue specificity of the αIIb/β3 receptor is determined by αIIb. Whereas β3 is expressed as part of the αv/β3 vitronectin receptor on many tissues, αIIb expression is limited to hematopoietic tissue and achieves high levels of expression only in developing megakaryocytes.5,6 In both the human and mouse genomes, there exists a single αIIb gene (ITGA2B), which contains 30 exons and spans a distance of about 18 kb along chromosomes 17 and 11, respectively.7,8 In vitro studies in megakaryocytic cell lines have defined important tissue-specific regulatory domains in the immediate 5′-flanking region, including proximal (−55 bp upstream from the transcriptional start site) and distal (−480 bp) GATA-1 DNA-binding sites, each associated with an adjacent Ets-binding site,9,10 as well as a potential silencer region located between the 2 GATA/Ets motifs.11
These in vitro studies, which determined that about 600 bp of 5′ flanking of the αIIb gene proximal promoter region is sufficient to support tissue-specific expression, have been consistent with in vivo transgenic murine studies. In those reports, as little as 787 bp of the 5′-flanking region of the αIIb gene was used to drive thymidine kinase (TK) toxigene expression in the megakaryocytes of transgenic mice.12 Using ganciclovir as the toxic agent, these studies demonstrated TK expression predominantly in the bone marrow. In these studies, the relative level of expression of the reporter gene was not determined, as even low levels of TK expression would make cells susceptible to ganciclovir. Hence, these in vivo studies, although indicating domains important for tissue and hematopoietic lineage–restricted expression, did not define the specific extent of regulatory domains required to constitute a complete, functional αIIb gene locus, capable of withstanding position effects and hence generating high mRNA and protein levels.
In this regard, we wished to pursue the molecular basis of αIIb gene expression in vivo, by focusing not only on the proximal promoter region, but also examining more distal intergenic regions in the αIIb gene locus. The difficulty in examining these intergenic regions lies in their broad size. One technique for localizing regulatory domains within a large expanse of DNA sequence is phylogenetic footprinting. In this approach, nucleotide sequences of 2 or more species are compared. Functionally important intergenic regulatory regions are less likely to diverge than the remaining intergenic sequence and hence are highlighted by their own conservation.13,14 A second approach for defining functional cis-regulatory elements takes advantage of the fact that regulatory regions are often embedded in less compacted chromatin as reflected in their greater susceptibility to digestion by general endonucleases such as deoxyribonuclease (DNase) I.15,16 For example, in the upstream region of the β-globin gene locus, there are multiple DNase I hypersensitive (HS) sites that comprise a region essential for high-level expression of the various β-globin genes.17-19 A third approach involves transgenic animals containing various lengths of a gene and its flanking regions. Such studies can define important distal elements that regulate tissue specificity, copy number dependency, and levels of expression relative to the native gene.18,20-22 These intergenic regions are often not only functionally important, but also conserved between species. For example, transgenic studies of the platelet basic protein (PBP)/platelet factor 4 (PF4) double gene locus, identified a region between 2.5 and 4.5 kb upstream of the human PBP gene that appears to be an important enhancer of PBP expression and that is also highly conserved between the human and mouse PBP/PF4 gene loci.22
In the studies described below, we use phylogenetic footprinting to compare the human and mouse flanking sequences and define a series of intergenic conserved regions. Several of these regions also coincide with tissue-specific, DNase I HS sites. Using these potential functional domains as a guide, transgenic mice were made with segments of the human (h) αIIb gene locus, encompassing increasing 5′ and 3′ lengths. These studies collectively define a limited domain surrounding the hαIIb gene that is required for achieving consistent high levels of hαIIb message levels and detectable surface expression of the hαIIb receptor on murine platelets. Thus, these studies represent the first localization of important distal regulatory elements flanking the megakaryocyte-specific αIIb gene.
Materials and methods
Phylogenetic footprinting and sequence analysis
Sequence determination and analysis of the human and murine αIIb gene locus were performed as previously described.8,23 Generated sequences were submitted to the GenBank public database at www.ncbi.nih.gov, and include the following accession numbers: AF170316, AF169829, AF160252, and AF489555. Using these original sequence files together with both complete and incomplete BAC genomic sequence files available at GenBank (accession numbers: AC007722, AC003043, AC025326, a murine chromosome 11 BAC clone-RP11-621L13) and Celera Databases (scaffold number: GA_x5J8B7W82RK: 6500001.6874571; a partially complete mαIIb locus scaffold), we compiled composite sequences of both the human and murine sequences, encompassing for each a 200-kb sequence domain. These files were then used for a homology comparison of orthologous regions using the nucleotide sequence comparison program, Visual Tools for Alignment (VISTA/AVID [www-gds.lbl.gov/vista]). Length of comparison was set to 100 bp with a minimum of 50% match. Specific regions within the 200-kb region were compared using the Basic Local Alignment Search Tool (BLAST).24 Specific subregions were also compared to the public expressed sequence tag (EST) database at www.ncbi.nih.govusing BLAST. A 10-kb subregion extending from +7.8 kb within the αIIb 3′-intergenic domain was also analyzed using GeneMachine gene prediction software available athttp://genome.nhgri.nih.gov/genemachine. This program allows users to query multiple exon and gene prediction programs in an automated fashion and incorporates BLAST analysis as well in assessing final predictions. Additionally, the Transcript Assembly Program (TAP) available at http://sapiens.wustl.edu/∼zkan/TAP/ was used to delineate gene structures using genomically aligned EST sequences. Analysis of aligned conserved sequence regions for consensus transcription factor–binding sites, was done using both the rVISTA program (www.gds.lbl.gov/vista) and the Transcription Element Search Software (TESS) developed by the University of Pennsylvania (PENN) Computational Biology Informatics Laboratory (http://www.cbil.upenn.edu/).
DNase I HS studies
Human erythroleukemia (HEL) and Children's Hospital Research Foundation (CHRF) 288-11 cells were the 2 megakaryocytic αIIb-expressing cell lines25,26 used in these DNase I HS site studies. The fibroblast line HeLa27 and the gastric carcinoma cell line SNU-128 were studied as non-αIIb–expressing lines. HEL, HeLa, and SNU-1 cells were obtained from American Type Culture Collection (Rockville, MD). CHRF cells were obtained from Dr Michael Liebman (University of Cincinnati, OH). HEL, SNU-1, and HeLa cells were cultured in RPMI 1640, 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and l-glutamine (Invitrogen, Carlsbad, CA). CHRF cells were cultured as above but with 20% FBS. All cell lines were split 24 hours before each DNase I HS experiment. Preparation of cell nuclei for DNase I treatment was as previously described.29 Nuclei from 1 × 108 cells were centrifuged for 10 minutes at 500g and were suspended in 4.5 mL DNase I buffer (10 mM Tris [tris(hydroxymethyl)aminomethane], pH 7.5, 10 mM NaCl, 5 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride, 5 mM sodium butyrate, and 1 mM CaCl2). Aliquots of 500 μL nuclei were added to 500 μL DNase I buffer containing DNase I enzyme (Invitrogen) varying from 0 to 3.4 μg/mL. The tubes were gently mixed, then placed at 37°C for 5 minutes, and then replaced back on ice. Genomic DNA was recovered and digested further with either EcoRI orBamHI restriction enzymes for Southern blot analysis as previously described.29,30 The probes used for detection were produced via polymerase chain reaction (PCR) amplification. Primers used to generate these probes were the following sense/antisense pairs: αIIb exon 1 (P1)7: 5′-CCTGTGGAGGAATCTGAA-3′/5′-TCCTGCTCTCTCCCAATAC-3′; αIIb exon 4 (P2)7: 5′-CAATCGGGGGCAGGGACAC-3′/5′-CAAGCCGTCGCGAGTGGG-3′; and αIIb exon 30 (P3)7: 5′-GAGTACAGTGGGCTTCATGTTCT-3′/5′-CCCTGGCAGTGACTCTCTCGTTCA-3′. A previously described genomic λ clone containing the hαIIb locus served as template for the reactions.23
Transgenic constructs
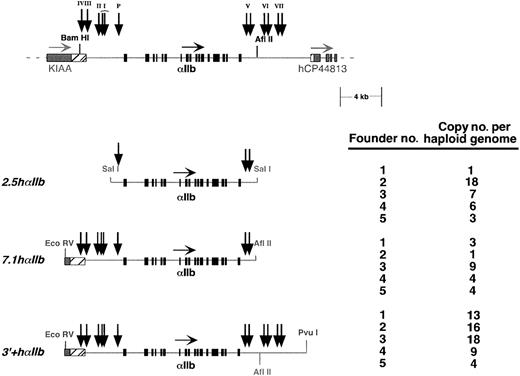
The SalI fragment used to make the2.5hαIIb transgenic animal lines was isolated from a λDash (Stratagene, La Jolla, CA) bacteriophage clone subcloned from P1(clone no. 1147), an hαIIb-containing P1 clone previously described.23 This construct extends from the λDash vector polylinker SalI site through 2.5 kb of the 5′-flanking region, 17.35 kb of coding exon/intron sequence, and 2.2 kb of the 3′-flanking sequence up to the 3′-polylinkerSalI site and has a total length of 22 052 bp. TheEcoRV/AflII restriction fragment used to make the7.1hαIIb transgenic animal lines was isolated from an hαIIb pWE15 cosmid clone. This fragment extends from a vector pWE15 polylinker EcoRV site through 7.1 kb of the 5′-flanking region to 2.1 kb downstream of the hαIIb gene stop codon, ending at an endogenous AflII site with a total length of 26 559 bp. The final transgenic construct3′+hαIIb is an EcoRV/PvuI restriction fragment, derived from the same cosmid clone described above. It is identical with the 7.1hαIIbconstruct, except that the 3′-flanking region is 5.2 kb longer, extending to a polylinker PvuI site in pWE15 with a total length of 31 759 bp.
Generation and initial analysis of transgenic animals
Transgenic mice were generated by pronuclear injections following standard methods at the PENN Transgenic Mice Core Facility. Positive founder animals were detected by polymerase chain reaction (PCR) analysis of tail genomic DNA using multiple human-specific primer pairs for hαIIb.7 Murine PF4 genomic primers22 were used as a control. These sense/antisense primer pairs were as follows: hαIIb-exon 14: 5′-AGGCCTCTGTCCAGCTAC-3′/5′-GCCATTCCAGCCTCCGTG-3′; andmPF4: 5′-GTCCAGTGGCACCCTCTTGA-3′/5′-AATTGACATTTAGGCAGC-3′.
Positive founder lines with intact hαIIb genes by Southern and PCR analysis were further characterized for copy number by Southern blot. Tail genomic DNA was digested with EcoRI and size separated on a 0.8% (wt/vol) agarose gel. The Southern blot and copy number determination procedures were as described before.22 23
Tissue and platelet RT-PCR analysis
RNA isolation and reverse transcriptase (RT)–PCR procedures for all mRNA expression analyses, as well as a list of the 11 tissues examined, have been described before in our previous transgenic studies.22 Briefly, using about 0.1 μg total platelet RNA, or 1 μg RNA from other tissues, RT-PCR was done using the SuperScript II Reverse Transcriptase Kit (Invitrogen) with the following sets of sense/antisense primer pairs:hαIIb7: 5′-AGGCCTCTGTCCAGCTAC-3′/5′-G CCATTCCAGCCTCCGTG-3′;mαIIb8: 5′-TCAAGACTCCCTGAATCCAACAC-3′/5′-GGGCTCCTCCAGTCTCTTCT-3′;mPBP22: 5′-GCCTGCCCACTTCATAACCTC-3′/ 5′-GGGTCCAGGCACGTTTTT-3′;mPF422: 5′-GTCCAGTGGCACCCTCTTGA-3′/5′-AATTGACATTTAGGCAGCTGA-3′;mHPRT31: 5′-CACAGGACTAGAACACCTGC-3′/5′-GCTGGTGAAAAGGACCTCT-3′;mKIAA05538: 5′-GACCCAAAGGTGCTTGTAAT-3′/5′-GAAAACTACCTCCAGGATGG-3′.
Control experiments included studies where no RT was added and others treated with RNase A (Sigma, St Louis, MO) prior to the RT step, were done to ensure that PCR bands seen in the experiments were not generated via pre-RT sources of DNA.22 Also, control experiments to control for genomic DNA contamination were done where the RNA was treated with DNase I (Invitrogen), as previously described.22 Quantitation results from DNase I–treated versus DNase I–untreated samples did not vary significantly; hence, all RT-PCR data presented below used untreated RNA samples.
To quantitate the level of hαIIb transgene mRNA expression in platelets relative to endogenous mαIIb, we used methods previously described.18,20,22 Briefly, PCRs from first-strand cDNA of platelet RNA were set up using the primers described above and the cycle number at which both genes were in their linear range of amplification. Antisense primers were 5′ labeled with a fluorescent dye “Cy-5” (Oligos Etc, Wilsonville, OR). Preliminary experiments were done to determine the detectable linear range of amplification for the native mαIIb gene and the hαIIb transgene for each founder line. Thereafter, a 100-μL PCR mixture was divided into six 15-μL aliquots, one for each cycle beginning 3 cycles below midpoint of the detectable linear range and extending to 2 cycles above it, prior to initiating the PCR. PCR was performed at 94°C for 2 minutes, followed by cycles at 94°C for 25 seconds, 62°C for 36 seconds, 72°C for 55 seconds in a PTC-100 Programmable Thermal Controller (MJ Research, Waltham, MA). For each consecutive single cycle change, one of the PCR tubes for hαIIb and mαIIb reactions was removed from the thermocycler and placed on ice. All of the samples were then run on a 12-well 10% acrylamide Ready Gel (Bio-Rad Laboratories, Hercules, CA), and bands were detected with a STORM imaging system red light laser (Molecular Dynamics, Sunnyvale, CA). The detected bands were then analyzed using ImageQuant PhosphorImager software (Molecular Dynamics).22 The log of the signal intensity of each band was calculated. The log value difference for each band between 2 consecutive cycles was calculated, with the expected log difference being 0.3 units (log102). Those values, which fell into a range of 0.2- to 0.4-log difference units/cycle, were considered within the linear range of amplification. The raw signal intensity of those human PCR bands within the linear range was normalized to that of the mouse product, also within the linear range at the same cycle. The signals were further normalized for differences in the ability of the primer/PCR conditions to amplify hαIIb and mαIIb from equal molar amounts of the appropriate cDNA control template as described before.22These normalized values were used to compare transgene expression level among the different founder lines.
Protein detection
Platelet-rich plasma (PRP) from human and mice blood was isolated as described.22 For these studies, prostaglandin E1 (Sigma) was added to a final concentration of 1 μM prior to spinning down the platelets at 800g for 10 minutes at room temperature. The pellets were washed twice in platelet buffer (PB; 134 mM NaCl, 3 mM KCl, 0.3 mM NaH2PO4, 2 mM MgCl2, 5 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 5 mM glucose, 0.1% NaHCO3, pH 6.5) plus 1 mM EGTA (ethyleneglycoltetraacetic acid) and resuspended in PB without EGTA. The platelets were lysed by freezing and thawing twice. The protein concentration of each lysate was determined by the Pierce BCA Protein Assay kit according to the manufacturer's instructions (Pierce, Rockford, IL). Twenty micrograms of the total platelet protein was electrophoresed on a 4% to 12% gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue. A duplicate SDS-PAGE gel was transferred to a polyvinylidene difluoride (PVDF) Immobilon-P Transfer Membrane (Millipore, Bedford, MA). The hαIIb protein was detected using MAB1990 (Chemicon, Temecula, CA), a mouse anti-hαIIb monoclonal antibody. Western blot signals were visualized by phosphoimaging of the enhanced chemiluminescence (Perkin-Elmer, Wellesley, MA) signal on the STORM and quantitated using Imagequant PhosphorImager software. For flow cytometric studies, 1 × 107 wild-type and transgenic mice platelets and human platelets were prepared as previously described,32using the monoclonal MAB1990 antibody32 as the primary antibody and a fluorescein isothiocyanate (FITC)–labeled, goat antimouse IgG (Sigma) as the secondary antibody.
Results
Phylogenic footprinting of the αIIb gene locus
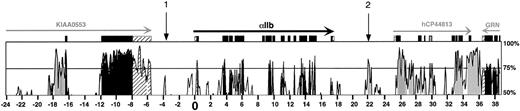
To search for upstream and downstream distal, intergenic regulatory elements in the αIIb gene locus, we used a cross-species sequence comparison to define evolutionarily constrained regions that may reflect important functional domains. We previously reported on the sequencing of an approximate 30-kb region surrounding the human and murine αIIb gene loci7,8 23 (GenBank accession numbers:AF170316, AF169829, AF160252, and M33320). In the present study, we have extended our sequencing of the murine locus (accession number:AF489555) and used our collective sequences together with partially complete public and private database sequences (accession numbers: human αIIb locus, AC007722 and AC003043; murine αIIb locus,AC025326; and Celera-scaffold number, GA_x5J8B 7W82RK:6500001.6874571) to compile an ordered 200-kb stretch of both the human and murine sequences surrounding their αIIb gene loci. We then used the VISTA/AVID global alignment program to examine this extended region for conserved domains. Figure 1 shows a subregion of this original 200-kb analysis centered on a 62-kb region surrounding the αIIb gene. (The complete 200-kb comparison is included in the supplemental Figure 1 online.) The data clearly show that conserved homologous regions exist not only within the coding regions of the multiple genes found within the compared domains, but also intergenically in the noncoding domains upstream and downstream of these genes.
VISTA/AVID global alignment of the αIIb gene locus.
The homology between the human and mouse αIIb gene locus is shown for homology 50% or more using the VISTA program. The x-axis defines 62 kb of human sequence, whereas the vertical axis indicates the percentage homology between this and the orthologous murine sequence. The 75% homology level, a point halfway between the minimal and maximal homology shown, is denoted. The αIIb gene is located near the center of the figure in a 5′ → 3′ orientation. Annotations for gene nomenclature are indicated and also have their 5′ → 3′ transcriptional direction and exon structure shown. Numbering in kilobase pairs is shown beginning at the transcriptional start site of the αIIb gene (position zero, “0”). The sequences upstream of the αIIb genes' start site are indicated as negative numbers and those downstream as positive numbers. Sequences that are 70% or more identical over a continuous 100-bp stretch have shaded peaks. Stretches of homology extending less than 100 bp are shown but are not shaded. Homology within coding regions is shown in black, within the 5′- or 3′-untranslated regions is striped, and within the intergenic and intronic, noncoding region is shown in gray. The 2 vertical arrows, “1” and “2,” highlight homology regions of potential physiologic importance discussed in this paper. Predicted exons for the newly defined hCP44813 gene are shown above the gray-shaded homology matches, formerly thought to be areas of noncoding sequence. Gray-filled exons within hCP44813 denote predicted alternatively spliced exon locations. GRN indicates Granulin gene.
VISTA/AVID global alignment of the αIIb gene locus.
The homology between the human and mouse αIIb gene locus is shown for homology 50% or more using the VISTA program. The x-axis defines 62 kb of human sequence, whereas the vertical axis indicates the percentage homology between this and the orthologous murine sequence. The 75% homology level, a point halfway between the minimal and maximal homology shown, is denoted. The αIIb gene is located near the center of the figure in a 5′ → 3′ orientation. Annotations for gene nomenclature are indicated and also have their 5′ → 3′ transcriptional direction and exon structure shown. Numbering in kilobase pairs is shown beginning at the transcriptional start site of the αIIb gene (position zero, “0”). The sequences upstream of the αIIb genes' start site are indicated as negative numbers and those downstream as positive numbers. Sequences that are 70% or more identical over a continuous 100-bp stretch have shaded peaks. Stretches of homology extending less than 100 bp are shown but are not shaded. Homology within coding regions is shown in black, within the 5′- or 3′-untranslated regions is striped, and within the intergenic and intronic, noncoding region is shown in gray. The 2 vertical arrows, “1” and “2,” highlight homology regions of potential physiologic importance discussed in this paper. Predicted exons for the newly defined hCP44813 gene are shown above the gray-shaded homology matches, formerly thought to be areas of noncoding sequence. Gray-filled exons within hCP44813 denote predicted alternatively spliced exon locations. GRN indicates Granulin gene.
In the 5′-intergenic region between the αIIb and theKIAA0553 genes, there are 2 conserved domains. The first is a double peak between positions O and −2 kb (Figure 1), and this double peak is consistent with the previously reported homology between the rat and human αIIb immediate promoter regions.33Further upstream is a small conserved domain, positioned at about 4 kb (Figure 1, vertical arrow “1”). Analysis of this 5′-intergenic human sequence for Alu consensuslike sequences34demonstrated that there were several Alu repeat elements within this 5′-intergenic region, but these do not overlap with the conserved domains described above (data not shown).
A homology comparison of the 3′-intergenic region between the αIIb gene and the previously reported downstream gene,Granulin,8,23 defined several noncoding conserved loci (Figure 1). The region most proximal to the αIIb gene is positioned 18.0 to 18.6 kb downstream of the hαIIb start site (Figure 1) and is about 65% conserved. Further downstream from this homologous region is a conserved domain extending from 21.5 to 23.3 kb downstream of the hαIIb start site or 4.3 to 6.1 kb downstream of the αIIb stop codon (Figure 1, vertical arrow “2”). This domain is conserved up to 85% between human and murine sequences. Downstream of this conserved region is a highly conserved domain between +26 to +36 kb. Analysis using the GeneMachine program, a gene prediction program,35 suggested that this homologous region defines another gene. BLAST comparison of this about 10-kb region against the public EST database yielded multiple matches for mRNAs expressed in diverse tissues, but predominantly brain and heart tissue in the human, rat, and mouse databases. BLAST comparison of the highest scoring ESTs from the public database against the Celera human protein database yielded a 99% match with hCP44813, a Celera-defined protein of unknown function that corresponds to a 1500-bp Celera human mRNA transcript hCT21474. Using the TAP36 to analyze this domain, we showed that the Celera human transcript subdivides with perfect identity into 9 separate domains in the human αIIb sequence, each flanked by canonical splice junctions (data not shown) and each conserved in the murine sequence. Hence, we conclude that a previously unrecognized gene coding for the hCP44813 protein lies 8.5 kb downstream of the αIIb gene in the same 5′ → 3′ orientation as the αIIb gene.
DNase I HS mapping of the 5′- and 3′-intergenic regions of αIIb
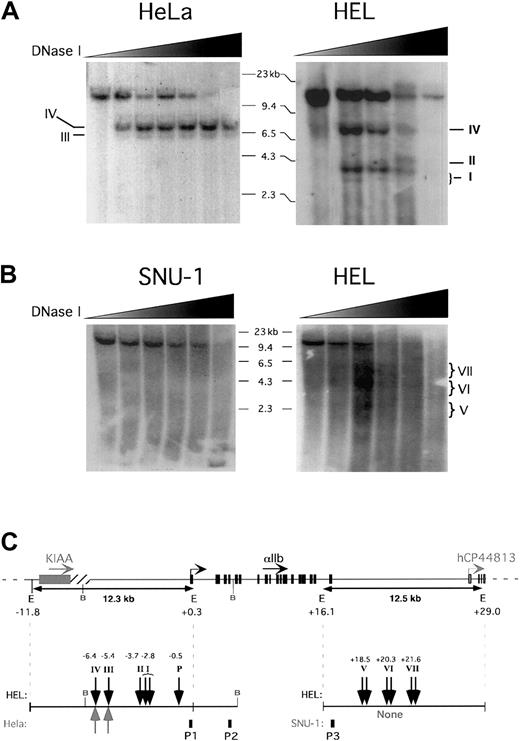
As an additional method for defining distal cisregions that could potentially be involved in controlling the platelet-specific expression of the αIIb gene, we also carried out DNase I sensitivity mapping on chromatin from active (megakaryocytic) hαIIb-expressing tissue versus those from inactive (nonmegakaryocytic) nonexpressing tissue. Nuclei were isolated and subjected to limited DNase I digestion followed by EcoRI restriction and analysis by Southern blot. A series of bands were seen on genomic Southern blots with a probe P1, covering αIIb exon 1 (Figure 2A,C). The 12.3-kb, full-lengthEcoRI band disappears at increasing DNase I concentrations in both the megakaryocytic and nonmegakaryocytic cell lines (Figure2A). A band representing a DNase I HS at position −6.4 kb upstream of the αIIb gene (HS IV), that was common to both cell lines, was identified, and increased in signal strength with higher concentrations of DNase I and then was digested away at the highest concentrations. The band for HS III, located directly below HS IV in the HeLa sample, is absent at the lowest DNase I concentration then subsequently appears and persists up to the highest DNase I concentration. Note also that HS III observed in the HeLa cell blot and not in the megakaryocytic HEL cell blot was actually also observed in HEL cell studies in other experiments (data not shown). An additional series of bands, ranging from about 3.1 to about 4.0 kb in length (HS I-II), appeared in the HEL cell study in a DNase I–dependent fashion, but were absent in the HeLa study (Figure 2A,C). Here, we describe HS I as a composite of 2 closely localized bands. The lowest molecular weight band is 3.1 kb and appears only transiently in HEL (second lane from left), but not at the lowest DNase I concentration, indicating its dependence on DNase I. The second band, which is slightly longer and more intense (∼3.7 kb), appears at the same point as the 3.1-kb band and is eventually digested away at the higher DNase I concentrations. Consistent results were observed using an additional restriction enzyme, BamHI, with HEL, CHRF (megakaryocytic), and HeLa cell lines and using a probe P2, covering exon 4 (data not shown, but see Figure 2C). TheBamHI studies also showed an additional tissue-specific HS site in the immediate promoter region (HS “P” in Figure 2C). It is of interest that the tissue-specific HS domains I/II and “P” overlap with the conserved 5′-intergenic regions at about 3.7 and 0.5 kb, respectively, shown in Figure 1. Figure 2, panels B and C, show DNase I HS mapping of the 3′-intergenic domain of the αIIb gene. HEL nuclei and nuclei from SNU-1, a human gastric, non-αIIb–expressing cell line, were treated as above. SNU-1 cells were used over HeLa cells because they were suspension rather than adherent cells. Analysis by Southern blot studies using probe P3 based on hαIIb exon 30 (Figure 2B) revealed in the SNU-1 cells the expected 12.5 kb. This band decreased in intensity at increasing DNase I concentrations, but no additional bands were seen (Figure 2B-C). In HEL cells, the 12.5-kb EcoRI fragment also was visualized, but there were, in addition, a series of HS segments appearing at the mid-concentration range of DNase I, which correspond to regions of increased DNase I susceptibility centered around 18.5 kb (HS V), 20.3 kb (HS VI), and 21.6 kb (HS VII; Figure 2B-C). Because these HS regions were absent in the SNU-1 cells, these HS domains appear to be tissue specific. Although HS region V is very faintly represented, we believe it is an HS domain. First, it appears in both lanes 3 and 4 (from left side of the blot). Second, in other experiments using a lower concentration range of DNase I it also appeared with even greater presence (data not shown). This suggests that it may be a short-lived series of fragments present only within a narrow time period. It appears that HS domains V to VII do not exist as clear distinct bands, but rather as a broader series of DNase I/EcoRI bordered fragments within each HS domain. In contrast to restriction sites that have a single cleavage point within a 4- to 8-nucleotide recognition sequence, HS sites can vary in size from 200 bp to more than 800 bp,37 and are nonspecifically cleaved. Hence, it is likely that HS V-VII in Figure 2B represent larger extended HS domains. These domains are not present at the lowest DNase I concentration, then faintly appear within the second lane from the left of the HEL blot reaching maximum intensity in the third lane. Note also that none of the broad bands are present at the highest DNase I level. These types of broad, tissue-specific HS domains have been reported to be present in enhancer-containing regions of other genes such as the MyoD muscle-specific enhancer,38 which is localized about 20 kb upstream of the gene. In this enhancer the broad HS domains corresponded to a series of tandem transcription factor–binding sites. Perhaps related to this, it appears that some of the tissue-specific HS domains of our αIIb 3′ locus overlap with conserved intergenic domains in Figure 1. In particular, the HS region VII overlaps with the conserved domain at +22 kb (Figure 1, vertical arrow “2”).
DNase 1 HS studies of the 5′- and 3-intergenic regions
. (A) Nuclei from megakaryocytic HEL and nonmegakaryocytic HeLa cells were digested with DNase I nuclease at increasing concentrations (wedges). The genomic DNA was then isolated, digested with EcoRI restriction enzyme, and analyzed by Southern blotting. The blots shown were hybridized to probe P1 covering hαIIb exon 1. The size marker is indicated as well as the detected HS sites, which are denoted with Roman numerals. (B) Same as in panel A but for SNU-1 and HEL cells, and the probe P3 covered hαIIb exon 30. (C) The hαIIb gene locus is shown schematically at the top with its transcriptional start site indicated as well as the orientation of the genes shown. Restriction sites of importance are indicated with E indicating EcoRI and B, BamHI. The full-length fragment anticipated after EcoRI digestion for the studies in panels A and B and a schema of the observed HS sites (vertical arrows) within the cell lines tested are shown. P1 indicates probe forEcoRI blot in panel A; P2, probe for BamHI blot (not shown); and P3, probe for EcoRI blot in panel B. Similar results were seen on at least 2 independent studies for each probe, cell line, and endonuclease restriction conditions.
DNase 1 HS studies of the 5′- and 3-intergenic regions
. (A) Nuclei from megakaryocytic HEL and nonmegakaryocytic HeLa cells were digested with DNase I nuclease at increasing concentrations (wedges). The genomic DNA was then isolated, digested with EcoRI restriction enzyme, and analyzed by Southern blotting. The blots shown were hybridized to probe P1 covering hαIIb exon 1. The size marker is indicated as well as the detected HS sites, which are denoted with Roman numerals. (B) Same as in panel A but for SNU-1 and HEL cells, and the probe P3 covered hαIIb exon 30. (C) The hαIIb gene locus is shown schematically at the top with its transcriptional start site indicated as well as the orientation of the genes shown. Restriction sites of importance are indicated with E indicating EcoRI and B, BamHI. The full-length fragment anticipated after EcoRI digestion for the studies in panels A and B and a schema of the observed HS sites (vertical arrows) within the cell lines tested are shown. P1 indicates probe forEcoRI blot in panel A; P2, probe for BamHI blot (not shown); and P3, probe for EcoRI blot in panel B. Similar results were seen on at least 2 independent studies for each probe, cell line, and endonuclease restriction conditions.
Human αIIb transgenic mice
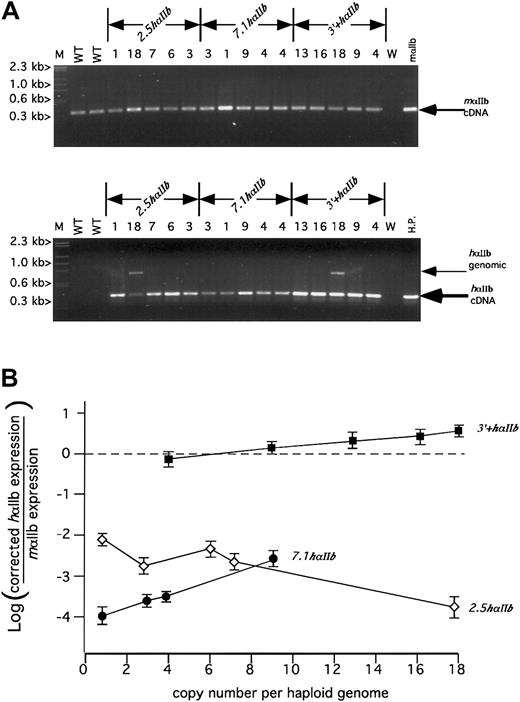
The identification of at least 5 tissue-specific DNase I HS domains located in the flanking intergenic regions of αIIb, some of which coincide with conserved domains, suggests the presence of corresponding control elements that might be essential for establishing appropriate transcriptional activation and critical for generating high levels of hαIIb on the surface of murine platelets. To test this, a series of transgenic mice containing one of 3 different hαIIb constructs were made (Figure 3). These include the shortest construct 2.5hαIIb, which extends from 2.5 kb upstream of the hαIIb gene to 2.2 kb downstream of the stop codon; 7.1hαIIb, which extends from 7.1 kb upstream of the hαIIb gene to 2.1 kb downstream of the stop codon; and the longest construct, 3′+hαIIb, which is identical to the7.1hαIIb construct at the 5′ end, but extends an additional 5.2 kb downstream. Five founder lines for each construct were evaluated. Copy number was determined and covered a wide range from low to high copy number per haploid genome for each construct (Figure 3). To begin our expression analysis, we investigated our founder lines to determine if any or all of the transgenic lines were expressing hαIIb mRNA at some level. Because our genome copy number range varied extensively, we anticipated similarly varied levels of mRNA expression. Hence, we initially used nonquantitative PCR at a high cycle number to detect the presence of any platelet hαIIb mRNA. All 15 founder lines had detectable hαIIb mRNA in platelets by nonquantitative RT-PCR analysis after 33 PCR cycles, well beyond the linear/exponential range of amplification (Figure4A). In the linear range, at 20 PCR cycles, only the endogenous mαIIb mRNA and the 3′+hαIIbmRNA samples produced visible bands on ethidium gel analysis (Figure4A, top panel, and data not shown). Interestingly, the3′+hαIIb line produced visible bands even as low as 14 PCR cycles, suggesting its expression level was significantly greater than the 2.5 hαIIb and 7.1 hαIIb lines (data not shown). None of these lines expressed hαIIb mRNA in any tissue other than platelets, bone marrow, and spleen, consistent with restricted expression of high levels of αIIb expression to developing megakaryocytes (data not shown), and consistent with published transgenic mice toxigene reporter studies.12
Transgenic mice constructs studied.
The hαIIb gene locus is shown at the top with the restriction sites of interest and the HS sites shown in Figure 2 are indicated as vertical arrows. Below that are the 3 constructs from which transgenic lines were made. For each, 5 founder lines were established, and the copy number of the transgene per haploid genome for each line is shown to the right.
Transgenic mice constructs studied.
The hαIIb gene locus is shown at the top with the restriction sites of interest and the HS sites shown in Figure 2 are indicated as vertical arrows. Below that are the 3 constructs from which transgenic lines were made. For each, 5 founder lines were established, and the copy number of the transgene per haploid genome for each line is shown to the right.
RT-PCR analysis of platelet RNA for human and mouse αIIb.
(A) Agarose gel of size-fractionated nonquantitative RT-PCR products for mαIIb (top) or hαIIb (bottom) from total platelet RNA for the various transgenic lines. The mαIIb samples were amplified for 20 PCR cycles within the linear range of amplification for comparison of relative cDNA levels. The hαIIb samples were amplified for 33 PCR cycles, beyond the linear range of amplification, to facilitate detection of hαIIb within all the diversely expressed transgenic lines. Size markers (M) are indicated to the left and the observed cDNA or genomic bands indicated at the right of each figure. The observed genomic bands were variably present and only seen with the 2 lanes indicated where there appears to be little hαIIb message, but high transgene copy number, was present. Copy numbers are listed as numbers above each lane. WT indicates wild-type mouse platelet RNA control; W, water; mαIIb, cDNA plasmid RT amplified; HP,human platelet RNA control. (B) Analysis of semiquantitative RT-PCR comparing relative expression of human to mouse αIIb message versus the copy number for a particular line. The open diamonds are the2.5hαIIb lines, the closed circles are the7.1hαIIb lines, and the closed squares are the3′+hαIIb lines. The experiments were done in triplicate and repeated at least 3 times. A single SD is indicated for each point.
RT-PCR analysis of platelet RNA for human and mouse αIIb.
(A) Agarose gel of size-fractionated nonquantitative RT-PCR products for mαIIb (top) or hαIIb (bottom) from total platelet RNA for the various transgenic lines. The mαIIb samples were amplified for 20 PCR cycles within the linear range of amplification for comparison of relative cDNA levels. The hαIIb samples were amplified for 33 PCR cycles, beyond the linear range of amplification, to facilitate detection of hαIIb within all the diversely expressed transgenic lines. Size markers (M) are indicated to the left and the observed cDNA or genomic bands indicated at the right of each figure. The observed genomic bands were variably present and only seen with the 2 lanes indicated where there appears to be little hαIIb message, but high transgene copy number, was present. Copy numbers are listed as numbers above each lane. WT indicates wild-type mouse platelet RNA control; W, water; mαIIb, cDNA plasmid RT amplified; HP,human platelet RNA control. (B) Analysis of semiquantitative RT-PCR comparing relative expression of human to mouse αIIb message versus the copy number for a particular line. The open diamonds are the2.5hαIIb lines, the closed circles are the7.1hαIIb lines, and the closed squares are the3′+hαIIb lines. The experiments were done in triplicate and repeated at least 3 times. A single SD is indicated for each point.
To determine the relative level of platelet hαIIb transgene expression to the levels of endogenous mαIIb, semiquantitative RT-PCR was performed using platelet total RNA.18,20 22Species-specific primer pairs were used to detect both species' αIIb transcripts. Samples from 6 consecutive PCR cycles taken during the exponential range of amplification were obtained for both mouse and human primer sets. In total, the levels of 180 RT-PCR amplification products were measured for each experiment (3 experiments collectively) using fluorescent-modified antisense primers. Additionally, values obtained were corrected for differences in primer efficiency for the detection of their respective transcripts. The summary of these studies are shown in Figure 4B, which indicates the platelet hαIIb mRNA relative expression versus native mαIIb gene expression, for each founder line plotted against its corresponding copy number.
The range of expression for the 2.5 hαIIb constructs was from 7.5 × 103-fold to 1.5 × 102 less than that of the endogenous mRNA level. These values were also not copy number dependent. Indeed, the lowest copy line had the highest expression. For the 7.1hαIIb line, the range of expression was 9.6 × 103- to 6.5 × 102-fold less than that of the native mαIIb mRNA level. Unlike the2.5hαIIb construct, the 7.1 hαIIb constructs appears to show copy-number dependency, suggesting that the additional 5′ sequence between 7.1 and 2.5 kb upstream of the hαIIb gene protects the transgenic gene locus from local chromosomal influences at the site of insertion. Addition of more 3′ sequence in the3′+hαIIb constructs led to a marked increase in absolute expression levels, which ranged from 1.3-fold lower to 3.5-fold higher than that of the endogenous message level. These expression levels were linear with respect to change in copy numbers, again demonstrating position-independent expression. For the 3+hαIIbconstructs, each copy of the hαIIb gene was expressed at about 30% of the expression level of a single copy of the native mαIIb gene.
Expression of hαIIb protein by the transgenic lines
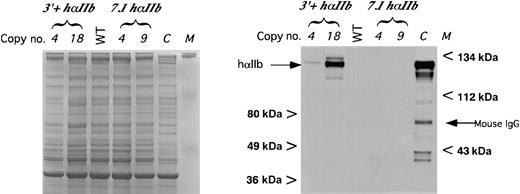
We examined the protein level of platelet hαIIb in these transgenic animals as a secondary measurement of expression. Immunoblot studies of low and high hαIIb-expressing 7.1hαIIb and3′+hαIIb lines were done with MAB1990, a murine monoclonal antibody directed specifically against hαIIb32 (Figure5). Only in protein extracts from platelets derived from the 3′+hαIIb lines was hαIIb detectable. The relative level of protein expression in the 2 tested3′+hαIIb lines was comparable to their 4.5-fold difference in hαIIb mRNA, after accounting for lane loading differences (Figure5, Coomassie panel). Not shown was that protein levels for all of the2.5hαIIb lines, as with the 7.1hαIIblines, was undetectable even for the highest expressing one copy founder line whose mRNA level was about 1% of native levels (data not shown). Compared with the human platelet control in Figure 5, it appears that the highest 3′+hαIIb transgenic mice platelets expressed only about 10% of the hαIIb expressed by human platelets on a per milligram basis, although it is unclear from these studies how this level compares with the level of mαIIb expressed. Thus, it appears that although these platelets have about 3.5 times the level of hαIIb mRNA compared with mαIIb mRNA, the protein level of hαIIb/mβ3 was fairly low as compared with human platelets.
Immunoblot of platelet protein for hαIIb.
Immunoblot of size-fractionated platelet proteins using the monoclonal anti-hαIIb antibody MAB1990 as the primary antibody (right panel). Size markers are indicated on the sides of the blot as well as the expected hαIIb band and the mouse IgG band. C indicates human platelet control. Bands below the expected band were due to partial sample degradation. Equal loading into each lane was confirmed on a parallel gel, where a comparable loading of each sample was done and the gel stained using Coomassie blue (left panel).
Immunoblot of platelet protein for hαIIb.
Immunoblot of size-fractionated platelet proteins using the monoclonal anti-hαIIb antibody MAB1990 as the primary antibody (right panel). Size markers are indicated on the sides of the blot as well as the expected hαIIb band and the mouse IgG band. C indicates human platelet control. Bands below the expected band were due to partial sample degradation. Equal loading into each lane was confirmed on a parallel gel, where a comparable loading of each sample was done and the gel stained using Coomassie blue (left panel).
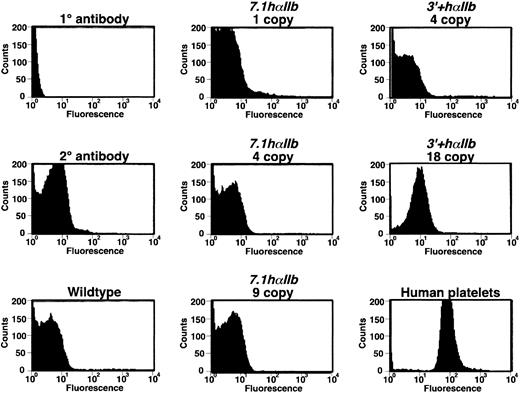
We also measured platelet hαIIb/mβ3 surface levels by flow cytometry. Consistent with the immunoblot studies, hαIIb/mβ3 complex was detected only on the surface of the highest expressing3′+hαIIb line platelets and not on the surface of platelets obtained from the lower expressing 3′+hαIIb line or from the 7.1hαIIb lines (Figure6) or the 2.5hαIIb lines (data not shown). Also consistent with the immunoblots in Figure 5, the level of hαIIb/mβ3 surface expression on the 3′+hαIIbplatelets was about 10% of that on the human platelet control.
Flow cytometric analysis of hαIIb/mβ3 on the surface of platelets.
The mixed species hαIIb/mβ3 receptors were detected by flow cytometry using MAB1990 as the primary antibody. Except for the 18 copy3′+hαIIb platelets, none of the other animals had platelets showing demonstrable hαIIb/mβ3 receptors on their surface, and the intensity in this highest line was still only about 10% of the human platelet control.
Flow cytometric analysis of hαIIb/mβ3 on the surface of platelets.
The mixed species hαIIb/mβ3 receptors were detected by flow cytometry using MAB1990 as the primary antibody. Except for the 18 copy3′+hαIIb platelets, none of the other animals had platelets showing demonstrable hαIIb/mβ3 receptors on their surface, and the intensity in this highest line was still only about 10% of the human platelet control.
Discussion
In vitro studies of the αIIb gene, using transient reporter systems, previously demonstrated that the proximal promoter was sufficient to direct tissue-specific expression.9,10,33 Studies in transgenic mice of the human and mouse αIIb proximal promoter driving the expression of a toxigene have been consistent with these in vitro studies, but did not examine relative αIIb expression levels nor the effects of the site of chromatin integration on transgene expression. Our studies had a different focus. We were interested in understanding the molecular basis of αIIb gene expression, in vivo, without concentrating on its immediate proximal promoter region, but rather searching for additional regulatory regions. We were prompted to do this because our previous structural studies indicated that the platelet-specific αIIb gene was surrounded by 2 ubiquitously expressed genes, KIAA0553, positioned about 5.8 kb upstream and the Granulin gene located about 18 kb downstream.23 The proximity of these ubiquitous genes suggested to us the need for intergenic regulatory constraints to act as boundaries to prevent inappropriate cross-regulation between genes. Our current observation, that the widely expressed hCP44813gene is located about 8.5 kb downstream of the αIIb gene and before the Granulin gene (Figure 1), is still in agreement with our original premise, but further narrows the downstream intergenic region between the αIIb gene and its nearest downstream neighbor.
The studies presented here demonstrate that there are phylogenetically conserved, noncoding, intergenic domains both upstream and downstream of the αIIb gene, that these regions overlap with DNase I HS sites and that the inclusion of the upstream region in a transgenic mice construct confers copy number–dependent expression to the transgene, whereas inclusion of the downstream intergenic region confers an approximate 103-fold increase in expression. These data extend what was previously known about the proximal αIIb promoter described above with its ability to drive tissue-specific expression in vitro and in vivo. Our studies with the shortest2.5 hαIIb lines are in agreement with those studies and demonstrate that a minimal 5′ promoter can restrict αIIb expression to megakaryocytes and platelets. But, in addition, we now add to those previous studies, by analyzing transgene expression relative to native αIIb mRNA levels. Our data reveal that 2.5 kb of 5′-flanking region, though tissue specific, is insufficient to direct high levels of αIIb in vivo. We estimate that the transgenic mice in the earlier toxigene reporter system studies using either 780 bp of the hαIIb promoter or about 2.7 kb of the 5′ region of the mαIIb promoter to drive expression,12,39 most likely expressed their transgenes at an approximate 0.1% to 1% of the native mαIIb gene expression. Perhaps that is why a toxigene reporter model was successful for monitoring expression, as even low expression levels could be phenotypically detected. We would also predict that in those models expression levels would not have been copy number dependent (see below). Whereas the 2.5 hαIIb transgene lines did not show any correlation between copy number and the relative hαIIb to mαIIb message level, the 7.1hαIIb transgene lines demonstrated a clear correlation (Figure 4B). These data suggest that there may be a regulatory element(s) between 2.5 and 7.1 kb upstream, consistent with properties similar to an insulator element, which by definition protects a gene from both positive and negative influences of nearby chromatin, but may also impede enhancer action in a directional fashion.40 41 It is possible that either the tissue-specific HS I-II sites at −3.1 to −4.0 kb upstream of the hαIIb gene or the constitutive HS III and IV sites at −5.4 and −6.4 kb, respectively, upstream of the hαIIb gene are involved. Both are phylogenetically conserved and both are contained in the 7.1hαIIb transgenic construct. Further studies will define which of these regions are needed to observe consistent expression in transgenic mice studies.
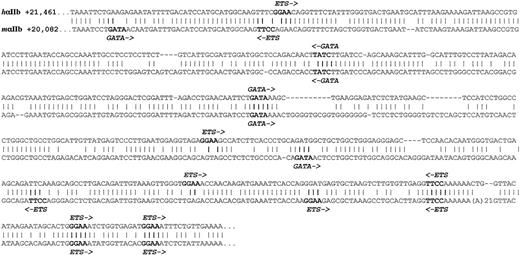
The addition of 5.2 kb of downstream sequence in the3′+hαIIb construct led to an approximate 103-fold increase in hαIIb expression above that from the7.1hαIIb construct. Within this 3′ domain, we found tissue-specific HS sites that overlap with a conserved domain and this domain is a strong candidate for containing an enhancer, because other enhancer elements for other genes have been shown to share these structural features.42,43 The alignment of this conserved domain for the human and murine loci is shown in Figure7, and was analyzed for consensus-binding sites of transcription factors known to be involved in megakaryopoiesis. Unlike the β-globin upstream HS sites, which contain a number of NF-E2–binding sites,44,45 there are no NF-E2–binding sites in the analyzed αIIb regions. Mice lacking NF-E2 have considerable megakaryocytic defects; however, these studies did not examine αIIb expression levels.45 The only previously defined megakaryocyte-specific enhancer is upstream of thePBP gene22 and it too has no NF-E2 sites. In addition, neither region has conserved-binding sites for the AML-1 transcription factor, which is also involved in megakaryopoiesis.46 However, both the αIIb and PBP enhancers have conserved GATA and Ets consensus-binding sites. Whether these are of biologic importance and the mechanism(s) by which these binding sites might lead to high level tissue-specific αIIb expression are unclear, but what is clear is that for the erythroid-specific β-globin gene LCR, functionally important GATA-binding sites are abundantly present in the upstream DNase I-HS regions.47 48
Conserved intergenic regions sequences.
Downstream intergenic, conserved sequences are compared with the human sequence on top and the mouse below. A “∣” refers to an identical nucleotide. Conserved elements that may represent important nuclear factor–binding sites are indicated in bold and the nuclear factor and orientation of binding are shown.
Conserved intergenic regions sequences.
Downstream intergenic, conserved sequences are compared with the human sequence on top and the mouse below. A “∣” refers to an identical nucleotide. Conserved elements that may represent important nuclear factor–binding sites are indicated in bold and the nuclear factor and orientation of binding are shown.
One interesting observation from these studies was that the3′+hαIIb transgenic mice expressed only about 30% hαIIb message compared with native mαIIb per genome copy. Although it is possible that species-specific factors may limit the relative expression of hαIIb in mice models, an alternative explanation is that there are additional upstream or downstream sequences that would further increase the relative level of hαIIb expression. Our studies indicate that there are additional noncoding, phylogenetically conserved sequences both upstream and downstream of the areas investigated in this paper (Figure 1; also see supplemental Figure 1 provided online). Further, examples of genes in which regulatory regions are found within and beyond neighboring genes have been described.14,49 50 For instance, the flanking sequence of the interleukin (IL) IL4, IL13, IL5 gene locus contains a coordinate regulation element that exists between IL4and IL13; however, the third gene, IL5, is actually not continuous with these 2, having the RAD50 gene between itself and IL13. Larger transgenic constructs containing more distal conserved flanking regions of the αIIb locus would have to be tested to see if expression level of the transgene would be further increased.
The relative hαIIb to mαIIb message steady-state level in circulating platelets would have suggested that in the highest-expressing 3′+hαIIb transgenic lines that platelet surface level expression of hαIIb/mβ3 receptors would have been higher than that observed in Figures 5 and 6. If one assumes that the density of the αIIb/β3 receptor is similar on mice and human platelets, and that these mice have continued expression of mαIIb at normal levels, then the highest expressing hαIIb line should have had hαIIb protein levels 3 times that of the endogenous protein. The hαIIb/mβ3 heterodimers should have accounted for about 75% of total surface αIIb/β3. Instead, our flow cytometry studies showed these platelets had only about 10% of hαIIb/mβ3 on its surface compared with human platelets. This would suggest that there was an approximate 30-fold lower level of hαIIb/mβ3 to mαIIb/mβ3 from expected. This deficit in hαIIb/mβ3 receptor level may be due to a decrease in translational efficiency of hαIIb mRNA or in posttranslational hαIIb or hαIIb/mβ3 protein stability compared with its mouse counterparts. The αIIb pairs with β3 as it is trafficked through the Golgi. The misparing or nonpairing with its heterodimer partner leads to retention and ubiquitin-mediated destruction.51 One intriguing possibility is that human and mouse αIIb compete for a limited supply of mβ3 chains.52 The hαIIb may be out-competed by mαIIb for mβ3, and the unpaired hαIIb is removed. Such a model would fit with the proposed limited supply of β3, which selectively binds αIIb over αv.52 In thrombasthenic patients without αIIb, the level of αv more than doubles. If the mβ3 is in limited supply, then decreased levels of mαIIb in the megakaryocytes would theoretically increase detectable hαIIb/β3 expression, because there would be less competing mαIIb. Consistent with this model, we have found in preliminary flow cytometry data, that the level of hαIIb/mβ3 increased 3- to 4-fold in transgenic animals that were also heterozygotes for an mαIIb gene targeted disruption53 (data not shown).
The formation of αIIb/β3 heterodimer appears to involve the N-terminal β-propeller of αIIb and the βA domain of β3.54 These domains show about 80% amino acid cross-species homology. Whether any of the remaining amino acid differences account for the dearth of hαIIb/mβ3 seen on the surface of the transgenic mice platelets remains to be tested.
In summary, our studies have extended the analysis of the regulated expression of the αIIb gene to the intergenic flanking regions between the αIIb gene and its nearest gene neighbors. These studies were consistent with others showing that the proximal αIIb promoter is sufficient to drive megakaryocyte-specific expression, but also defined 2 previously unrecognized distal regulatory regions. The first is an upstream region that allows for consistent position-independent αIIb gene expression. And the second is a downstream region that drives high-level αIIb gene expression in vivo. Inclusion of these regulatory regions appears to account for at least 30% of total αIIb expression levels. Further studies of these regions and comparison with similar elements regulating other megakaryocyte-specific genes may provide important insights into the mechanism(s) by which these tissue-specific genes are highly expressed during megakaryopoiesis.
We would like to thank Dr Edward Rubin and his colleague Jan F. Cheng, both at the Lawrence Berkeley National Laboratory (Berkeley, CA), who assisted us in setting up the VISTA analysis. The hαIIb pWE15 cosmid clone was generously provided by Dr Susan L. Neuhausen at the University of Utah. CHRF cells were provided by Dr M. Liebman at the University of Cincinnati. P1 clones used to generate hαIIb λ clones were provided by screening a P1 library made available through Dr Nat Sternberg at Dupont (Glenolden, PA).
Prepublished online as Blood First Edition Paper, July 25, 2002; DOI 10.1182/blood-2002-05-1307.
Supported in part by grant HL40387 (to M.P.), a grant from the Schulman Foundation (to M.P.), and a gift from the Plummer Family (to M.P.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Mortimer Poncz, Children's Hospital of Philadelphia, 34th Street and Civic Center Boulevard, Philadelphia, PA 19104; e-mail: poncz@emailchop.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal