The convertase furin is involved in the maturation of key growth/aggregation mediators synthesized by the platelet producers, megakaryocytes, but the regulation of furin in these cells remains unknown. Computer-assisted search of the furin promoter sequence revealed multiple potential binding motifs for GATA-1, suggesting that furin is expressed and regulated in these cells. Using megakaryoblastic Dami cells, we observed that fur mRNA expression increased gradually on phorbol 12-myristate 13-acetate–induced differentiation, reaching maximum levels (8.3-fold increase) at 10 days. Transient transfections with P1, P1A, or P1B fur-LUC–promoter constructs revealed that in Dami cells, the P1 promoter is the strongest and the most sensitive to forced expression of GATA-1. Coexpression of GATA-1 and its comodulator, Friend of GATA-1 (FOG-1), resulted in a cooperative increase in P1 activity. Deletion analysis indicated that important GATA-1–regulated sequences are located in the most proximal region of the P1 promoter. Further analysis revealed 2 potential GATA-binding motifs at positions −66 and +62. Point mutation of each of the 2 motifs indicated that the intactness of the first GATA site is required for full basal and GATA-1–stimulated promoter activity. Finally, the inhibition of furin activity through gene transfer of the inhibitor α1-AT-PDX led to a block in maturation of the furin substrates transforming growth factor-β1 and platelet-derived growth factor. Taken together, these results indicate that the most proximal GATA element in the P1 promoter is needed forfur gene expression in megakaryoblastic cells. They also suggest that proper regulation of the fur gene in megakaryocytes has an impact on the activation of furin substrates involved in megakaryocyte maturation and platelet functions.
Introduction
Platelet generation implies a set of well-regulated processes that take place during megakaryocyte differentiation. These processes are mediated by bioactive proteins; among them are the growth/differentiation factors transforming growth factor-β1 (ΤGF-β1)1,2 and platelet-derived growth factor (PDGF),3 the key platelet adhesion molecules αIIb4,5 and von Willebrand factor (VWF),6 and the cell-surface receptor Notch-1,7 which are first synthesized as larger, inactive precursor molecules. Their activation occurs through limited endoproteolytic cleavage after a sequence of 2 or more basic residues (K or R). In the 1990s, 7 closely related mammalian subtilisin/kexin-like serine proteases with this cleavage specificity were discovered. They are grouped under the generic name proprotein convertases (PCs) and include PC1/PC3, PC2, PC4, PC5/PC6, PC7, PACE 4, and furin.8 Among the convertases, furin represents the first and so far the best-characterized enzyme. The biologic importance of this convertase stems from the large number and variety of bioactive proteins and peptides that can be generated through its activity. These include key elements involved in normal and physiopathologic conditions, as exemplified by growth/differentiation processes. In contrast to PC1/PC3, PC2, and PC4, which display tissue-specific expression pattern, the fur gene is ubiquitously expressed,9-11 but its levels of expression vary depending on cell type and degree of cell differentiation.
For instance, high levels of furin mRNA were observed in liver and kidney, whereas lower levels were detected in brain, spleen, and thymus and even lower levels in heart muscle, lung, and testis.2,12,13 In addition, the amounts of furin mRNA varied throughout development; they are found coordinately expressed with known furin substrates such as TGF-β1, TGF-β2, bone morphogenic protein-4 (BMP4), BMP7,14-17 and insulinlike growth factor.18 In particular, by embryonic day 12 (E12) and the midgestational stage, there is remarkable temporal correlation between the expression of furin and TGF-β1 in developing liver.19 This corresponds to the establishment of hematopoiesis in this tissue, in particular for the megakaryocyte lineage.20 It is well known that megakaryocytes and platelets are a predominant source of the TGF-β1 isoform involved in a variety of processes associated with megakaryocyte differentiation and platelet reactivity.21
The mechanisms by which the fur gene is differentially expressed and regulated are still poorly defined. It is known that at least 3 distinct promoters mediate transcription of the furgene. The fur mRNAs generated differ in their 5′ end but are all translated from the same AUG in exon 2, giving rise to identical furin proteins.22 The P1A and P1B promoters resemble housekeeping genes with multiple Sp1-binding sites. On the other hand, the P1 promoter has TATA and CCAAT elements in the proximal region and has been shown to be transactivated by C/EBP-β.22 Differential expression of the P1 and P1A promoters has been observed in hepatocytes, where the P1 promoter was found to be the strongest; in endocrine cells, the P1A promoter showed the strongest activity.22 Recent observations indicated that in HepG2 hepatic cells, the P1 promoter is the strongest and the most sensitive to TGF-β.23 Studies of the transcriptional activity involved indicate that the Smad signal transducers are central participants of TGF-β–induced furregulation in these cells. However, the transcriptional machinery involved in the regulation of furin in other cell types, including megakaryocytes, remains unknown. Interestingly, analysis of thefur P1, P1A, and P1B promoter sequences reveals the presence of putative recognition elements for the transcription factor GATA-1 (A/T)GATA(A/G), including several closely spaced sites characteristic of GATA-1–regulated genes. This suggests that GATA-1 may regulatefur promoter activity in megakaryocytes.
Megakaryocyte differentiation leading to platelet generation is characterized by successive and highly controlled stages coordinated by the expression of transcription factors.24 Among the transcription factors, GATA-1 is known to be essential for the expression of several characterized megakaryocyte genes. For example, the cis-regulatory promoter regions of VWF,25αIIb,26 and platelet factor 4,27which are expressed in megakaryocytes, have critical GATA-1 recognition motifs. Gene disruption study involving a loss of GATA-1 in the megakaryocyte lineage resulted in developmental arrest at a late differentiation stage, firmly establishing the role of GATA-1 in the differentiation process.28
GATA-1 is the first recognized member of the GATA family, which now comprises 6 members named GATA-1 to -6 (for reviews, see Orkin et al29 and Charron and Nemer30). In hematopoietic tissues, the transcription factor GATA-1 is expressed in multipotent progenitors, erythrocytes,31 megakaryocytes, mast cells,32 and eosinophils,33 whereas GATA-2 and GATA-3 are mostly expressed in early hematopoietic progenitors and T lymphocytes, respectively.34 Even if their pattern of expression differs, these transcription factors can recognize the same consensus target sequence (T/A)GATA(A/G) through their highly conserved zinc fingers. A characteristic feature of many GATA-1–sensitive promoter regions is the presence of recognition motifs in repeats or in a palindromic configuration that may stabilize GATA-1 anchoring.35 GATA-1 activity can also be modulated by combinatorial association with other zinc finger proteins. Recently, a GATA-1 cofactor, named Friend of GATA-1 (FOG-1), was identified using a yeast 2-hybrid screen for GATA-1–interacting proteins.36 FOG is a large zinc finger protein that interacts with GATA-1 through binding to its n-terminal zinc finger.37 FOG-1 and GATA-1 cooperate to drive megakaryocyte differentiation, and they have been shown to synergistically activate the p45 NF-E2 and αIIbpromoters.36,38 FOG-1 is largely expressed in hematopoietic tissue and in the liver.36 Like GATA-1, FOG-1 is essential for megakaryopoiesis, but unlike GATA-1, which acts late in megakaryocyte differentiation, the disruption of FOG leads to the ablation of megakaryocyte lineage at an early stage.39
The finding that furin is involved in the maturation/activation of several growth factors and integrins synthesized by megakaryocytes, coupled with the observation that the fur promoters contain multiple GATA-1 motifs, prompted us to investigate the expression of the fur gene in these cells. In this report, we demonstrate that the expression of the fur gene is rapidly induced on differentiation of the human megakaryocytic Dami cells, and we identified a GATA-binding sequence within the proximal region of thefur P1 promoter that is required for full basal and GATA-1–induced activation. Blockage of furin activity in differentiated cells has an impact on the maturation of furin substrates TGF-β1 and PDGF. These findings provide insight into the mechanisms by which furin is expressed in differentiating megakaryocytes and suggest that furin is involved in megakaryocyte differentiation and platelet functions.
Materials and methods
Cell lines
Megakaryoblastic Dami cells were routinely grown in Iscove medium (Gibco BRL, Burlington, ON, Canada) supplemented with 10% fetal bovine serum (FBS; Biomedia Canada, Drummondville, PQ, Canada). For differentiation experiments, 5 × 106 cells were cultured in 100-mm plates (Costar, Cambridge, MA) containing 5 mL complete culture medium and were treated with 100 nM on phorbol 12-myristate 13-acetate (PMA; Sigma, Oakville, ON, Canada) for various time periods, as indicated in the figure legends. In Petri dishes, PMA treatment increased the main ploidy of the culture, a unique developmental feature of megakaryocytes.40 The other human cell lines used in this study are the K562 cells (American Type Culture Collection [ATCC], Manassas, VA), Meg-01 megakaryoblastic cells (ATCC), HL-60 promyelocytic cells (ATCC), and Jurkat T cells (ATCC). The murine myeloid leukemia clonal lines M1/pMGS (control vector) and M1GATA-Y22 (enforced GATA-1 expressing), generously provided by Dr Y. Yamaguchi (Kumamoto University, Japan), have been characterized elsewhere.41 The murine thymoma cell line EL-4 and the pre-B cell line 70Z/3 were provided by Dr H. R. MacDonald (Ludwig Institute for Cancer Research, Basel, Switzerland), and the pre-B cell line 70Z/3 was from ATCC.
Northern blot analysis
Total cellular RNA was extracted from Dami cells with guanidine isothiocyanate according to the previously described TRI-reagent protocol.42 Northern blot analysis was performed as previously described using a rat cDNA probe (generously provided by Dr Robert Day, University of Sherbrooke, Canada).2 23
Immunocytochemistry
Dami cells were grown on Goldline microscope slides (VWR Canlab, ville Mont-Royal, QC, Canada) in the presence or absence of 100 nM PMA for 3 days. Cells were fixed in cold methanol and permeabilized in cold phosphate-buffered saline–0.1% Triton. For immunolabeling, cells were incubated overnight with antihuman furin (1/1000; Chiron, Emeryville, CA), antihuman GATA-1 (1/100; Santa Cruz Biotechnology, Santa Cruz, CA), or monoclonal anti–Golgi 58K protein (1/100; Sigma). Cells were washed and incubated for 30 minutes with fluorescein-conjugated antibodies (antirabbit or antimouse, 1/200) or rhodamine-conjugated antibody (antimouse, 1/800). Negative controls were incubated with either preimmune rabbit serum or mouse immunoglobulin G (IgG; Sigma).
Western blot analysis
Western blotting was performed on total cell lysates transferred to nitrocellulose membranes (Roche Molecular Biochemicals, Québec, PQ, Canada) using antibodies directed against αIIb (1/1000; Immunotech, Marseilles, France), FOG-1 (1/1000; a generous gift from Dr Stuart H. Orkin), GATA-1 (1/500; Santa Cruz Biotechnology), or actin (1/200; Sigma). Secondary antibodies were peroxidase-conjugated antimouse IgG (1/2500; Amersham Pharmacia Biotech), antigoat IgG (1/8000; Sigma), or antirabbit IgG (1/5000; Amersham Pharmacia Biotech). Blots were developed using ECL Western blotting detection reagent (Amersham Pharmacia Biotech).
Plasmids for transient transfections
The human fur promoter luciferase constructs pGL2-P1, pGL2-P1-SacI, pGL2-P1-NheI, pGL2-P1-KpnI, pGL2-P1A, and pGL2-P1B were generously provided by Dr Torik A. Y. Ayoubi (University of Leuven and Flanders Interuniversity, Belgium). The plasmid pMGS-GATA-1 was kindly given by Dr Toshio Suda (University School of Medicine, Japan). As a control vector, we used the pMGS plasmid from which GATA-1 cDNA was previously excised with the restriction enzyme XhoI. FOG cDNA, kindly provided by Dr Stuart H. Orkin, was inserted in pCDNA3.
Luciferase assays
Cells were transiently transfected by CaPO4precipitation technique using a Mammalian Cell Transfection Kit (Specialty Media, Lavallette, NJ) as previously described.43 Briefly, 24 hours before transfection, Dami cells were plated at a density of 500 000 cells/well in 6-well plates (Falcon Labware, Missassauga, ON, Canada) and were differentiated with 100 nM PMA in 2 mL Iscove medium supplemented with 10% FBS. Cells were fed with fresh complete media 3 to 4 hours before transfection. Dami cells were transfected with 1 to 5 μg total plasmid/well, as indicated in the figure legends. Plates were incubated overnight, and cell lysates were assayed for luciferase activity as previously described.23 Data were from at least 3 independent experiments performed in duplicate. Values were normalized for transfection efficiency with either green fluorescence protein (GFP) mean fluorescence or β-galactosidase activity.
Elimination of the GATA-1 recognition motifs on thefur P1-KpnI promoter
The following oligonucleotides were used to engineer the different fur P1-KpnI mutants: 5′-GCATTCTAGTTGTGGTTTGTCC-3′ (sense), 5′-GTGCGACCATCTATGTCACCACCAC-3′ (sense), 5′-CCTGTGAAGGTCTCTGAGCCTGACTG-3′ (sense), 5′-GTGGTGGTGACATAGATGGTCGCA-3′ (antisense), 5′-CAGTCAGGCTCAGAGACCTTCACAGG-3′ (antisense), and 5′-CATAGCCTTATGCAGTTGCTCTCCA-5′ (antisense). First, the furP1-KpnI-Mut1 was obtained by polymerase chain reaction (PCR) using the pGL2-P1-KpnI plasmid with primers 1 and 4 or 2 and 6. After amplification, the 2 PCR products were mixed together, and a subsequent PCR was performed with primers 1 and 6. The obtained fragment was excised KpnI and HindIII and subcloned into pGL2. Fur P1-KpnI-Mut2 was performed using primers 1 and 5 or 3 and 6 in separate reactions. The 2 distinct products were pooled together and subjected to another PCR with primers 1 and 6. The resultant sequence was excised withKpnI and HindIII and subcloned into pGL2.Fur P1-KpnI-Mut1/2 was obtained by replacement of the fur P1-KpnI-Mut1ApaI/HindIII region by the fur P1-KpnI-Mut2 ApaI/HindIII portion. All mutants were sequenced before transfection.
Electrophoretic mobility shift assays
Gel mobility retardation assays were performed as described.44 45 Synthetic nucleotides used in electrophoretic mobility shift assay (EMSA) experiments were designed to include the −66 GATA sites of the P1 promoter with the respective sequences (−74) 5′-GTGCGACCAGATATGTCACCACCACATCACTTTTAG-3′. Other nucleotides used for cold-competition corresponded to −66 GATA oligonucleotide with the following mutations (bold italic letters) within the GATA site sequence (−66) 5′-GTGCGACCATCTATGTCACCACCACATCACTTTTAG-3′ or a GATA consensus oligonucleotide featuring 2 GATA sites in tandem 5′-CACTTGATAACAGAAAGTGATAACTCT-3′ (Santa Cruz Biotechnology).
Adenoviral vector construction
The gene encoding full-length α−1antitrypsin-Portland (PDX; kindly provided by Jeff Lipps, Hedral Therapeutics, Portland, OR), human TGF-β1 (ATCC), or humanfur (from Dr Gary Thomas, Vollum Institute) was inserted into the multiple cloning site of the transfer vector pAd-TR5F-DC-GFP and was placed under the control of a modified cytomegalovirus (CMV) promoter containing a tetracycline (tet)–regulated expression cassette46,47 and expressed together with the GFP tracer. Adenoviral vectors were produced as described47 and were titered by flow cytometry using GFP as a marker of infection. AdCMVtTA, expressing the transactivator tTA under the control of a constitutive CMV promoter, was obtained from Dr Bernard Massie (Biotechnology Research Institute of Montreal, QC, Canada).
Measure of mature TGF-β1 and PDGF-AB
Supernatants were assayed for TGF-β1 and PDGF-AB using a commercially available enzyme-linked immunosorbent assay specific for bioactive TGF-β1 or mature PDGF-AB (R&D Systems, Minneapolis, MN). Supernatants were activated for 10 minutes at 80°C before TGF-β1 detection. The detection limit for TGF-β1 is 30 pg/mL and 9 pg/mL for PDGF-AB.
Results
Increased expression of endogenous furin in differentiating Dami cells
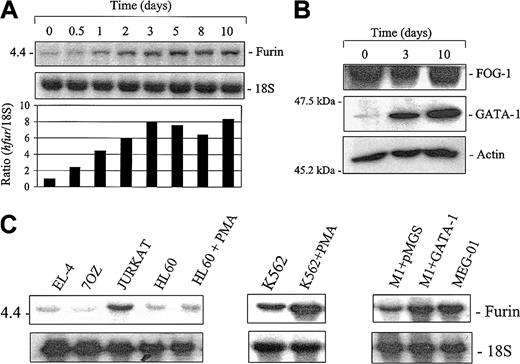
To define the pattern of furin expression during megakaryocyte differentiation, we used the megakaryoblastic Dami cell line. These cells exhibit many morphologic and biochemical characteristics of human megakaryocytes and can be induced to further differentiation along megakaryocyte/platelet lineage on PMA treatment.48 To investigate the expression of furin in megakaryocytes, Dami cells were differentiated for different time periods with 100 nM PMA. Northern blot analysis revealed a rapid increase in furin mRNA levels, with a 7.9-fold increase observed at 3 days and maximum levels (8.3-fold) reached at 10 days (Figure 1A). As previously demonstrated, differentiation of these cells with PMA results in cell adhesion, DNA ploidy, and augmentation in structures characteristic of proplatelet formation48 (data not shown). This was correlated with an increase in the transcription factor GATA-1 (Figure 1B), a molecule known to be up-regulated on megakaryocyte differentiation.45,31 In contrast, the expression levels of FOG-1, a GATA-1 cofactor known to act early in megakaryopoiesis,39 remained essentially unchanged.
Expression of furin in differentiating megakaryocytic cells.
Megakaryoblastic Dami cells were cultured with 100 nM PMA for various time periods as indicated. (A) Kinetics of fur mRNA accumulation. Total mRNA (5 μg/lane) was probed with rat riboprobe specific for furin or 18S. The autoradiogram and the densitometry ratio of furin/18S (control set at 1) is represented. (B) Western blot analysis of GATA-1 and FOG-1 in differentiated Dami cells. Cell lysates were separated on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose membranes. Immunoblotting of GATA-1 and FOG-1 were performed using GATA-1(1/500) or FOG-1 (1/1000)–specific antibodies. Equal loading was confirmed using an antibody against actin (1/200). (C) Fur expression in megakaryocytic cells. Total mRNA was extracted from unstimulated murine and human hematopoietic cell lines and from HL-60 and K562 cells cultured with 100 nM PMA for 3 days; mRNA was probed with rat riboprobe specific for furin or 18S.
Expression of furin in differentiating megakaryocytic cells.
Megakaryoblastic Dami cells were cultured with 100 nM PMA for various time periods as indicated. (A) Kinetics of fur mRNA accumulation. Total mRNA (5 μg/lane) was probed with rat riboprobe specific for furin or 18S. The autoradiogram and the densitometry ratio of furin/18S (control set at 1) is represented. (B) Western blot analysis of GATA-1 and FOG-1 in differentiated Dami cells. Cell lysates were separated on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose membranes. Immunoblotting of GATA-1 and FOG-1 were performed using GATA-1(1/500) or FOG-1 (1/1000)–specific antibodies. Equal loading was confirmed using an antibody against actin (1/200). (C) Fur expression in megakaryocytic cells. Total mRNA was extracted from unstimulated murine and human hematopoietic cell lines and from HL-60 and K562 cells cultured with 100 nM PMA for 3 days; mRNA was probed with rat riboprobe specific for furin or 18S.
We next examined the regulated expression of the fur gene in other megakaryocytic cell differentiation systems. For this, Northern blot analysis was performed using mRNA from the human K562 cells differentiated along megakaryocyte/erythroid lineage with PMA, the murine myeloid cell line M1, a murine myeloid cell that undergoes megakaryocytic differentiation on enforced GATA-1 expression,41 and MEG-01 cells, a human megakaryoblastic cell line with features characteristic of differentiated megakaryocytes.49,50 As a control, we used human promyelocytic leukemia HL-60 cells, which are known to differentiate along the monocytic lineage in the presence of PMA.51 As observed with the Dami cell differentiation system, treatment of K562 cells with PMA for 3 days resulted in a marked increase in furexpression (Figure 1C). In contrast, similar treatment did not significantly affect furin mRNA levels in myeloid HL-60 cells. These results suggest that fur regulation is associated with the cell differentiation process toward the erythroid/megakaryocyte (K562) and megakaryocyte (Dami) lineages rather than direct PMA stimulation. Further supporting this possibility, fur expression levels were also elevated in myeloid M1 cells transfected with GATA-1, a process known to induce a change in the phenotype of these cells from myeloid to erythroid/megakaryocyte lineage.41
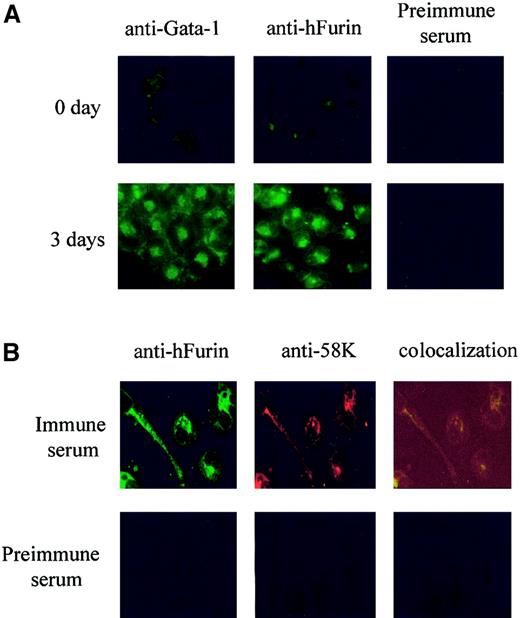
To ensure that the augmentation in furin mRNA levels resulted in an increase in the furin protein, we assessed the relative levels of immunoreactive furin in Dami cells that were or were not differentiated with 100 pM PMA for 3 days. Results expressed in Figure2A indicated that PMA treatment resulted in a strong increase in furin immunoreactivity compared with untreated cells. Furin immunostaining was found to overlap with the staining of the 58K Golgi marker (Figure 2B), a feature consistent with the main localization of furin within the trans-Golgi compartment.52 53
Localized expression of furin convertase in differentiated Dami cells.
Immunofluorescence was performed using Dami cells undifferentiated or differentiated for 3 days with 10−7 M PMA. Cells were fixed, permeabilized, and stained with (A) mouse anti–GATA-1 (1/100) or rabbit anti-furin (1/1000) and then visualized by fluorescence microscopy, using a secondary antibody coupled to fluorescein isothiocyanate. (B) Three-day differentiated cells were costained with rabbit anti-furin (FITC) and mouse anti-58K (1/100; rhodamine). Fluorescence colocalization corresponds to Golgi structures around the nuclei. Preimmune serum was used as negative control. Magnification, × 400.
Localized expression of furin convertase in differentiated Dami cells.
Immunofluorescence was performed using Dami cells undifferentiated or differentiated for 3 days with 10−7 M PMA. Cells were fixed, permeabilized, and stained with (A) mouse anti–GATA-1 (1/100) or rabbit anti-furin (1/1000) and then visualized by fluorescence microscopy, using a secondary antibody coupled to fluorescein isothiocyanate. (B) Three-day differentiated cells were costained with rabbit anti-furin (FITC) and mouse anti-58K (1/100; rhodamine). Fluorescence colocalization corresponds to Golgi structures around the nuclei. Preimmune serum was used as negative control. Magnification, × 400.
Differential sensitivity of the fur promoters to the transcription factor GATA-1
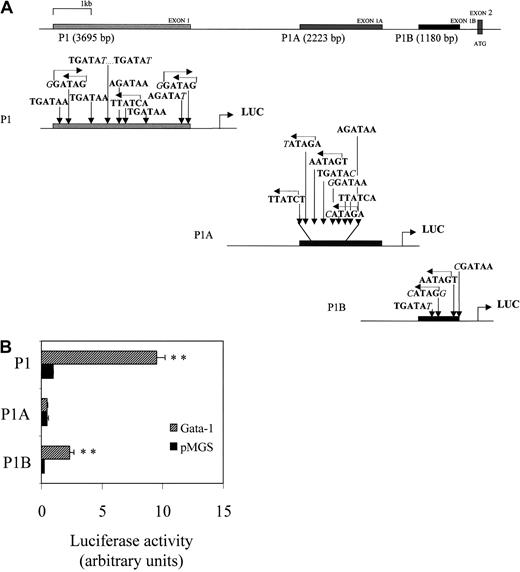
Furin transcription is mediated by at least 3 distinct promoters—P1, P1A, and P1B. As mentioned, the P1 promoter has features of regulated promoters, whereas P1A and P1B have the characteristics of promoters for constitutive/housekeeping genes. Computer-assisted search for the presence of the consensus GATA-binding motif 5′-(A/T) GATA (A/G)-3′54 within these promoters identified 10, 8, and 4 potential GATA sites for P1, P1A, and P1B promoters, respectively (illustrated in Figure 3A). To determine the capacity of each promoter to be transactivated by GATA-1, Dami cells were transiently transfected with P1, P1A, or P1B promoter-Luc construct with or without construct-encoding GATA-1. As indicated in Figure 3B, enforced expression of GATA-1 increased fur P1 and P1B promoter activity by 9.54 ± 0.68-fold and 6.36 ± 0.34-fold, respectively. In the same set of experiments, the P1A promoter did not express any significant sensitivity to exogenous GATA-1. Further studies, therefore, were performed with the most sensitive P1 promoter of GATA-1.
Basal and GATA-1–induced transactivation of the fur P1, P1A, and P1B promoters.
(A) Schematic representation of localization and sequence of the GATA-1 recognition motifs present in the 5′ noncoding exons 1, 1A, and 1B of the human fur gene. (B) Transient cotransfection of Dami cells, with fur P1, P1A, or P1B promoters and either pMGS control vector or pMGS–GATA-1 vector. Before transfection, Dami cells were differentiated overnight with 100 nM PMA. Luciferase activity is expressed as fold-increase relative to P1 promoter activity (set at 1) cotransfected with pMGS control vector. Data are expressed as the mean ± SEM; n = 3. **P < .001, compared with promoter constructs without GATA-1.
Basal and GATA-1–induced transactivation of the fur P1, P1A, and P1B promoters.
(A) Schematic representation of localization and sequence of the GATA-1 recognition motifs present in the 5′ noncoding exons 1, 1A, and 1B of the human fur gene. (B) Transient cotransfection of Dami cells, with fur P1, P1A, or P1B promoters and either pMGS control vector or pMGS–GATA-1 vector. Before transfection, Dami cells were differentiated overnight with 100 nM PMA. Luciferase activity is expressed as fold-increase relative to P1 promoter activity (set at 1) cotransfected with pMGS control vector. Data are expressed as the mean ± SEM; n = 3. **P < .001, compared with promoter constructs without GATA-1.
FOG acts in cooperation with GATA-1 to amplify furP1 promoter transactivation
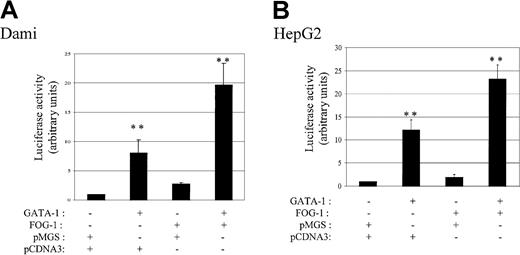
In addition to contacting DNA, GATA-1 was reported to interact with transcriptional cofactors such as the multitype zinc finger FOG-1.37 FOG-1 and GATA-1 can cooperate to activate or to repress promoter activities, depending on the cell type or the promoters studied.36,38 55 To define the impact of FOG on fur P1 promoter transactivation, Dami cells were transfected with FOG-1 in the presence or absence of GATA-1. As demonstrated in Figure 4A, GATA-1 alone increased by 8.1 ± 2.3-fold the levels of fur P1 promoter transactivation compared to control P1 vector, whereas FOG-1 expression resulted in a milder 2.8 ± 0.2-fold increase in activity. Coexpression of FOG-1 with GATA-1 induced an additional amplification (2.4-fold) of fur P1 promoter activity. Therefore, FOG-1, used in the fur P1 promoter context, acts as a potent coactivator of fur expression in Dami cells. Coactivation by FOG-1 and GATA-1 was not dependent on other megakaryocyte-specific transcription factors because similar results were observed using the nonhematopoietic HepG2 cell line (Figure 4B).
Transactivation of the fur P1 promoter by GATA-1 and FOG-1.
(A) Dami cells were differentiated overnight with 100 nM PMA, or (B) HepG2 cells were transiently cotransfected with fur P1 promoter and either pMGS control vector or pMGS-GATA-1, in combination with pCDNA3 control vector or pCDNA3–FOG-1. Luciferase activity is expressed as fold-increase relative to the activity of P1 promoter cotransfected with pMGS control vector (value set at 1). Data are expressed as the mean ± SEM; n = 3. **P < .001, compared with P1 promoter activity.
Transactivation of the fur P1 promoter by GATA-1 and FOG-1.
(A) Dami cells were differentiated overnight with 100 nM PMA, or (B) HepG2 cells were transiently cotransfected with fur P1 promoter and either pMGS control vector or pMGS-GATA-1, in combination with pCDNA3 control vector or pCDNA3–FOG-1. Luciferase activity is expressed as fold-increase relative to the activity of P1 promoter cotransfected with pMGS control vector (value set at 1). Data are expressed as the mean ± SEM; n = 3. **P < .001, compared with P1 promoter activity.
Analysis by 5′ deletion of P1 promoter activity in Dami cells
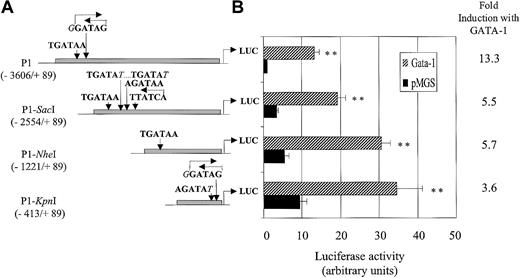
To delineate the promoter regions implicated in the constitutive and GATA-1–induced regulation of the fur P1 promoter, 5′ deletion constructs were tested in transient transfection assays. As the promoter sequence was progressively deleted in 5′, constitutive promoter activity gradually increased in Dami cells, reaching 9.5 ± 1.7-fold more luciferase activity for the shorter 502–base pair (bp) KpnI fragment (Figure5). In contrast, the sensitivity of the P1 promoter fragment to overexpressed GATA-1 gradually decreased with truncation of the distal promoter region with 3.6 ± 0.6-fold stimulation for the KpnI fragment compared to 13.3 ± 1.3-fold for the intact P1 promoter. These results indicate that even though several of the putative GATA sites dispersed throughout the P1 promoter are presumably functional, 27% of the P1 sensitivity to GATA-1 remains within the shorter 502-bp region between position −413 to +89 of the fur P1 promoter. They also suggest the existence of repressor region(s) upstream of the most proximal −413 promoter region.
Basal and GATA-1–induced transactivation of P1 5′ deletion constructs.
(A) Schematic representation of GATA recognition sequences present infur P1 promoter fragments shortened in 5′ accordingly to endogenous SacI, NheI, or KpnI restriction sites. Base positions are numbered relative to the TATA box. (B) Dami cells were incubated overnight with 100 nM PMA and were cotransfected with fur P1, P1-SacI, P1-NheI, or P1-KpnI constructs and either pMGS control vector or pMGS–GATA-1 vector. Luciferase activity is expressed as fold-increase relative to the P1 promoter cotransfected with the pMGS control vector. Data are expressed as the mean ± SEM; n = 3. **P < .001, compared with promoter constructs without GATA-1.
Basal and GATA-1–induced transactivation of P1 5′ deletion constructs.
(A) Schematic representation of GATA recognition sequences present infur P1 promoter fragments shortened in 5′ accordingly to endogenous SacI, NheI, or KpnI restriction sites. Base positions are numbered relative to the TATA box. (B) Dami cells were incubated overnight with 100 nM PMA and were cotransfected with fur P1, P1-SacI, P1-NheI, or P1-KpnI constructs and either pMGS control vector or pMGS–GATA-1 vector. Luciferase activity is expressed as fold-increase relative to the P1 promoter cotransfected with the pMGS control vector. Data are expressed as the mean ± SEM; n = 3. **P < .001, compared with promoter constructs without GATA-1.
Functional analysis of GATA-binding sites within theKpnI- P1 promoter fragment
Analysis of the nucleotide sequence of the 502-bp DNA fragment extending upstream of the transcription start site of thefur gene reveals the presence of 2 potential GATA-binding sites (Figure 6A). One site, at position −66, exhibited the sequence 5′-AGATAT- 3′, with one mismatch with the consensus GATA-binding motif (5′-(T/A)(GATA)(A/G)-3′54 at its 3′end (T instead of A/G). The second site, 5′-GGATAG-3′ located +62 bp downstream of the first site, also diverges slightly from the consensus sequence, with one mismatch at the 5′ end (G instead of T/A).
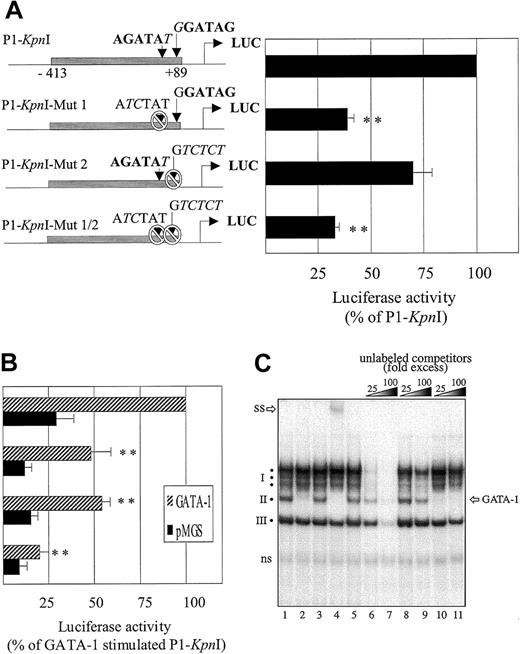
Functional analysis of GATA-binding sites within
fur P1-Kpn1 promoter region. Dami cells were incubated overnight with 100 nM PMA and transiently transfected with the fur P1-KpnI mutant promoter constructs illustrated on the left side of the figure. (A) Measure of basal promoter activity (cotransfection with pMGS control vector); data are expressed as percentage of nonmutated P1-KpnI construct. (B) GATA-1–induced activity (cotransfection with pMGS or pMGS–GATA-1 vectors). Data in panels A and B are expressed as percentage of the nonmutated P1-KpnI construct. Results are expressed as the mean ± SEM; n = 4. **P < .001, compared with nonmutated P1-KpnI promoter construct. (C) Binding of GATA-1 to the −66 P1 GATA site. Nuclear extracts (1.5 μg) from PMA-differentiated Dami cells were incubated (1 hour at 4°C) in binding buffer containing 0.8 μg poly (dI-dC) before further incubation (10 minutes at room temperature) with a 32P-labeled oligonucleotide spanning the −66 GATA motif (bp −74 to −39) of the fur P1 promoter. The specificity of complex formation was tested by the inclusion of unlabeled competitors in the binding buffer (cold probe, lanes 6, 7; cold P1 probe with mutated −66 GATA sites, lanes 8, 9; or unlabeled consensus GATA probe, lanes 10, 11), or by including antibodies to GATA-1 (antibodies N6, sc-265 and C-20, sc-1233; Santa Cruz Biotechnology), lanes 2 and 4, respectively) or isotype-matched controls (rat IgG or goat IgG, lanes 3 and 5, respectively). SS indicates supershift complex; ns, nonspecific band; I, cluster of slowly migrating bands; II and III, faster migrating complexes.
Functional analysis of GATA-binding sites within
fur P1-Kpn1 promoter region. Dami cells were incubated overnight with 100 nM PMA and transiently transfected with the fur P1-KpnI mutant promoter constructs illustrated on the left side of the figure. (A) Measure of basal promoter activity (cotransfection with pMGS control vector); data are expressed as percentage of nonmutated P1-KpnI construct. (B) GATA-1–induced activity (cotransfection with pMGS or pMGS–GATA-1 vectors). Data in panels A and B are expressed as percentage of the nonmutated P1-KpnI construct. Results are expressed as the mean ± SEM; n = 4. **P < .001, compared with nonmutated P1-KpnI promoter construct. (C) Binding of GATA-1 to the −66 P1 GATA site. Nuclear extracts (1.5 μg) from PMA-differentiated Dami cells were incubated (1 hour at 4°C) in binding buffer containing 0.8 μg poly (dI-dC) before further incubation (10 minutes at room temperature) with a 32P-labeled oligonucleotide spanning the −66 GATA motif (bp −74 to −39) of the fur P1 promoter. The specificity of complex formation was tested by the inclusion of unlabeled competitors in the binding buffer (cold probe, lanes 6, 7; cold P1 probe with mutated −66 GATA sites, lanes 8, 9; or unlabeled consensus GATA probe, lanes 10, 11), or by including antibodies to GATA-1 (antibodies N6, sc-265 and C-20, sc-1233; Santa Cruz Biotechnology), lanes 2 and 4, respectively) or isotype-matched controls (rat IgG or goat IgG, lanes 3 and 5, respectively). SS indicates supershift complex; ns, nonspecific band; I, cluster of slowly migrating bands; II and III, faster migrating complexes.
Site-directed mutagenesis was performed with the AGATAT site changed for ATCTAT and the GGATAG site changed for a GTCTCT sequence.27 This resulted in 3 distinct mutants—P1-KpnI-Mut1, P1-KpnI-Mut2, and P1-KpnI-Mut1/2—that correspond to the elimination of the first, the second, or both GATA recognition sites. Expression of the P1-KpnI-Mut1 in Dami cells, which constitutively express endogenous GATA-1, reduced the activity to 39% ± 3% of the intact fragment, indicating that this site is critical for promoter activity. In contrast, expression of P1-KpnI-Mut2 resulted in a more modest decrease in luciferase activity (to 70% ± 9%), suggesting that this site has a minor contribution to P1 promoter transactivation. Finally, mutation of the 2 GATA sites (P1-KpnI-Mut1/2) decreased Luc activity to a level (33% ± 2%) similar to the one observed with P1-KpnI-Mut1 (Figure 6A) but resulted in an additive reduction in GATA-induced transactivation (Figure 6B). In parallel, results from coexpression of the transcription factor GATA-1 indicated that the first AGATAT motif of the KpnI P1 fragment is indeed the most responsive to GATA-1 (Figure 6B). From these results, we conclude that the intactness of the −66 AGATAT site is critical for high-level expression of the fur gene in Dami cells and that the second +62 site can act in cooperation, especially when GATA-1 is highly expressed.
We also tested whether the −66 GATA site of the fur P1 promoter was likely to be occupied by its cognate transcription factors in PMA-differentiated Dami cells. To this end, nuclear extracts were analyzed in EMSA using an oligonucleotide probe spanning the −66 GATA site. This revealed the existence of 3 groups of DNA-binding activities (Figure 6C, lane 1): a cluster of slowly migrating bands (complex 1) and 2 faster-migrating complexes (2 and 3). All complexes showed specificity in their binding to the P1 region used as a probe because they were effectively competed by an excess of cold probe (lanes 6, 7) but mostly were unaffected by equivalent quantities of a similar oligonucleotide featuring mutated GATA sites (lanes 8, 9). By contrast, only the middle complex (complex 2) was competed using a commercially available oligonucleotide featuring a consensus GATA sequence (lanes 10, 11). More direct evidence that complex 2 contains GATA proteins was obtained using an antibody that interferes with the DNA-binding activity of GATA-1, which displaced complex 2 (lane 2). Similarly, an antibody recognizing the C-terminal domain of GATA-1 supershifted complex 2 (lane 4). Because neither antibody displayed cross-reactivity with other GATA family members, we conclude that endogenous GATA-1 is a component of the DNA-binding complex interacting with thefur P1 promoter in PMA-differentiated Dami cells.
Impact of furin regulation in megakaryocytes on the production of mature furin substrates
Megakaryocyte differentiation is accompanied by sequential expressions of growth factors/receptors and adhesion molecules characterized by the presence of a consensus furin recognition motif at the maturation site.1-7 Among them are the key growth factors TGF-β and PDGF and the αIIb chain component of the integrin αIIbβ3 complex. As a first step to define the biologic relevance of regulated furin expression in megakaryocytes, we investigated whether this convertase is coordinately regulated with furin substrates in Dami cells. Results expressed in Figure7A indicate that PMA-induced Dami cell differentiation is associated with a gradual increase in the accumulation of TGF-β1 and PDGFAB in the culture medium. Interestingly, the production and the maturation of the αIIb chain of the integrin αIIbβ3 are also up-regulated, as evidenced by the increased abundance of the precursor and the mature forms, combined with a gradual augmentation in conversion ratios.
Role of furin in the production of mature integrin and growth factors.
(A) Megakaryoblastic Dami cells were cultured with 100 nM PMA for various time periods, as indicated, supernatants were collected, and TGF-β1 and PDGF AB were measured by ELISA. Corresponding cell lysates were separated by SDS-PAGE and immunoblotted using αIIb-specific antibodies (1:1000). Western blots were scanned, and differences in staining intensity were measured using NIH image software. Western blot results are expressed as a ratio of mature to precursor forms, which is an estimation of conversion efficiency. (B) Dami cells were incubated overnight in the presence of 100 nM PMA. Cells were infected with the indicated recombinant adenovirus. Forty-eight–hour supernatants were collected for TGF-β1 and PDGFAB determination. Data are expressed as the mean ± SEM; n = 3. **P < .001, compared with control adenovirus. (C) Cells were coinfected with the indicated recombinant adenovirus. 48 hours after infection, supernatants were concentrated, separated on SDS-PAGE gels, and immunoblotted using an anti–TGF-β–specific antibody (1:1000; R&D Systems).
Role of furin in the production of mature integrin and growth factors.
(A) Megakaryoblastic Dami cells were cultured with 100 nM PMA for various time periods, as indicated, supernatants were collected, and TGF-β1 and PDGF AB were measured by ELISA. Corresponding cell lysates were separated by SDS-PAGE and immunoblotted using αIIb-specific antibodies (1:1000). Western blots were scanned, and differences in staining intensity were measured using NIH image software. Western blot results are expressed as a ratio of mature to precursor forms, which is an estimation of conversion efficiency. (B) Dami cells were incubated overnight in the presence of 100 nM PMA. Cells were infected with the indicated recombinant adenovirus. Forty-eight–hour supernatants were collected for TGF-β1 and PDGFAB determination. Data are expressed as the mean ± SEM; n = 3. **P < .001, compared with control adenovirus. (C) Cells were coinfected with the indicated recombinant adenovirus. 48 hours after infection, supernatants were concentrated, separated on SDS-PAGE gels, and immunoblotted using an anti–TGF-β–specific antibody (1:1000; R&D Systems).
In parallel experiments, we tested whether blockage of furin activity impacts the production of mature forms of furin substrates. For this, we used a modified adenovirus vector (AdTR5) for cell delivery of α1-AT-PDX, a potent furin inhibitor.56 In this system, the α1-AT PDX gene is placed under the negative control of a tet/doxycycline-regulated promoter sensitive to low concentrations of doxycycline in cell cultures.46 As observed in Figure 7B, the infection of differentiated Dami cells with AdTR5PDX significantly reduced the amounts of mature and active TGF-β1 and PDGFAB released in the cell supernatants. Similarly, complete blockage in the processing αIIb chain of integrin αIIbβ3 was observed on adenovirus-induced transduction of α1-ATPDX in Dami cells (data not shown). The addition of 1 μg/mL Dox to the cultures abolished AT-PDX production and its effect on the production of mature growth factors. To confirm that, in our system, PDX overexpression indeed resulted in an inhibition of furin-dependent processing, coinfection experiments were performed using recombinant adenovirus encoding for the TGF-β1 substrate or for α1-ATPDX, and concentrated supernatants were analyzed for TGF-β1 processing by immunoblotting. As illustrated in Figure 7C, supernatants from TGF-β1–infected cells produced approximately 60% of processed TGF-β1 as seen by the relative intensity of the proregion and the pro–TGF-β1 bands. Coinfection of cells with 100 multiplicity of infection (MOI) of adenovirus encoding α1-ATPDX abrogated pro–TGF-β1 proteolytic processing mediated either by endogenous furin activity or by overexpressed furin. Taken together, these results indicate that the TGF-β1, PDGFAB, and αIIb substrates that are associated (integrin) or involved (growth factors) in megakaryocyte differentiation undergo furin-dependent cleavage.
Discussion
The mammalian convertase furin is responsible for the maturation of key platelet aggregation/coagulation mediators synthesized by the platelet producers, megakaryocytes.1-6 To date, however, there has been no study on the expression/regulation of thefur gene in these cells. In this report, we demonstrate that the fur gene and furin-converting activities are expressed at low levels in human megakaryocytic cells, and their expression is rapidly induced on cell differentiation with PMA. By promoter deletion and mutation analysis, we identified a region within the humanfur gene that regulates fur transcription in megakaryocytes. This site contains the core-binding (A/T)GATA(A/G)55 sequence for GATA zinc finger transcription factors and is required for constitutive and GATA-1–induced transactivation of the proximal P1 promoter region.
Three alternative promoters can drive transcription of thefur gene. The P1A and P1B promoters resemble housekeeping genes with multiple Sp1-binding sites. On the other hand, the P1 promoter has TATA and CCAAT elements in the proximal region and has been shown to be transactivated by C/EBP-β22 and members of the Smad family.23 Herein, we show that even though each of the 3 fur promoters has several potential GATA recognition sequences, in Dami cells, the P1 promoter is the strongest and the most sensitive to forced expression of GATA-1, whereas little or no induction was observed for P1B or P1A. The exact reason for this discrepancy is unclear, but it has been reported for several GATA-1–controlled genes (including the β maj-globin57and the mpl receptor58) that GATA motifs located proximal to the translation initiation site are critical for promoter transactivation. As demonstrated in Figure 3A, proximal GATA elements are found within the P1 and P1B promoters only, and this correlates with their capacity to be transactivated by GATA-1. In addition, for the P1 promoter, the GATA sites (−66 and +62) were found proximal to CCAAT (−129) and CACCC (−28) elements, a promoter context that might contribute to its robust transcriptional activation by GATA-1—a situation that has been described for the PECAM-1 promoter.59 In contrast to other megakaryocyte-restricted genes, such as the integrin αIIbβ3, the fur P1 promoter more likely qualifies as a megakaryocyte-sensitive promoter because it is expressed in other cell types, such as HepG2 and COS cells. In the nonhematopoietic HepG2 cell, for example, the transcriptional activity of the P1 promoter is in part under the control of endogenous C/EBP-β and of members of the Smad transducers.22 23 The known capacity and yet to be uncovered capacity of thefur promoters to respond to various transcriptional contexts would likely help explain the temporal and spatial expression of furin in megakaryocytes and in other tissues and organs.
The proximal region of the fur P1 promoter contains 2 potential GATA-binding sites at positions −66 and +62. The relative contribution of each GATA-1–binding site to the transcriptional activity of the promoter was examined. Ablation by mutation of the first AGATAT site resulted in a strong (61%) reduction in constitutive promoter activity and a 52% inhibition in overexpressed GATA-1–induced P1-Mut1 promoter construct. The importance of its integrity for fur promoter activity in Dami cells clearly indicates that this GATA site represents a major cis-acting element for the human fur gene in these cells. Even though the AGATAT site diverges slightly from the well-known optimal (A/T)GATA(A/G) consensus sequence, EMSA indicated that it does interact with GATA-1. In contrast to the −66 site, destruction by mutation of the second GATA site resulted in milder 30% reduction in constitutive promoter activity and 46% inhibition in GATA-1–induced promoter activation. This site, GGATAG, slightly differs from the consensus GATA recognition site and is located downstream of the TATA box; neither situation may be favored within thefur promoter context. Finally, mutation of the 2 GATA sites (P1-KpnI-Mut1/2) decreased basal promoter activity to a level similar to that observed with mutation of the first GATA site only, but it resulted in an additive reduction in GATA-induced transactivation. From these results, we conclude that the intactness of the −66 AGATAT site is critical for high-level expression of thefur gene in Dami cells and that the second +62 site may have lower affinity for GATA-1 because it is involved in conditions in which GATA-1 is overexpressed.
Several studies have shown that GATA-1 site repeats found in 5′ or 3′ orientation or high-affinity palindromic sites, composed of one complete (A/T)GATA(A/G) and one partial (GAT) canonical motif found on the opposite DNA strand, are the hallmark of erythroid/megakaryocyte cell-expressed genes, including GATA-1 and the human γ-globin promoter.35 It has been demonstrated that the palindromic sites can bind one molecule of GATA-1 with high affinity because they are able to engage the N-terminal and C-terminal zinc fingers of the protein.35 60 Interestingly, the prominent P1 GATA site is flanked by a partial GATA motif, TGATGT, found in a 3′ position, 9 bp on the opposite strand of the AGATAT site. Such positioning evokes the reported high-affinity palindromic GATA site, with more distance between the minor and the major GATA sites (9 bp instead of 3 bp).
Deletion studies of the P1 promoter indicated that as the promoter sequence was progressively truncated in 5′, constitutive promoter activity gradually increased in Dami cells, reaching 9.5 ± 1.7-fold more luciferase activity for the more proximal 502-bp KpnI fragment. This could indicate the existence of a repressor region upstream of the most proximal −2554 to −413 region. The exact nature of this repressor region is unknown, but analysis of the P1 sequence indicates the presence of a series of CATGAG sequences upstream of the more responsive GATA-1 region. One site is located between theNheI-KpnI restriction sites and 3 sites betweenSacI and NheI. Because CATGAG sites have been shown to repress cis-acting elements of the αIIb promoter,61 it is conceivable that such sites act in a similar fashion for fur P1 promoter regulation. In this context, temporal expression of furin during megakaryocyte differentiation would involve a block in the activity of these repressor regions.
In addition, we provide evidence that FOG-1, a cofactor for GATA-binding proteins, acts as a positive cofactor for furP1 transactivation. FOG, in association with GATA-1, consistently resulted in more than a 2-fold increase in P1 transactivation than GATA-1. Similar cooperation between FOG and GATA-1 had been observed for the transactivation of the megakaryocytic gene αIIb38 and the erythromegakaryocytic genep45 NF-E2,36 among others. This is consistent with its essential participation in the maturation of erythroid cells and the early development of thrombocytic lineage.39 On the other hand, FOG recruited by GATA-1 acts as a repressor of the eosinophil lineage and down-regulates the eosinophil-specific markers EOS47, C/EBPβ, and Mim-1.55 In contrast to GATA-1, which is expressed at low levels in Dami cells and is up-regulated on cell differentiation, the FOG-1 protein remained at a relatively high level of expression throughout the differentiation process (Figure 1B). In this context, cooperative interaction between GATA-1 and FOG-1 might be important for the rapid elevation of furin observed at an early stage of megakaryocyte cell differentiation, a situation in which others62 and we have detected only threshold levels of endogenous GATA-1.
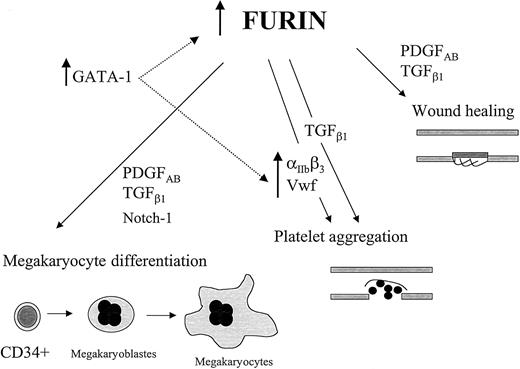
The observed increase of furin expression in megakaryocytes suggests the implication of this convertase in the bioavailability of selected mediators. Among the known furin substrates, the potent regulator of cell growth/differentiation PDGF and TGF-β, and the platelet adhesion molecules to VWF, integrin subunits α6 and αIIb were observed to be increased along megakaryocyte differentiation.48,63-66 A body of literature has firmly established that VWF and αIIb are essential for thrombus formation at the vascular injury site, whereas platelet α-granule–derived TGF-β and PDGF are major players of endothelial cell proliferation and vessel repair.67-70 In addition, TGF-β1 has been shown to enhance platelet aggregation through a nontranscriptional effect on the fibrinogen receptor.71Furin has also been identified as the convertase responsible for the maturation of Notch-1, a cell-surface receptor involved in multiple developmental processes.7 Interestingly, a recent study using a constitutively active intracellular domain of Notch-1 in bipotent hematopoietic cells indicated that Notch-1 is permissive for the differentiation of megakaryocytes while it suppresses erythroid maturation.72 Therefore, the proper expression/regulation of furin in megakaryocytes would likely be important for correct processing of mediators involved in megakaryocyte differentiation and platelet function (Figure 8).
Proposed impacts of fur expression/regulation in megakaryocytes.
The events depicted here are initiated with hematopoietic cell differentiation into megakaryocytes. In this scheme, we extrapolate the results observed with various megakaryocytic cell lines to the events proposed to occur in primary cells. Fur regulation in megakaryocytes is, in part, under the control of the transcription factor GATA-1, and the relevance of other transcription factors in this regulation is not excluded. The role of furin in the maturation/activation of the integrin αIIb will be published elsewhere (M.-H. L. et al, mansucript in prepration).
Proposed impacts of fur expression/regulation in megakaryocytes.
The events depicted here are initiated with hematopoietic cell differentiation into megakaryocytes. In this scheme, we extrapolate the results observed with various megakaryocytic cell lines to the events proposed to occur in primary cells. Fur regulation in megakaryocytes is, in part, under the control of the transcription factor GATA-1, and the relevance of other transcription factors in this regulation is not excluded. The role of furin in the maturation/activation of the integrin αIIb will be published elsewhere (M.-H. L. et al, mansucript in prepration).
We thank David Bouchard for performing the site-directed mutagenesis, Dr Jean Luc-Parent for his help in mutagenesis design, Penny Rudd for her assistance in luciferase assays, and Tayna Bardati for performing the gel shift assays.
Supported by the Canadian Institutes of Health Research grant MT-13222.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Claire M. Dubois, Department of Pediatrics, Immunology Division, Faculty of Medicine, University of Sherbrooke, Québec, QC, Canada J1H 5N4; e-mail:claire.m.dubois@usherbrooke.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal