Abstract
FLT3 length mutation (FLT3-LM) is a molecular marker potentially useful for the characterization of acute myeloid leukemia (AML). To evaluate the distribution of FLT3-LM within biologic subgroups, we screened 1003 patients with AML at diagnosis for this mutation. FLT3-LM was found in 234 (23.5%) of all patients and thus is the most frequent mutation in AML described so far. Of all positive patients, 165 (70.5%) revealed a normal karyotype. Of the 69 patients with chromosome aberrations, 24 (34.8%) had a t(15;17). The mutation was rare in AML with t(8;21), inv(16) 11q23 rearrangements, and complex karyotypes. FLT3-LM was not distributed equally within different French-American-British (FAB) subtypes and was correlated with a high peripheral blood count in FAB M1, M2, and M4 (P < .0001). In addition, the median age of patients with the mutation was lower (54.9 vs 57.6 years;P = .043), and, at a ratio of 1.36:1 (P = .023), the mutation was more frequent in females than in males. Within the AMLCG study, FLT3-LM was of intermediate prognostic significance. The complete remission rate of 70.3% in patients with FLT3-LM was similar to that (70.4%) in patients without FLT3-LM. Overall survival was not different between patients with or without FLT3-LM. In contrast, patients with FLT3-LM had a significantly shorter event-free survival (7.4 vs 12.6 months;P = .0072) because of a higher relapse rate. Besides the importance of FLT3-LM for biologic and clinical characterization of AML, we show its value as a marker for disease monitoring based on 120 follow-up samples of 34 patients.
Introduction
The detection and characterization of chromosome alterations in acute myeloid leukemia (AML) has provided the means to identify distinct biologic and prognostic subgroups and to establish a new pathogenesis-oriented classification of the disease (World Health Organization classification).1 Based on karyotype analysis, 3 different groups of AML can be distinguished: (1) AML with balanced chromosomal aberrations and favorable clinical course—this group mostly comprises specific recurrent chromosomal translocations that target and deregulate genes encoding for transcription factors that are important in hematopoiesis2-4; (2) AML with nonbalanced chromosomal aberrations and poor clinical outcome—this group includes nonrandom chromosomal losses or deletions, suggesting that antioncogenes are involved in the pathogenesis of this AML subgroup2,3,5; (3) 45% of AML patients had normal karyotype and were pooled together in the prognostically intermediate group—this AML group lacked markers indicative of prognosis and may be useful for molecular follow-up analysis. The pathogenesis of these AML subgroups is poorly understood. Within the cytogenetically healthy group, partial tandem duplication of the MLL gene has been described in 8% to 10% of patients and identifies a prognostically very poor risk group.6-11 This rearrangement leads to the fusion of a portion of the putative proto-oncogene MLL with itself and seems to represent a new genetic mechanism for leukemogenesis. In addition, a length mutation of the juxtamembrane domain-coding sequence of the FLT3 gene caused by an internal tandem duplication (ITD) has been described in 20% of AML patients.12,13 In most patients, the duplicated sequence includes those from exon 11 and, in rare cases, from intron 11 or exon 12, and all retain the reading frame. Because this mutation is not always a simple duplication but often contains foreign sequences or additions of extra nucleotides, we will not use the term ITD but will refer to it here as FLT3-length mutation (FLT3-LM). FLT3-LM has been shown to lead to ligand-independent autophosphorylation of the receptor and results in the proliferation of AML cells in vitro, because it appears to stimulate proliferation and to inhibit apoptosis.14,15 In addition, it has been shown that FLT3-LM induces factor-independent growth and leukemogenesis of 32D cells that are mediated by the Ras and STAT5 pathways.16,17 The leukemogenic potential of FLT3 activation was further supported by a study that shows the effects of an artificial TEL-FLT3 receptor in an animal model.18 In addition to FLT3-ITD, a point mutation of D8315 in exon 17 of theFLT3 gene has recently been described that also leads to constitutive tyrosine phosphorylation.19,20 Some studies indicate that the presence of FLT3-LM is associated with a poor outcome in adults21,22 and in children.23 24 These studies are restricted to certain ethnologic groups or study groups and are hampered by a limited number of analyzed cases and a limited association with cytogenetic subgroups.
Thus, the main aims of this study were to analyze the frequency of FLT3-LM in a large group of consecutive, unselected patients with AML; to correlate FLT3-LM with karyotype, French-American-British (FAB) subtype, and other biologic characteristics; to investigate the outcome of patients with FLT3-LM in AML who entered the German AML Cooperative Group (AMLCG) study and thus received standardized diagnostic work-up and therapy; and to assess the applicability of this mutation as a marker for minimal residual disease in follow-up studies.
Patients, materials, and methods
Patient samples
Fresh blood or bone marrow samples from 1003 consecutive patients were analyzed. All diagnoses indicated AML according to standard FAB criteria,1,25 26 and all patients were referred to the laboratory for leukemia diagnostics between July 1997 and December 2000 for cytomorphologic, cytogenetic, and molecular analyses. All patients were adults. Of the 1003 patients, 873 had de novo AML, 77 had secondary AML after MDS prephase (s-AML), and 52 had AML after treatment of a previous malignant disease (t-AML). Informed consent according to the Declaration of Helsinki was approved by the local ethics committee of the Ludwig-Maximilians-University of Munich.
Treatment protocol of the German AMLCG Study
Treatment consisted of randomized comparison of TAD9/TAD9 versus TAD9/HAM (AMLCG92)27 or of TAD9/HAM versus HAM/HAM (AMLCG99) double induction followed by TAD9 consolidation (AMLCG92) (TAD, thioguanine–cytosine arabinoside–daunorubicin; HAM, high-dose cytosine arabinoside–mitoxantrone). In the AMLCG92 study, patients achieving complete remission were subsequently randomized for monthly maintenance therapy or S-HAM for a second consolidation.
Complete remission (CR) was assumed when there were less than 5% blasts in a normo-cellular bone marrow with normal levels of peripheral neutrophil and platelet counts. Overall survival (OS) was calculated from the first day of therapy to death. Disease-free survival (DFS) was measured from the date of CR to relapse or death.
Cytogenetics
Cytogenetic G-banding analysis was performed with standard methods. The definition of a cytogenetic clone and descriptions of karyotypes followed the International System for Human Cytogenetic Nomenclature.28
Nucleic acid isolation
DNA was extracted with a salting-out procedure29from fresh bone marrow or peripheral blood cells after Ficoll separation of mononucleated cells. From the same specimens total RNA was isolated with RNeasy (Qiagen, Hilden, Germany) following the manufacturer's instructions.
Genomic polymerase chain reaction
One hundred nanograms genomic DNA was amplified specifically for exon 11 to exon 12, including intron 11, using primers 11F and 12R.30 Amplification was performed for 35 cycles (1 minute at 94°C, 1 minute at 60°C, 1 minute at 72°C), in 50 μL with 10 pmol each primer, 10 mmol dNTP, and 1.25 U Taq polymerase (Gibco/BRL, Eggenstein, Germany) in the buffer shipped by the supplier.
Reverse transcription–polymerase chain reaction
One microgram total RNA was reverse transcribed with 300 U Superscript (Gibco/BRL) in a 40-μL reaction using random hexamers as primers. An equivalent quantity of 25 ng RNA was amplified as described above.
For each sample, ABL-specific reverse transcription–polymerase chain reaction (RT-PCR) was performed to control the integrity of DNA or RNA using primers abl5′: 5′-GGCCAGTAGCATCTGACTTTG-3′ andabl3′: 5′-ATGGTACCAGGAGTGTTTCTCC-3′. Water instead of cDNA was included as a blank sample in each experiment. Amplification products were analyzed on 2% agarose gels stained with ethidium bromide.
Semiquantitative analysis of the mutations
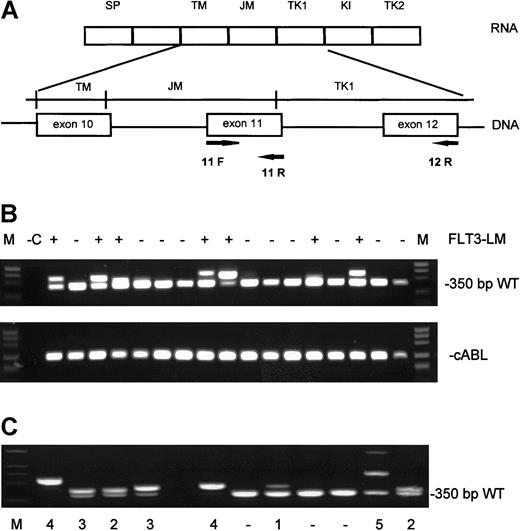
Analysis of the amplification fragments on agarose gels revealed that the band representing the mutation was not always of the same intensity as the wild-type allele. Thus, we divided FLT3-LM into 5 categories according to densitometric estimations of ethidium bromide–stained agarose gels with the gelpro32-software (INTAS, Göttingen, Germany), as follows: (1) mutant fragment less intense than the wild-type band, (2) mutant fragment equally intense as the wild-type band, (3) mutant fragment more intense than the wild-type band, (4) only mutant fragment with loss of the wild-type band, and (5) presence of more than 1 mutant fragment (Figure1C).
Analysis of the amplification fragments on agarose gels.
(A) Genomic and mRNA structure of the FLT3 gene with position of the primers F11, R12, R5, and R6. (B; top) Ethidium bromide–stained agarose gel with RT-PCR products. (Bottom) cABL control PCR. (C) Classification of FLT3-LM according to intensities of the amplified fragments. Category 1, low intensity; category 2, as intense as wild type; category 3, more intense than wild type; category 4, only mutation; category 5, more than one mutated fragment.
Analysis of the amplification fragments on agarose gels.
(A) Genomic and mRNA structure of the FLT3 gene with position of the primers F11, R12, R5, and R6. (B; top) Ethidium bromide–stained agarose gel with RT-PCR products. (Bottom) cABL control PCR. (C) Classification of FLT3-LM according to intensities of the amplified fragments. Category 1, low intensity; category 2, as intense as wild type; category 3, more intense than wild type; category 4, only mutation; category 5, more than one mutated fragment.
Sequencing
All PCR products larger than the wild-type allele were sequenced to identify the type and extent of duplication. To this end, amplified fragments were cut from agarose gels and were isolated with Quiaex II (Qiagen) following the manufacturer's instructions. Approximately 100 ng purified PCR products were directly sequenced with 3.3 pmol primers as described above with the Big Dye Terminator Cycle Sequencing Kit (Perkin Elmer, Weiterstadt, Germany). After initial denaturation at 95°C for 5 minutes, 25 cycles at 94°C for 15 seconds and at 60°C for 4 minutes were performed. Sequence analysis was performed on an ABI 310 Sequence Detection System (Perkin Elmer).
Statistical analysis
Survival curves were calculated for OS, event-free survival (EFS), and DFS according to Kaplan-Meier.31 Survival curves were compared using a 2-sided log-rank test; results were considered significant at the P < .05 level on both sides. Pearson χ2 analysis and Student t test were used to test for differences in the distribution of dichotomous variables and in the means of continuous distributions. Multivariate analyses were performed for the respective dependent variables using length mutations of the FLT3 gene, favorable cytogenetics, unfavorable cytogenetics, and secondary etiology of AML as dichotomous covariates, respectively, and age as a continuous covariate.
Results
Genomic and RT-PCR of the FLT3 gene
Genomic DNA and total RNA were obtained from the bone marrow of all 1003 patients at diagnosis. In addition, follow-up samples were obtained at 2 to 8 time points from 34 patients carrying the FLT3 mutation. Exons 11 to 12 of the FLT3 gene were amplified by genomic PCR, and exon 11 was amplified by RT-PCR. The FLT3-LM amplification yielded a higher molecular weight product on a 2% agarose gel stained with ethidium bromide (Figure 1). Of the 1003 samples analyzed, 234 (23.3%) revealed FLT3-LM. The sizes of the length mutation varied from 3 bp to more than 400 bp and comprised different parts of the juxtamembrane domain. In 70% of the samples, genomic and RT-PCR were performed in parallel. There were no discrepant results between the methods. In addition, there were no significant intensity variations when RT-PCR products were compared with genomic amplifications. Thus, transcription levels of wild-type and mutant FLT3 appeared to be in the same range.
Correlation to cytogenetics
Cytogenetic analyses were available from all 1003 analyzed patients. They were grouped into 9 categories according to cytogenetics: group 1, normal karyotype (n = 428); group 2, t(15;17) (n = 68); group 3, t(8;21) (n = 69); group 4, inv(16)/ t(16;16) (n = 47); group 5, t(Mq23) (n = 36); group 6, rare recurrent translocations—t(6;9), t(1;3), inv(3)/t(3;3), t(3;12), t(3;21), t(8;16) (n = 23); group 7, complex karyotypes (n = 120); group 8, −5/−7/7q− (n = 28); group 9, all others (n = 184). In the AMLCG study, the different categories are defined as prognostically good (groups 2, 3, and 4), intermediate (groups 1 and 9), and poor (groups 7 and 8). Group 6 is a mixed group.
Ordinal χ2 analysis shows that FLT3-LM is not randomly distributed within cytogenetic subgroups (P < .0001). Of the 234 patients with FLT3-LM, 165 (70.5%) had a normal karyotype (Table 1). This was significantly more than in the group without the mutation, in whom only 244 of 769 (31.7%) were cytogenetically normal (P < .0001). The mutation is most common in the normal karyotype group (38.6%) and in the t(15;17) group (35.3%). In contrast, compared with the total cohort, FLT3-LM was rare in patients with t(8;21) (6 of 69 patients) (P = .0063), 11q23 translocations (1 of 36 patients) (P = .0062), −5/5q−/7q− (1 of 28 patients) (P = .014), rare recurrent translocations (1 of 23 patients), and complex rearrangements (3 of 120 patients) (P < .0001). The mutation was not detected in the 47 patients with inv(16)/ t(16;16). Interestingly, FLT3-LM was relatively frequent in groups with other than the above-mentioned karyotypic anomalies (14.1%). With the exception of one patient with 5q− and 3 patients with complex karyotypes, all were so-called other aberrations such as +8, +11, 9q−, 11q−, or were nonrecurrent rearrangements known as secondary changes in different cytogenetic subgroups and are thus far of unknown prognostic significance.
Correlation of the FLT3 status to cytogenetics
| Karyotype . | Total (%) . | FLT3-LM negative (%) . | FLT3-LM positive (%) . | FLT-LM in karyotype group (%) . | P . |
|---|---|---|---|---|---|
| — | 1003 (100) | 769 (76.7) | 234 (23.3) | 23.3 | — |
| Normal | 428 (42.7) | 244 (31.7) | 165 (70.5) | 38.6 | < .0001 |
| t(15;17) | 68 (6.8) | 41 (5.3) | 24 (10.2) | 35.3 | .0131 |
| t(8;21) | 69 (6.9) | 61 (7.9) | 6 (2.6) | 8.7 | .0063 |
| inv(16) | 47 (4.7) | 42 (5.5) | — | — | .0002 |
| t(11q23) | 36 (3.6) | 32 (4.2) | 1 (0.4) | 2.8 | .0062 |
| Rare (t) | 23 (2.3) | 17 (2.2) | 1 (0.4) | 4.3 | .0758 |
| Complex | 120 (12.0) | 105 (13.7) | 3 (1.3) | 3.7 | < .0001 |
| − 5/− 7/7q− | 28 (2.8) | 27 (3.5) | 1 (0.4) | 3.6 | < .0001 |
| Others | 184 (18.3) | 151 (24.8) | 33 (14.1) | 17.9 | .1071 |
| Karyotype . | Total (%) . | FLT3-LM negative (%) . | FLT3-LM positive (%) . | FLT-LM in karyotype group (%) . | P . |
|---|---|---|---|---|---|
| — | 1003 (100) | 769 (76.7) | 234 (23.3) | 23.3 | — |
| Normal | 428 (42.7) | 244 (31.7) | 165 (70.5) | 38.6 | < .0001 |
| t(15;17) | 68 (6.8) | 41 (5.3) | 24 (10.2) | 35.3 | .0131 |
| t(8;21) | 69 (6.9) | 61 (7.9) | 6 (2.6) | 8.7 | .0063 |
| inv(16) | 47 (4.7) | 42 (5.5) | — | — | .0002 |
| t(11q23) | 36 (3.6) | 32 (4.2) | 1 (0.4) | 2.8 | .0062 |
| Rare (t) | 23 (2.3) | 17 (2.2) | 1 (0.4) | 4.3 | .0758 |
| Complex | 120 (12.0) | 105 (13.7) | 3 (1.3) | 3.7 | < .0001 |
| − 5/− 7/7q− | 28 (2.8) | 27 (3.5) | 1 (0.4) | 3.6 | < .0001 |
| Others | 184 (18.3) | 151 (24.8) | 33 (14.1) | 17.9 | .1071 |
P values: frequency of the mutation within the cytogenetic subgroup compared with the total cohort. Ordinal χ2 analysis shows that FLT3-LM is not randomly distributed within cytogenetic subgroups (P < .0001).
In total, FLT3-LM was more common in patients with de novo AML (24.5%) than in patients with s-AML (15.6%) or in t-AML (11.5%) (Table2). However, as in de novo AML, FLT3-LM was highly correlated to normal and “other” karyotypes in patients with s-AML and t-AML (Table 2).
Comparison of FLT3-LM incidence in de novo AML, t-AML, and s-AML
| Karyotype . | De novo n = 213 of 871 (24.5%) . | s-AML n = 12 of 77 (15.6%) . | t-AML n = 6 of 52 (11.5%) . |
|---|---|---|---|
| Normal | 149 of 379 (39.3%) | 8 of 33 (24.2%) | 5 of 13 (38%) |
| t(15;17) | 24 of 66 (36.4%) | — | −/2 |
| t(8;21) | 6 of 65 (9.2%) | — | −/4 |
| inv(16) | 0 of 46 | — | −/1 |
| t(11q23) | 1 of 34 (2.9%) | — | −/2 |
| Rare (t) | 1 of 19 (5.2%) | −/1 | −/3 |
| Complex | 2 of 94 (2.1%) | 1 of 14 (7.1%) | −/12 |
| − 5/− 7/7q− | 1 of 16 (6.3%) | −/7 | −/5 |
| Others | 29 of 152 (19.1%) | 3 of 22 (9.1%) | 1 of 10 (10%) |
| Karyotype . | De novo n = 213 of 871 (24.5%) . | s-AML n = 12 of 77 (15.6%) . | t-AML n = 6 of 52 (11.5%) . |
|---|---|---|---|
| Normal | 149 of 379 (39.3%) | 8 of 33 (24.2%) | 5 of 13 (38%) |
| t(15;17) | 24 of 66 (36.4%) | — | −/2 |
| t(8;21) | 6 of 65 (9.2%) | — | −/4 |
| inv(16) | 0 of 46 | — | −/1 |
| t(11q23) | 1 of 34 (2.9%) | — | −/2 |
| Rare (t) | 1 of 19 (5.2%) | −/1 | −/3 |
| Complex | 2 of 94 (2.1%) | 1 of 14 (7.1%) | −/12 |
| − 5/− 7/7q− | 1 of 16 (6.3%) | −/7 | −/5 |
| Others | 29 of 152 (19.1%) | 3 of 22 (9.1%) | 1 of 10 (10%) |
The incidence of FLT3-LM according to karyotype was ranked as follows: normal (38.6%) > t(15;17) (35.3%) > others (17.9%) > t(8;21) (8.7%) > rare recurrent translocations (4.3%) > complex karyotypes (3.7%) > −5/−7/7q− (3.6%) > t(Mq23) (2.8%) > inv(16) (0%).
Correlation with cytomorphology
For 864 patients cytomorphologic analysis was available. No correlation with a single, specific FAB subtype was found (Table3). Significant differences were found among the M3 and M5 subtypes with an FLT3-LM frequency of 23.4% for M3 versus 65% for M3v and of 6.4% for M5a versus 34.4% for M5b. The incidence of FLT3-LM was ranked as follows: M3v (65%) > M1 (36.1%) > M5b (34.4%) > M4 (26.8%) > M3 (23.4%) > M2 (19.6%) > M0 (17.2%) > M5a (6.4%). No patient was positive for the duplication in M4eo, M6, or M7. However, patients with M6 or M7 were encountered only as small groups (n = 17 and n = 10, respectively), and no definite conclusion is possible for these subtypes.
Correlation of FLT3-LM to cytomorphology
| FAB . | Total n = 864 (%) . | FLT3-LM negative n = 664 (%) . | FLT3-LM positive n = 200 (%) . | FLT-LM in FAB-group (%) . |
|---|---|---|---|---|
| M0 | 29 (3.4) | 24 (3.6) | 5 (2.5) | 17.2 |
| M1 | 133 (15.4) | 85 (12.8) | 48 (24.0) | 36.1 |
| M2 | 312 (36.1) | 251 (37.8) | 61 (30.5) | 19.6 |
| M3 | 47 (5.4) | 36 (5.4) | 11 (5.5) | 23.4 |
| M3v | 20 (2.3) | 7 (1.1) | 13 (6.5) | 65.0 |
| M4 | 138 (16.0) | 101 (15.2) | 37 (18.5) | 26.8 |
| M4eo | 47 (5.4) | 47 (7.1) | — | — |
| M5a | 47 (5.4) | 44 (6.6) | 3 (1.5) | 6.4 |
| M5b | 64 (7.4) | 42 (6.3) | 22 (11.0) | 34.4 |
| M6 | 17 (2.0) | 17 (2.6) | — | — |
| M7 | 10 (1.2) | 10 (1.5) | — | — |
| FAB . | Total n = 864 (%) . | FLT3-LM negative n = 664 (%) . | FLT3-LM positive n = 200 (%) . | FLT-LM in FAB-group (%) . |
|---|---|---|---|---|
| M0 | 29 (3.4) | 24 (3.6) | 5 (2.5) | 17.2 |
| M1 | 133 (15.4) | 85 (12.8) | 48 (24.0) | 36.1 |
| M2 | 312 (36.1) | 251 (37.8) | 61 (30.5) | 19.6 |
| M3 | 47 (5.4) | 36 (5.4) | 11 (5.5) | 23.4 |
| M3v | 20 (2.3) | 7 (1.1) | 13 (6.5) | 65.0 |
| M4 | 138 (16.0) | 101 (15.2) | 37 (18.5) | 26.8 |
| M4eo | 47 (5.4) | 47 (7.1) | — | — |
| M5a | 47 (5.4) | 44 (6.6) | 3 (1.5) | 6.4 |
| M5b | 64 (7.4) | 42 (6.3) | 22 (11.0) | 34.4 |
| M6 | 17 (2.0) | 17 (2.6) | — | — |
| M7 | 10 (1.2) | 10 (1.5) | — | — |
Correlation of FLT3-LM with leukocyte count in FAB subgroups
For 810 patients, peripheral leukocyte counts at diagnosis were available. Among the total cohort, the leukocyte count was significantly higher in the group with the FLT3-LM than in the group without the mutation (P < .0001). When the leukocyte counts within single cytomorphologic subgroups were regarded, we found significantly elevated leukocyte counts only in FAB M1, M2, and M4 (Table 4).
Correlation between FLT3-LM and leukocyte count in different FAB subgroups
| FAB . | Numbers . | Median leukocyte count . | P . | ||
|---|---|---|---|---|---|
| FLT-LM− . | FLT3-LM+ . | FLT-LM− . | FLT3-LM+ . | ||
| Total | 620 | 190 | 34 029 | 77 120 | < .001 |
| M0 | 22 | 9 | 37 979 | 64 551 | .453 |
| M1 | 68 | 37 | 35 575 | 107 659 | < .001 |
| M2 | 193 | 52 | 24 089 | 65 542 | < .001 |
| M3 | 30 | 8 | 8 351 | 17 113 | .171 |
| M3v | 5 | 9 | 79 560 | 40 589 | .518 |
| M4 | 89 | 30 | 42 866 | 86 746 | .001 |
| M5a | 36 | 3 | 45 445 | 40 167 | .564 |
| M5b | 39 | 17 | 64 657 | 88 359 | .243 |
| FAB . | Numbers . | Median leukocyte count . | P . | ||
|---|---|---|---|---|---|
| FLT-LM− . | FLT3-LM+ . | FLT-LM− . | FLT3-LM+ . | ||
| Total | 620 | 190 | 34 029 | 77 120 | < .001 |
| M0 | 22 | 9 | 37 979 | 64 551 | .453 |
| M1 | 68 | 37 | 35 575 | 107 659 | < .001 |
| M2 | 193 | 52 | 24 089 | 65 542 | < .001 |
| M3 | 30 | 8 | 8 351 | 17 113 | .171 |
| M3v | 5 | 9 | 79 560 | 40 589 | .518 |
| M4 | 89 | 30 | 42 866 | 86 746 | .001 |
| M5a | 36 | 3 | 45 445 | 40 167 | .564 |
| M5b | 39 | 17 | 64 657 | 88 359 | .243 |
Correlation of FLT3-LM with age and sex
The median age of patients with FLT3-LM was 54.9 years versus 57.6 years in the group without the mutation (P = .043). As shown by χ2 analysis, the mutation was more frequent in women than in men, with a 1.36:1 ratio (P = .023).
Prognostic significance of FLT3-LM within the German AMLCG study
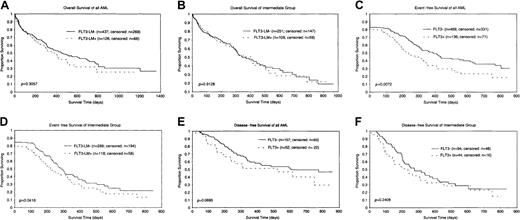
Analysis of the prognostic significance of FLT3-LM was restricted to 563 patients enrolled in the German AMLCG study. The median follow-up time was 11.1 months. Complete remission rates were similar between the groups with the mutation (70.3%) and without the mutation (70.4%). The group with FLT3-LM had a shorter OS than the group without the mutation (median, 12 months vs 15 months); however, this was not statistically significant (P = .3057) (Figure2A). In contrast, EFS was significantly shorter in the group with the mutation (7.4 months vs 12.9 months) (P = .0072) (Figure 2C). In addition, patients with normal karyotype and other karyotypes (n = 360) were analyzed separately because they represent most of the mutation-positive patients and belong to the prognostically intermediate group. OS (11.5 months vs 12.1 months) was almost identical in both groups (Figure 2B), whereas EFS (7.3 months vs 9.4 months) was shorter in the group with the mutation (P = .0416) (Figure 2D). Disease-free survival was 8.4 versus 12.8 months (P = .0695) in the total group (Figure 2E) and 6.9 versus 9.0 months (P = .2408) in the intermediate group (Figure 2F). Multivariate analysis including cytogenetics, age, and secondary etiology of AML as covariates showed that the FLT3-LM state is not an independent prognostic factor for OS, EFS, or DFS.
Kaplan-Meier analyses of the subgroup of patients treated within the German AMLCG study.
(A) OS of all patients included. (B) OS of patients with normal and other karyotypes (prognostically intermediate group). (C) EFS of all patients included. (D) EFS of patients from the prognostically intermediate group. (E) DFS of all patients included. (F) DFS of patients from the prognostically intermediate group.
Kaplan-Meier analyses of the subgroup of patients treated within the German AMLCG study.
(A) OS of all patients included. (B) OS of patients with normal and other karyotypes (prognostically intermediate group). (C) EFS of all patients included. (D) EFS of patients from the prognostically intermediate group. (E) DFS of all patients included. (F) DFS of patients from the prognostically intermediate group.
Mutations at diagnosis and at relapse
Twenty-five patients were analyzed at diagnosis and at relapse (Table 5). All patients with the mutation at diagnosis also carried it at relapse. Karyotype changes were found in 9 patients, 8 of whom had karyotype evolution. One patient experienced karyotype regression. All the chromosomal changes at relapse had to be interpreted as changes secondary to the FLT3 mutation because the FLT3 mutations were present in these patients before the chromosomal changes.
Karyotypes of 25 AML patients with FLT3-LM at diagnosis and at relapse
| No. patients . | Diagnoses . | Relapse . |
|---|---|---|
| 13 | Normal karyotype | Normal karyotype |
| 4 | Normal karyotype | Karyotype alterations (all belonging to other alterations) (karyotype evolution) |
| 1 | t(8;21) | t(8;21) + complex karyotype (karyotype evolution) |
| 3 | Rare or other alteration | Same aberrations |
| 3 | Aberrations | Same aberrations + others (karyotype evolution) |
| 1 | + 8, + 11 | Normal karyotype (karyotype regression) |
| No. patients . | Diagnoses . | Relapse . |
|---|---|---|
| 13 | Normal karyotype | Normal karyotype |
| 4 | Normal karyotype | Karyotype alterations (all belonging to other alterations) (karyotype evolution) |
| 1 | t(8;21) | t(8;21) + complex karyotype (karyotype evolution) |
| 3 | Rare or other alteration | Same aberrations |
| 3 | Aberrations | Same aberrations + others (karyotype evolution) |
| 1 | + 8, + 11 | Normal karyotype (karyotype regression) |
Semiquantitative analysis of the FLT3-LM
Semiquantitative analysis of the amplified fragments representing the mutation in relation to the intensity of the wild-type allele was performed in 180 patients (Table 6). A definition of mutated fragments according to quantity is given in the “Materials and methods” section. In 60% of the patients studied at diagnosis, the mutated fragments are in the range of the wild-type allele (type 2). Ten percent of the patients have weak fragments, which may indicate that the mutation is present in only a part of the leukemic cells (type 1). Six percent revealed only the mutated allele type 4, and 11% are of type 3. Two to 4 different altered bands (different mutations) were found in 13% of the patients at diagnosis. At relapse only 16% had type 2 mutations, and types 1 and 5 mutations were never detected. In contrast, 84% revealed type 3 or type 4 mutations, indicating that there is a tendency to lose the wild-type allele during leukemic evolution. Loss of the wild-type allele was confirmed in 2 patients by fluorescence in situ hybridization (FISH) using a full-length cDNA clone (data not shown). Only one patient showed regression of the mutated allele to the wild-type allele. This patient also showed regression of the karyotype to a normal karyotype, which may be interpreted as a very early stage of relapse.
Semiquantitative analysis of FLT3-LM at diagnosis and distribution in 5 categories according to fragment intensities of the mutated allele(s) (n = 180)
| Category . | At diagnosis n = 180 (%) . | At relapse n = 25 (%) . |
|---|---|---|
| 1 (mutated allele weak) | 18 (10) | — |
| 2 (mutated allele such as wild-type allele) | 108 (60) | 4 (16) |
| 3 (mutated allele strong) | 20 (11) | 8 (32) |
| 4 (only mutated allele) | 11 (6) | 13 (52) |
| 5 (more than one mutated allele) | 23 (13) | — |
| Category . | At diagnosis n = 180 (%) . | At relapse n = 25 (%) . |
|---|---|---|
| 1 (mutated allele weak) | 18 (10) | — |
| 2 (mutated allele such as wild-type allele) | 108 (60) | 4 (16) |
| 3 (mutated allele strong) | 20 (11) | 8 (32) |
| 4 (only mutated allele) | 11 (6) | 13 (52) |
| 5 (more than one mutated allele) | 23 (13) | — |
Evaluation of FLT3-LM as a follow-up marker
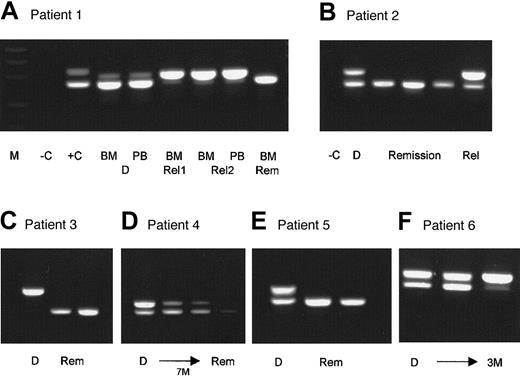
So far, 120 peripheral blood and bone marrow samples of 34 patients with FLT3-LM were evaluated at 2 (12 patients), 3 (14 patients), 4 (1 patient), 6 (1 patient), 7 (2 patients), 8 (1 patient), and 9 (1 patient) time points during therapy. Some examples are summarized in Figure 3. Simple one-step PCR was performed with a sensitivity of 1/100 to 1/1000 cells, depending on the initial FLT3-LM/wild-type ratio. In 2 patients, FISH analysis could be done in parallel because of trisomy 8 at 4 and 8 time points, respectively. In one patient AML1-ETO-PCR and FISH analysis could be conducted because of an initial t(8;21) at 3 time points. Six patients had rare chromosome aberrations at diagnosis that were not detectable by PCR or FISH. Twenty-five patients had normal karyotypes at diagnosis; thus, in 31 patients FLT3-LM was the only marker available for follow-up studies. We found good accordance between FLT3-LM status and clinical state, cytomorphology, cytogenetics, FISH, and PCR. Based on FLT3-LM status, a relapse could be predicted in 4 patients before any other method 2 or 3 months before clinical relapse. These results suggest that FLT3-LM is a useful marker for follow-up studies.
Examples of 6 patients with FLT3-LM at diagnosis who were evaluated for the mutation at different follow-up time points.
M, molecular weight standard; −C, blank control; +C, positive control; BM, bone marrow; PB, peripheral blood; D, sample at diagnosis; Rel, sample at relapse; Rem, sample from remission; M, months. Patient 1 had FLT3-LM at diagnosis in the bone marrow and in the peripheral blood. At follow-up (Rel1) he had a single mutated band in complete remission 3 weeks before morphologic relapse (Rel2). He lost the wild-type allele, which was confirmed by FISH analysis (data not shown). In second remission, only the wild-type allele could be amplified. Patient 2 was studied at 3 remission time points at 3-month intervals. At all remission time points, only the normal allele was detectable. Patient 3 was hemizygous for the mutation at diagnosis and is still in remission with only the wild-type allele detectable. Patient 4 needed 7 months of conventional chemotherapy to achieve clinical remission. Patient 5 was heterozygous for the mutation at diagnosis and is in continuing clinical and molecular remission for 6 months. Patient 6 was heterozygous at diagnosis, did not achieve clinical remission, and shifted to a hemizygous state for the mutation after 3 months.
Examples of 6 patients with FLT3-LM at diagnosis who were evaluated for the mutation at different follow-up time points.
M, molecular weight standard; −C, blank control; +C, positive control; BM, bone marrow; PB, peripheral blood; D, sample at diagnosis; Rel, sample at relapse; Rem, sample from remission; M, months. Patient 1 had FLT3-LM at diagnosis in the bone marrow and in the peripheral blood. At follow-up (Rel1) he had a single mutated band in complete remission 3 weeks before morphologic relapse (Rel2). He lost the wild-type allele, which was confirmed by FISH analysis (data not shown). In second remission, only the wild-type allele could be amplified. Patient 2 was studied at 3 remission time points at 3-month intervals. At all remission time points, only the normal allele was detectable. Patient 3 was hemizygous for the mutation at diagnosis and is still in remission with only the wild-type allele detectable. Patient 4 needed 7 months of conventional chemotherapy to achieve clinical remission. Patient 5 was heterozygous for the mutation at diagnosis and is in continuing clinical and molecular remission for 6 months. Patient 6 was heterozygous at diagnosis, did not achieve clinical remission, and shifted to a hemizygous state for the mutation after 3 months.
Discussion
According to cytogenetics, AML can be subdivided into certain prognostic subgroups. Because approximately 45% of all AML is cytogenetically normal and thus lacks markers that can be useful for subclassification and risk assessment, new molecular markers are urgently needed. We have analyzed the incidence of FLT3-LM in cytogenetic and cytomorphologic AML subgroups in great detail and have evaluated the prognostic significance within the German AMLCG study. The current study thus far comprises the largest series of patients in whom a systematic search for FLT3-LM has been performed and is the first one to assess FLT3-LM for distinct cytogenetic and cytomorphologic subgroups of AML. Its high frequency in patients without detectable cytogenetic aberrations or aberrations of currently unknown prognostic significance may provide the means to better understand the pathogenesis of these so far poorly characterized AML subgroups. The incidence of FLT3-LM in 23.3% of all unselected AML is in the range of that described before.12,13,21 FLT3-LM apparently is not independent of cytogenetics or of FAB subtype, as has been suggested by others.12,13 32
Incidence of FLT3-LM in different cytogenetic subgroups of AML
We could show that FLT-LM is highly correlated with a normal karyotype. It is also relatively common with so-called other chromosome aberrations. The high incidence of FLT3-LM in the latter group may be considered a further hint that these other aberrations are secondary events.33 In all other cytogenetic groups FLT3-LM was rare. Thus, FLT3-LM may be a central principle that characterizes a large subgroup within the cytogenetically normal AML.
Regarding the molecular mechanism of leukemogenesis, nonrandom chromosomal translocations, which usually target and deregulate genes coding for transcription factors and cause a differentiation block, are thought to mediate the initiation of leukemia.3,4 In addition, molecular mutations in the MLL, AML1, and CEBPα genes have been described in AML.6,10,34,35 On the other hand, gain-of-function mutations of the signal-transducing molecules, such as RAS and FLT3, have been described as frequent during AML progression.36The high frequency of FLT3-LM in AML with normal karyotype and the fact that all positive patients analyzed at relapse already carried the mutation at diagnosis implies that FLT3 may also play a role in leukemia initiation. However, our observation of FLT3-LM hemizygosity in a high percentage of relapses supports the hypothesis that loss of wild-type FLT3 has an impact on leukemia progression.
For reasons that are still unknown, AML with specific primary mutations tends to accumulate certain secondary mutations. For example, AML with t(8;21) tends to lose a sex chromosome, and with inv(16) it tends to accumulate chromosome 21, 22, or both.37 Moreover, in a subset of core-binding factor leukemia mutations within c-Kit, a receptor tyrosine kinase that belongs to the same family as FLT3 has been described.38 Now we have identified a mutation that seems to occur most commonly in cytogenetically normal AML. We speculate that at least one further mutation—for example, a mutation in genes for transcription factors—should be present in each of the patients with FLT3-LM to develop full-blown leukemia. It is still unknown what kind of molecular alterations may be second hits (primary or secondary) in leukemia with normal karyotype and FLT3-LM. We observed a partial tandem duplication (PTD) in the MLL gene in 23 of our FLT3-LM–positive patients, and one patient had a PTD and a C/EBPα mutation in addition to the FLT3-LM (data not shown). Thus, these are potential additional hits in FLT3-LM–positive AML.
Incidence of the FLT3-LM in different cytomorphologic subgroups of AML
FLT3-LM is not exclusively correlated with any certain FAB subtype, but it was not distributed equally within these groups. In addition, we found that FLT3-LM is specific for AML. Among 100 patients with acute lymphoid leukemia (ALL), we did not find a single patient with FLT3-LM (data not shown), confirming that FLT3-LM seems to be a rare event in ALL.2
FLT3-LM and leukocytosis
FLT3-LM has been associated with leukocytosis in acute promyelocytic leukemia.30 Because FLT3-LM has been shown to cause constitutive activation of the receptor tyrosine kinase,14 leading to autonomous, cytokine-independent cellular proliferation,16,22 it may be a causative factor for leukocytosis. Because FLT3-LM is particularly frequent in M3v, which is usually associated with elevated leukocyte counts, we speculate that FLT3-LM may be causative for the high leukocyte count in these patients. It still has to be analyzed whether those 35% of M3v patients in whom we did not detect FLT3-LM by PCR may carry a point mutation within the FLT3 gene, as has recently been shown to be a further common event in AML.19 20
Prognostic significance of FLT3-LM
The prognosis of AML depends on factors such as age, initial leukocyte count, FAB classification, karyotype, immune phenotype, and response to remission–induction therapy.39,40 Among them, cytogenetics is the most important prognostic factor.2 In most study groups, the good risk group is defined by t(15;17), t(8;21), and inv(16)/t(16;16); the intermediate group is defined by normal karyotypes and other chromosomal aberrations; and the poor risk group is defined by complex karyotypes, −5/5q−, −7/7q−, 3q− aberrations, and 11q23 translocations. Our study revealed that with the exception of patients with t(15;17), nearly all patients with FLT3-LM belong to the prognostically intermediate group, with a normal karyotype or “other” chromosomal aberrations. Thus, we concentrated on the prognostic evaluation of 184 patients from the intermediate group with normal cytogenetics and “other” aberrations. As could be shown in the German AMLCG study, CR and OS rates of FLT3-LM–positive patients did not differ significantly from those of FLT3-LM–negative patients. EFS was worse (P = .0072) and DFS was slightly worse (P = .0695) in patients with FLT3-LM; both were consistent with a higher relapse rate than in patients without the mutation. Previously, a high significance of FLT3-LM as an indicator for a bad prognosis has been shown in adults and in children.21-24 41 This difference between our data and findings from others may in part be an effect of a change in the prognosis because of an intensification of induction therapy in the AMLCG study. It could be speculated that double induction therapy with at least one course of high-dose ara C may overcome the otherwise poor prognosis of patients with FLT3-LM. Thus, within the AMLCG study, patients with FLT3-LM were not classified as having high-risk AML but were in the intermediate group.
In contrast to all AML patients, FLT3-LM has been shown not to correlate with prognosis in the AML M3 subgroup.30 Because of differentiation therapy with all-trans retinoic acid, the prognosis for AML M3 is favorable compared with other types of AML. Therefore, the M3/M3v group has to be analyzed separately. In our study of 68 patients with M3/M3v, it was not possible to compare the prognosis of the 24 patients with FLT-LM with those without the mutation, because death or relapses are too rare in the ongoing AMLCG trial to make these calculations reasonable.42
We found no reduced remission rates in our cohort of patients with the FLT3-LM, in contrast with findings of other studies.21 24The slightly worse prognosis of patients carrying FLT3-LM within the AMLCG was not due to a reduced remission rate but to a significantly higher risk for relapse than in the group without the mutation.
FLT3-LM at relapse
Relapse is a major cause of treatment failure in AML, and relapsed leukemia is generally resistant to chemotherapy. Thus, relapse does not simply mean the reappearance of leukemia. More than half of the patients with relapsed AML had karyotype changes,43-45frequently to a more complex karyotype and rarely to a normal or a clonally unrelated karyotype. These findings suggest that relapse is accompanied by clonal evolution. Few studies on molecular alterations are associated with relapse in AML. It may be speculated whether FLT3 mutations are responsible for disease progression and, therefore, are more common at relapse. We found the FLT3 mutation to be present at both time points in all 25 patients who could be analyzed at diagnosis and at relapse. This was in contrast to others, who described instability at diagnosis and relapse in 7 of 12 patients.46 In our study, 8 patients accumulated the mutation and 13 even proceeded to a hemizygous state of the mutation at relapse. This is in good agreement with the finding that the prognosis gets worse with an increasing ratio of FLT3-LM to the wild-type allele.47 48
FLT3-LM as a follow-up marker
Detection of minimal residual disease by highly sensitive PCR-based methods is of growing importance to monitor therapy responses and early relapses. However, only 25% of patients with AML carry fusion genes that are good targets for PCR detection. FLT3-LM now provides us with a PCR target for approximately another 20% of patients with AML. Because patients with FLT3-LM are prone to relapses, PCR detection of molecular relapses may be helpful in overcoming clinical relapse through early therapeutic intervention.
Future perspectives
Given the high relapse rates and the nature of the mutations characterized in the way it targets receptor tyrosine kinases, it may be appropriate to consider alternative therapies for FLT3-LM. Therefore, for patients with FLT3-LM who are refractory to primary induction therapy or for those with relapsed disease, novel strategies such as therapy with tyrosine kinase inhibitors may be useful. Hence, FLT3-LM may provide a novel molecular target for AML therapy.49
This is to date the largest study on FLT3-LM in adult patients with AML. Based on our findings, we conclude that FLT3-LM is most frequent in the subgroup with normal karyotypes. Thus, with the growing importance of minimal residual disease studies in AML, FLT3-LM provides a molecular marker in a large subset of patients with AML in whom no molecular monitoring was possible until now. Because therapy outcomes for patients with and without FLT3-LM did not differ significantly in our investigation, the prognostic significance of FLT3-LM in adult AML should be further evaluated before FLT3-LM is used as a possible marker for up-front risk stratification in adult AML.
We thank Gudrun Mellert and Sabine Koneberg for excellent technical assistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Susanne Schnittger, Department of Internal Medicine III, University Hospital Grosshadern, Ludwig-Maximilians-University, Marchioninistrasse 15, 81377 Munich, Germany; e-mail: susanne.schnittger@med3.med.uni-muenchen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal