Abstract
In primary chicken erythroblasts (stem cell factor [SCF] erythroblasts), transferrin receptor (TfR) messenger RNA (mRNA) and protein were hyperexpressed as compared to nonerythroid chicken cell types. This erythroid-specific hyperexpression was abolished in transformed erythroblasts (HD3E22 cells) expressing the v-ErbA and v-ErbB oncogenes of avian erythroblastosis virus. TfR expression in HD3E22 cells could be modulated by changes in exogenous iron supply, whereas expression in SCF erythroblasts was not subject to iron regulation. Measurements of TfR mRNA half-life indicated that hyperexpression in SCF erythroblasts was due to a massive stabilization of transcripts even in the presence of high iron levels. Changes in mRNA binding activity of iron regulatory protein 1 (IRP1), the primary regulator of TfR mRNA stability in these cells, correlated well with TfR mRNA expression; IRP1 activity in HD3E22 cells and other nonerythroid cell types tested was iron dependent, whereas IRP1 activity in primary SCF erythroblasts could not be modulated by iron administration. Analysis of avian erythroblasts expressing v-ErbA alone indicated that v-ErbA was responsible for these transformation-specific alterations in the regulation of iron metabolism. In SCF erythroblasts high amounts of TfR were detected on the plasma membrane, but a large fraction was also located in early and late endosomal compartments, potentially concealing temporary iron stores from the IRP regulatory system. In contrast, TfR was almost exclusively located to the plasma membrane in HD3E22 cells. In summary, stabilization of TfR mRNA and redistribution of Fe-Tf/TfR complexes to late endosomal compartments may contribute to TfR hyperexpression in primary erythroblasts, effects that are lost on leukemic transformation.
Introduction
All proliferating cells need iron but also have to cope with the extreme toxicity of free iron, which participates in redox reactions leading to radical formation. Therefore, iron metabolism has to be kept under tight surveillance.1 2This is especially true for erythroblasts with their increased demand for iron during terminal differentiation.
Vertebrate cells take up iron from iron-loaded diferric serum transferrin (Tf), which is internalized via transferrin receptors (TfRs) by receptor-mediated endocytosis. Inside the cell, the low pH in acidic endosomes liberates iron from Tf for cellular use or storage in ferritin (Fer). (Apo)–Tf/TfR complexes return to the cell surface for reutilization.3 In most cell types, coordinate regulation of TfR and Fer expression maintains iron homeostasis.2,4Cis-acting palindromic elements, termed iron responsive elements (IREs), and transacting factors, the iron regulatory proteins (IRP) IRP1 and IRP2, mediate this process. When iron is scarce, IRPs bind to IREs in the 3′ untranslated region (UTR) of TfR messenger RNA (mRNA), increasing its stability. IRP binding to the IRE in the 5′UTR of Fer mRNA decreases translation initiation. This leads to increased iron uptake via TfR and less capacity for iron storage in Fer.5-7 Conversely, high iron levels induce rapid TfR mRNA degradation and allow efficient Fer protein synthesis, inhibiting iron uptake and favoring storage. This type of regulation will be referred to here as the “standard” model of iron homeostasis. IREs have also been identified in several other mRNAs, including the erythroid isoform of δ-aminolevulinic acid synthase (e-ALAS),8 9 which catalyzes the first and rate-limiting step of heme biosynthesis.
Most studies addressing regulation of erythroid iron metabolism have been performed with established cell lines derived from leukemias10 or with spontaneously immortalized cells.11 All these cell types require no or nonphysiologic stimuli for proliferation and differentiation and usually fail to mature terminally as well as to express the correct pattern of erythroid proteins.12,13 To overcome these drawbacks, we used primary erythroblasts grown out from chicken bone marrow in the presence of the self-renewal factors stem cell factor (SCF), transforming growth factor α (TGF-α), estradiol (E2), and the synthetic glucocorticoid dexamethasone (Dex). In this medium, committed erythroblast colony-forming unit (CFU-E)–like erythroid cells (termed SCF erythroblasts) undergo 8 to 10 divisions, yielding sufficient material for biochemical analyses. SCF erythroblasts can be induced to differentiate synchronously by replacing self-renewal factors with erythropoietin (Epo) and insulin (Ins),14-17 resulting in mature cells virtually indistinguishable from erythrocytes isolated from peripheral blood. Hence, this primary chicken cell system represents a powerful tool to study the regulation of iron metabolism in committed erythroblasts.18
Our analyses focused on TfR expression and regulation by IRP1 in primary self-renewing erythroblasts compared with transformed erythroid cells and cells of nonerythroid origin. In particular, we used (1) primary SCF erythroblasts after 4 days of outgrowth from chicken bone marrow in the presence of self-renewal factors17; (2) erythroblasts stably transfected with a retroviral vector expressing the SCF receptor, c-Kit19 (these strictly factor-dependent cells undergo extended self-renewal in media containing Epo, SCF, and Dex for about 30 divisions before senescence); (3) immortalized HD3E22 erythroblasts, transformed by the leukemogenic avian erythroblastosis virus AEV (expressing the viral oncogenes v-ErbA and v-ErbB) and stably expressing the murine erythropoietin receptor (EpoR)20,21; (4) erythroblasts stably expressing either the v-ErbA alone22 (these cells share different aspects of the fully transformed erythroblast phenotype but both have an extended lifespan); (5) early hematopoietic progenitors expressing the v-Ski oncogene, capable of undergoing erythroid or myeloid differentiation depending on the growth factor combination23; (6) MC29-HD11, a monocytic cell line with phagocytic capacity24,25; (7) primary chicken embryo fibroblasts, CEF26; and the avian leghorn male hepatoma cell line, LMH.27
We show that iron metabolism is regulated differently in primary erythroblasts as compared to several other, constantly proliferating or transformed cell types. The physiologic demand for high iron uptake into committed erythroblasts is reflected by exceedingly high levels of TfR mRNA and protein expression which, in addition, cannot be modulated by variations in exogenous iron supply. This appears to be due to stabilization of TfR transcripts and redistribution of iron-loaded Tf/TfR complexes into intracellular compartments. Neither phenotype was observed in AEV-transformed erythroleukemic cells or erythroblasts overexpressing the v-ErbA oncogene alone. This suggests that TfR hyperexpression is an important feature of committed primary erythroblasts before the onset of differentiation, which can be abolished by v-ErbA-mediated leukemic transformation.
Materials and methods
Cell culture
The SCF erythroblasts were grown from the bone marrow of SPAFAS chicks as described.16,17 SCF erythroblasts and c-Kit–overexpressing erythroblasts19 were kept between 2 × 106 and 4 × 106cells/mL with CFU-E medium, consisting of Dulbecco modified Eagle medium (DMEM), 12% fetal calf serum (FCS), 4.4% chicken serum (chS), 15% double-distilled water, 1.9 mg/mL detoxified bovine serum albumin (BSA; Sigma, St Louis, MO), 2.0 mg/mL NaHCO3, and 0.13 mM 2-mercaptoethanol (2-ME). For SCF erythroblasts, CFU-E medium was supplemented with 100 ng/mL avian SCF,28 1 μM E2 (Sigma), and 1 μM Dex, (Sigma). The c-kit overexpressing erythroblasts were grown in the same medium plus 40 ng/mL insulinlike growth factor 1 (IGF-1) and 3% anemic chS as a source of avian erythropoietin. AEV-transformed HD3E22 erythroblasts (ectopically expressing murine recombinant erythropoietin receptor; v-ErbA/ v-ErbB/mEpoR) were grown in CFU-E medium with 40 μg human recombinant IGF-1.21CEFs26 received EBM-H medium composed of DMEM plus 8% FCS, 2% chS, and 20 mM HEPES (pH 7.1). Erythroblasts expressing v-ErbA29 were cultivated in CFU-E medium supplemented with 100 ng/mL SCF, 40 ng/mL IGF-1, 1 μM of the E2 antagonist ICI189.770 and 3 μM of the glucocorticoid antagonist ZK112.993. Primary v-Ski–transfected early hematopoietic progenitors23 were cultured in CFU-E medium plus 100 ng/mL SCF (see above), 10 ng/mL chicken myelomonocytic growth factor (cMGF)30 and 40 ng/mL IGF-1. MC29-HD11 monocytic cells25 received a 1:1 mixture of EBM-H and CFU-E medium. LMH cells were cultured in Waymouth medium plus 10% FCS and 2 mM glutamine.27 Murine Ltk− fibroblasts received DMEM plus 10% FCS.
Where indicated, iron-saturated chicken transferrin (Tf, 1 mg/mL = 12.5 μM Fe2Tf = 25 μM Fe; the physiologic concentration in serum, referred to as “high iron”), ferric ammonium citrate (FAC, 17.5% Fe saturation, 20 μg/mL = 63 μM Fe; referred to as “iron overload”) or desferrioxamine (DFO, 50 μM, Fe3+-chelator; referred to as “iron deprivation”) were added. Medium containing only the Tf from chS as an iron source is referred to as “low iron” (0.5-1.1 μM Fe, depending on the serum concentration in the medium).
Flow cytometry
Cells were stained in vivo for 30 minutes with a murine monoclonal antibody specific for chicken TfR, JS-8,31washed 3 times in 1% FCS/phosphate-buffered saline (PBS), incubated with fluorescein-labeled second antibody (goat antimouse; Dako, Glostrup, Denmark) for another 30 minutes, washed again, and subjected to flow cytometry (FACScan; Becton Dickinson). As controls, cells were analyzed without staining, or stained with a nonspecific antibody (51/3) and the fluorescein isothiocyanate (FITC)-conjugated secondary antibody, or solely with the secondary FITC-labeled antibody (Sigma).
RNA isolation and Northern blot analysis
Total RNA was prepared from 107 to 4 × 107 cells using 4 M guanidinium thiocyanate (GTC) lysis buffer and extraction with acid phenol/chloroform.32 RNA samples (10 μg) were separated in formaldehyde-3-[N-morpholino]-propanesulfonic acid (MOPS)-agarose gels and transferred to nylon membranes (GeneScreen; Du Pont, Wilmington, DE). After UV cross-linking (Stratalinker 2400; Stratagene, La Jolla, CA), transfer was checked by dyeing with methylene blue. Membranes were hybridized with random-primed32P-labeled probes specific for chicken TfR mRNA (nt1942-2277 cds cloned by reverse transcription-polymerase chain reaction [RT-PCR] according to the published sequence ACX55348)33 and for normalization with a probe specific for 18S ribosomal RNA (rRNA).34 Quantification of the32P-signals was performed by laser densitometry (Molecular Dynamics, Sunnyvale, CA) of autoradiographs or PhosphoImage analysis (Molecular Dynamics).
Electrophoretic mobility shift assay
Cytoplasmic extracts were prepared with a buffer containing 0.2% NP-40. Labeled IRE probes were prepared by in vitro transcription of pSPT-Fer (IRE of human ferritin H-chain mRNA)35 in the presence of α32P cytidine-tri-phosphate (CTP; 800 Ci/mM [2960 × 1010 Bq], NEN/Du Pont, Wilmington, DE). IRE/IRP binding reactions were carried out with 2 μg protein and 0.5 ng 32P-labeled IRE probes (specific activity: 1.3 × 109 dpm/μg) by incubation for 10 minutes at room temperature. After treatment with RNase T1 (20 U; Roche, Mannheim, Germany) and 5 μg/μL heparin for 10 minutes each, RNA-protein complexes were separated in 6% nondenaturing polyacrylamide gels. The total amount of IRP1 was assessed by in vitro reduction with 3% 2-ME prior to the binding reaction,36 which also served as internal loading control.37
Immunoelectron microscopy
Fixation, cryosectioning, and immunolabeling were performed as described.38 Briefly, cells were fixed by adding 16% paraformaldehyde in Pipes buffer (200 mM, pH 7.0) to a cell suspension to obtain a final concentration of 4% paraformaldehyde. Cells were centrifuged at 1000g for 6 minutes. The supernatant was removed and 8% paraformaldehyde carefully layered onto the pellet. After 1 hour, cells were centrifuged at 13 000g for 5 minutes and the pellets stored for 24 hours at 4°C. Pieces of the pellet were infused with 2.1 M sucrose and cryosectioned in an ultramicrotome (Leica, Solms, Germany). Ultrathin sections were immunolabeled with a monoclonal murine antibody against chicken TfR, JS-8,31 and visualized with rabbit antimouse Protein A/gold (10 nm) complex (Biocell, Helsinki, Finland). The samples were viewed in a transmission electron microscope (JEOL1210; JEOL USA, Peabody, MA) at 80 kV.
Results
Viral transformation with AEV abolishes hyperexpression of TfR in primary erythroblasts
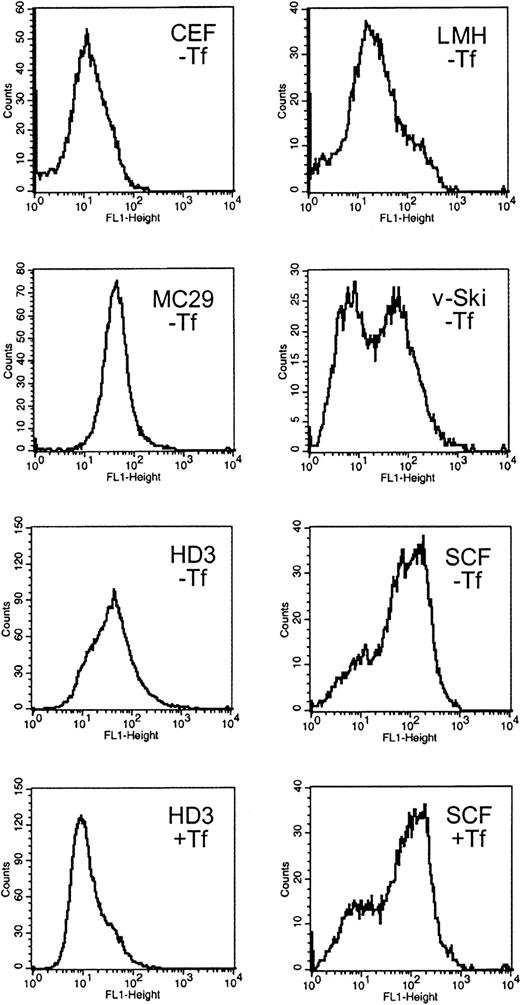
Expression of TfR on the cell surface was compared among several chicken cell types of erythroid and nonerythroid origin. Cytofluorometry after in vivo labeling with the antichicken TfR antibody JS-831 was performed with primary SCF erythroblasts, AEV-transformed HD3E22 erythroblasts expressing the viral oncogenes v-ErbA and v-ErbB, v-Ski oncogene-expressing hematopoietic progenitors, the monocytic cell line MC29-HD11, the hepatic cell line LMH, and primary CEF fibroblasts (see “Materials and methods”). All cell types were cultured under “low iron” conditions (ie, without additional iron except endogenous avian Fe-Tf contained in the chS of culture media, corresponding to 0.5-1.1 μM Fe; see “Materials and methods”). Under such conditions, TfR expression should be elevated due to limiting iron supply. HD3E22 and the SCF erythroblasts were also incubated in the presence of physiologic concentrations of iron-loaded conalbumin (1 mg/mL Fe2Tf = 25 μM Fe; “high iron”), which, according to the “standard model” of iron homeostasis, should lead to a reduction in TfR levels.
Under low iron, fibroblasts and hepatocytes (CEF, LMH) exhibited low TfR expression levels (12 and 20 fluorescence units, respectively; Figure 1), whereasmyc-transformed monocytes (MC29-HD11) had intermediate TfR levels (45 fluorescence units). Multipotent, v-Ski–expressing progenitors, which contain cells of both the myeloid and erythroid lineage,23 had an average TfR level of 30 units. Interestingly, the myeloid fraction of v-Ski progenitors exhibited TfR signals close to background levels (6 units), whereas the erythroid-committed v-Skicells exhibited 90 fluorescence units, indicating an erythroid-specific elevation of TfR expression.
Surface expression of functional TfRs in erythroid and nonerythroid cell types.
Cells were immunolabeled with the chicken TfR-specific antibody JS-831 and a secondary FITC-conjugated antibody immediately after harvesting. The cells were subjected to flow cytometry (FACScan; Becton Dickinson) and results analyzed using the CellQuest software package. The histograms represent the frequency distribution of fluorescence intensity (FL-1 height) directly correlating with the TfR expression levels on the cell surface. As controls, cells were analyzed without staining, stained with an unspecific antibody (51/3) and FITC or solely with FITC. The background fluorescence was less than 6 units (not shown). The nonerythroid cell types CEF, LMH, and MC29-HD11 and the hematopoietic v-Ski–expressing progenitors were cultured under standard conditions (without additional iron supply; labeled −Tf). The erythroleukemic HD3E22 cells and the primary erythroid SCF progenitors were cultured either under physiologic iron supply (1 mg/mL Tf; for details see “Materials and methods”; labeled +Tf), or without additional iron source (−Tf) for at least 24 hours. CEF indicates chicken embryo fibroblasts; LMH, leghorn male hepatoma cells; MC29, MC29-HD11 macrophagelike cells; v-Ski, hematopoietic cells expressing the v-Ski proto-oncogene; SCF, SCF progenitors; HD3, HD3E22 erythroblasts.
Surface expression of functional TfRs in erythroid and nonerythroid cell types.
Cells were immunolabeled with the chicken TfR-specific antibody JS-831 and a secondary FITC-conjugated antibody immediately after harvesting. The cells were subjected to flow cytometry (FACScan; Becton Dickinson) and results analyzed using the CellQuest software package. The histograms represent the frequency distribution of fluorescence intensity (FL-1 height) directly correlating with the TfR expression levels on the cell surface. As controls, cells were analyzed without staining, stained with an unspecific antibody (51/3) and FITC or solely with FITC. The background fluorescence was less than 6 units (not shown). The nonerythroid cell types CEF, LMH, and MC29-HD11 and the hematopoietic v-Ski–expressing progenitors were cultured under standard conditions (without additional iron supply; labeled −Tf). The erythroleukemic HD3E22 cells and the primary erythroid SCF progenitors were cultured either under physiologic iron supply (1 mg/mL Tf; for details see “Materials and methods”; labeled +Tf), or without additional iron source (−Tf) for at least 24 hours. CEF indicates chicken embryo fibroblasts; LMH, leghorn male hepatoma cells; MC29, MC29-HD11 macrophagelike cells; v-Ski, hematopoietic cells expressing the v-Ski proto-oncogene; SCF, SCF progenitors; HD3, HD3E22 erythroblasts.
In the HD3E22 erythroblasts, TfR levels were intermediate to high (45 units) when grown under low iron; however, the presence of high Tf caused the expected down-regulation of TfR expression (10 units). Compared with all the previously tested cell types, extremely high levels of TfR—here referred to as “hyperexpression”—were observed on the cell surface of primary SCF erythroblasts (120 units). Surprisingly, SCF erythroblasts cultured under high iron supply completely maintained this TfR hyperexpression, a result in sharp contrast to the iron-dependent modulation of TfR expression predicted by the standard model. These results suggested 2 distinct patterns of regulation: an iron-independent high-level expression of TfR in primary erythroblasts versus an iron-dependent modulation in erythroleukemic cells.
Hyperexpression of TfR mRNA in primary erythroblasts is independent of iron supply
Discrepancies between TfR protein and transcript abundance have been described previously. For example, TfR transcripts increased approximately 20-fold due to transcriptional activation and mRNA stabilization in J2E cells induced to partially differentiate by erythropoietin, whereas expression of surface TfR only doubled.39 Similar observations were also made in murine L-cell fibroblasts under different iron concentrations.40Therefore, we analyzed iron-dependent regulation of TfR mRNA levels in the cell types described above. In particular, we wanted to test whether iron would induce a modulation of TfR mRNA expression also in the apparently nonresponsive SCF erythroblasts.
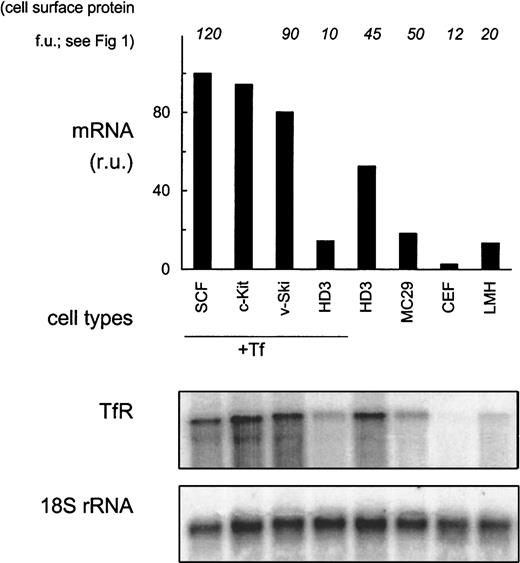
In general, expression of TfR mRNA correlated well with receptor levels on the cell surface. Primary erythroblasts showed hyperexpression of TfR mRNA 100 relative units (ru; Figure2) as compared with fibroblasts (3 ru; only visible after extended exposures), monocytic (18 ru) or hepatic (13 ru) cells. Whereas primary erythroblasts transfected with c-Kit or v-Ski maintained hyperexpression (94 ru and 80 ru, respectively), the immortalized leukemic HD3E22 cells lost high-level expression of TfR mRNA under physiologic concentrations of iron-loaded Tf (1 mg/mL; 14 ru). Under low iron, however, the HD3E22 cells displayed a 4-fold induction of TfR mRNA abundance (54 ru), which could be ascribed to IRP-dependent stabilization of TfR-mRNA (see below and data by others10).
TfR mRNA expression in erythroid and nonerythroid cells.
RNA samples from erythroid (SCF progenitors, c-Kit overexpressing erythroblasts, v-Ski progenitors, and HD3E22 cells) and nonerythroid cell types (MC29-HD11 macrophages, CEF, and LMH cells) were subjected to Northern blot analysis. Erythroid cell types were cultured in the presence of 1 mg/mL Tf (+Tf), HD3E22 erythroblasts (additionally for direct comparison), and the nonerythroid cells under standard conditions (without extra iron source). Northern blots were sequentially hybridized with a 32P-labeled probe specific for chicken TfR, and for normalization with 18S rRNA. The signals of the PhosphoImages were quantified by using ImageQuant software (Molecular Dynamics) and are expressed in relative units (ru; maximum value for each cell type set to 100). To facilitate comparison, the results of TfR protein expression measurements on the cell surface are indicated on top of the diagram. Abbreviations are the same as those in Figure 1.
TfR mRNA expression in erythroid and nonerythroid cells.
RNA samples from erythroid (SCF progenitors, c-Kit overexpressing erythroblasts, v-Ski progenitors, and HD3E22 cells) and nonerythroid cell types (MC29-HD11 macrophages, CEF, and LMH cells) were subjected to Northern blot analysis. Erythroid cell types were cultured in the presence of 1 mg/mL Tf (+Tf), HD3E22 erythroblasts (additionally for direct comparison), and the nonerythroid cells under standard conditions (without extra iron source). Northern blots were sequentially hybridized with a 32P-labeled probe specific for chicken TfR, and for normalization with 18S rRNA. The signals of the PhosphoImages were quantified by using ImageQuant software (Molecular Dynamics) and are expressed in relative units (ru; maximum value for each cell type set to 100). To facilitate comparison, the results of TfR protein expression measurements on the cell surface are indicated on top of the diagram. Abbreviations are the same as those in Figure 1.
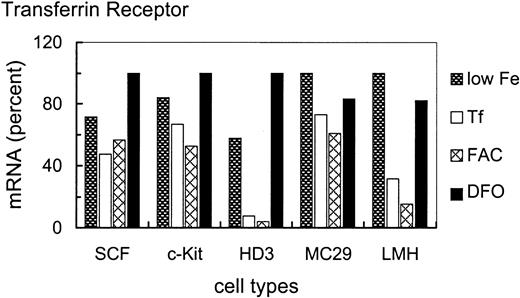
To assess the iron-dependent regulation potential of TfR mRNA in the various cell types in more detail, cells were treated for 24 hours with either no additional iron source, or 1 mg/mL Tf, or high concentrations (63 μM Fe) of the inorganic iron source FAC or 50 μM of the specific iron chelator DFO (Figure 3). Higher doses of or longer incubation times with FAC or DFO were not tolerated by the primary erythroblasts. These conditions were used to test whether the apparent nonresponsiveness of SCF erythroblasts to various concentrations of the physiologic iron donor Tf could be overcome by more extreme conditions of iron overload or deprivation, or by a Tf-independent route of iron administration.
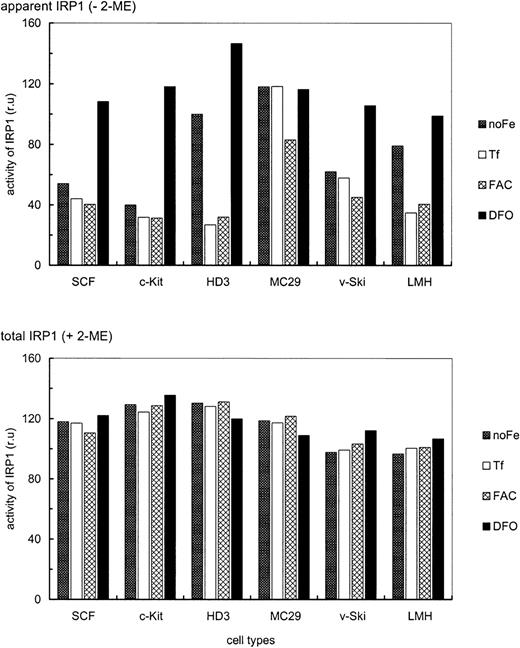
Iron-dependent modulation of TfR mRNA levels in primary SCF progenitors compared with erythroleukemic and somatic cell types.
Iron-dependent modulation of TfR mRNA was compared in primary and transformed erythroid (SCF, c-Kit, HD3E22) as well as nonerythroid (MC29-HD11 and LMH) cell types. The cells were cultured under different iron supply for 24 hours; no additional iron, 1 mg/mL Tf (= 25μM Fe), 20 μg/mL FAC (= 63 μM Fe), and 50 μM DFO. TfR mRNA levels were determined as described in the legend of Figure 2. To emphasize the iron-dependent changes in TfR mRNA levels between the different cell types, the respective maximal values were set to 100%. Note that the relatively “small” differences in mRNA abundance (as compared to other reports in the literature) are due to the linear quantification by PhosphoImage analysis used here as compared to densitometric evaluation of autoradiographs, which has a tendency to underestimate both very low as well as very high signal intensities.
Iron-dependent modulation of TfR mRNA levels in primary SCF progenitors compared with erythroleukemic and somatic cell types.
Iron-dependent modulation of TfR mRNA was compared in primary and transformed erythroid (SCF, c-Kit, HD3E22) as well as nonerythroid (MC29-HD11 and LMH) cell types. The cells were cultured under different iron supply for 24 hours; no additional iron, 1 mg/mL Tf (= 25μM Fe), 20 μg/mL FAC (= 63 μM Fe), and 50 μM DFO. TfR mRNA levels were determined as described in the legend of Figure 2. To emphasize the iron-dependent changes in TfR mRNA levels between the different cell types, the respective maximal values were set to 100%. Note that the relatively “small” differences in mRNA abundance (as compared to other reports in the literature) are due to the linear quantification by PhosphoImage analysis used here as compared to densitometric evaluation of autoradiographs, which has a tendency to underestimate both very low as well as very high signal intensities.
As with different Tf concentrations (Figure 1), primary SCF erythroblasts and erythroblasts overexpressing c-Kit exhibited only a very minor increase of TfR mRNA levels (1.5- to 2-fold) under low iron or iron deprivation compared with iron overload (Figure 3). This suggests that these primary erythroid cells maintained TfR hyperexpression independent of the amount or route of iron uptake. Interestingly, the v-myc–transformed monocytic cells also displayed a similarly small modulation of TfR mRNA abundance (1.5-fold). In contrast, under iron deprivation, the erythroleukemic HD3E22 cells increased TfR transcripts approximately 12-fold compared with FAC-induced iron overload. This regulation pattern closely resembled the situation in the liver-derived LMH cells (about 7-fold induction), which can be seen as a prototype for regulation according to the standard model of iron homeostasis. It should be noted that the maximum levels of TfR mRNA attainable in the different cell types were similar (revealed by multiple cross-correlations and direct comparisons between HD3E22 cells and primary erythroblasts; also compare with Figures 2 and 7). Thus, the extent of iron-dependent modulation of TfR mRNA depends both on the cell type and on leukemic transformation within the erythroid lineage.
Cell type–specific differences in TfR mRNA abundance correlate only partially with IRP1 activity
According to the standard model of iron homeostasis, opposite regulation of Fer and TfR expression, mediated by iron-dependent modulation of IRP mRNA binding activity, leads to a tight surveillance of intracellular iron levels.7 This model, however, falls short in explaining regulation of iron metabolism in specialized cell types such as macrophages, which have to temporarily store iron from phagocytosed erythrocytes for reutilization,41 or committed and terminally differentiating erythroid cells, which need a high capacity for iron uptake (TfR) and utilization for heme synthesis (e-ALAS), simultaneously.10 Therefore, we tested the correlation between IRP1 activity and TfR expression and the corresponding iron-dependent regulatory potential in the cell types described above.
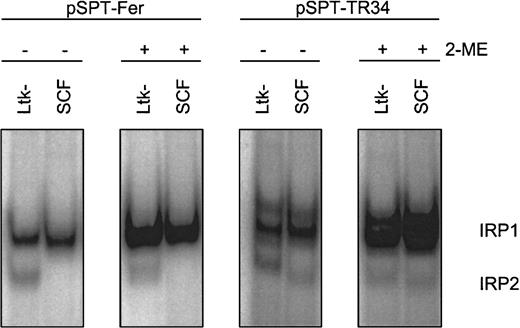
First, appropriate assay conditions had to be defined for the chicken system. As shown previously, IRP1 has similar structural and functional properties in different species, including chicken.42However, compared to the well-studied counterparts of human and murine origin,7 characteristics of chicken IRP2 versus IRP1 were only recently reported in the literature.22 Therefore, we determined the amounts and properties of the IRPs in primary SCF erythroblasts in relation to the well-characterized murine thymidine kinase deficient L-cell fibroblasts (Ltk−).35IRE/IRP electrophoretic mobility shift assays (EMSAs) were performed with crude cytosolic extracts and probes derived from the human Fer H-chain IRE (pSPT-Fer) and IRE “E” of human TfR mRNA (pSPT-TR34).35 Chicken IRE/IRP complexes displayed migration properties similar to the corresponding murine samples (Figure 4). The murine fibroblasts exhibited a comparable level of “apparent” IRP1 as the SCF erythroblasts, and this level could be increased to a similar activity by in vitro reduction with 2-ME, a measure for “total” IRP1 abundance.36 IRP1 association kinetics to the IRE probe was similar with samples from both species, suggesting similar affinities between chicken and mouse IRP1 and the IRE of the human Fer H-chain transcript (data not shown).
RNA-binding properties of IRP1 in chicken versus murine cells.
Apparent (−2-ME) and total (+2-ME) IRP activities in chicken SCF progenitors and murine L cells (Ltk−) were compared by IRE/IRP EMSAs using IRE probes derived from pSPT-Fer and pSPT-TR34 (see “Materials and methods”). The PhosphoImages show the respective migration properties of IRP1 and IRP2. In multiple measurements, the accuracy of this assay was at least ± 12% provided a linear quantification method of the 32P signals, that is, PhosphoImage analysis, was used.
RNA-binding properties of IRP1 in chicken versus murine cells.
Apparent (−2-ME) and total (+2-ME) IRP activities in chicken SCF progenitors and murine L cells (Ltk−) were compared by IRE/IRP EMSAs using IRE probes derived from pSPT-Fer and pSPT-TR34 (see “Materials and methods”). The PhosphoImages show the respective migration properties of IRP1 and IRP2. In multiple measurements, the accuracy of this assay was at least ± 12% provided a linear quantification method of the 32P signals, that is, PhosphoImage analysis, was used.
In chicken, IRE/IRP2 complexes could not be observed with the Fer H probe but were detectable at low levels with pSPT-TR34. These IRE/IRP2 complexes (1) migrated similarly to their murine counterparts; (2) were insensitive toward in vitro reduction by 2-ME, a characteristic property of IRP243; and (3) exhibited the same regulatory pattern and similar association kinetics to the IRE probe in both species (data not shown). Due to the apparently low level of IRP2 in the primary chicken erythroblasts (< 5% that of IRP1), however, IRP2 could not be held responsible for the regulation of iron homeostasis and will not be discussed further. Thus, taken together, the human Fer H-chain IRE transcript appeared to be appropriate for testing apparent and total activity of IRP1 in chicken cells.
Next, modulation of IRP1 mRNA binding activity by alterations in iron supply was compared between the erythroid and nonerythroid chicken cell types described above. To reach equilibrium, all cells were maintained for 24 hours under the various conditions regarding iron supply. EMSAs revealed no significant cell type specific differences in total IRP1 levels, as assessed by in vitro reduction with 3% 2-ME (Figure5). Apparent IRP1 activity, however, showed a distinct, iron-dependent regulation, depending on the cell type. HD3E22 cells showed a 3.7-fold increase in IRP1 activity under low iron (only Tf from the chS in the medium) and a 4.6-fold elevation under complete iron deprivation (DFO), similarly to hepatic cells (LMH). In contrast, primary SCF and c-Kit erythroblasts exhibited essentially no difference in IRP1 activity under low versus high iron (additional exogenous Tf or ferric ammonium citrate). Only after nonphysiologic iron deprivation with DFO was an intermediate response observed (2.5- and 3.7-fold elevation, respectively). Interestingly, this phenotype resembled the regulation in v-Skiprogenitors, which contain a significant fraction of erythroid progenitors resembling SCF erythroblasts.23 In contrast, the v-myc–transformed monocytic cells (MC29-HD11) showed high IRP1 activity under all conditions, in line with their iron-independent elevated level of TfR mRNA (Figure 3). This may explain the discrepancy between the rather high TfR expression level on the cell surface as compared with the relatively low mRNA abundance in this cell type (Figures 1 and 2).
Iron-dependent modulation of IRP1 activity in erythroid versus nonerythroid cells.
IRE/IRP EMSAs were performed with cytoplasmic extracts of the following erythroid and nonerythroid cells: SCF progenitors (SCF), c-Kit progenitors (c-Kit), HD3E22 erythroblasts (HD3), MC29-HD11 macrophages (MC29), v-Ski progenitors (v-Ski), and leghorn male hepatocytes (LMH). The cells were cultured in the presence of 1 mg/mL Tf (Tf), 20 μg/mL FAC (FAC), 50 μM DFO (DFO) or in the absence of any additional iron source (no Fe). Apparent (−2-ME) and total IRP1 (+2-ME) activities were quantified as described in the legend of Figure 2 and in “Materials and methods” and are expressed in relative units, which are directly comparable within this data set.
Iron-dependent modulation of IRP1 activity in erythroid versus nonerythroid cells.
IRE/IRP EMSAs were performed with cytoplasmic extracts of the following erythroid and nonerythroid cells: SCF progenitors (SCF), c-Kit progenitors (c-Kit), HD3E22 erythroblasts (HD3), MC29-HD11 macrophages (MC29), v-Ski progenitors (v-Ski), and leghorn male hepatocytes (LMH). The cells were cultured in the presence of 1 mg/mL Tf (Tf), 20 μg/mL FAC (FAC), 50 μM DFO (DFO) or in the absence of any additional iron source (no Fe). Apparent (−2-ME) and total IRP1 (+2-ME) activities were quantified as described in the legend of Figure 2 and in “Materials and methods” and are expressed in relative units, which are directly comparable within this data set.
The presence or absence of changes in apparent IRP1 activities, induced by high versus low iron in transformed versus primary erythroblasts, respectively, correlated well with the presence or absence of respective iron-dependent changes in the expression levels of TfR mRNA and protein (Figures 1 and 3). To attribute elevated levels of TfR mRNA under iron scarcity to stabilization by IRP1 would of course conform well to the standard model of iron homeostasis. On the other hand, we had observed significant absolute differences in TfR mRNA abundance and protein levels between the erythroid and nonerythroid cell types even under conditions of low iron (Figures 1 and 2). These lineage-specific differences in absolute TfR expression levels were not at all reflected by the corresponding IRP activities. Thus, additional mechanisms besides IRP-dependent stabilization of TfR mRNA must account for the up-regulation of TfR mRNA and protein in primary erythroid cells.
Under high iron, TfR mRNA is stable in primary erythroblasts but destabilized in v-ErbA–transformed leukemic cells
To assess how an increase of TfR mRNA in SCF erythroblasts could occur essentially independent of IRP, we asked whether the specific endonuclease involved in TfR mRNA degradation44 might be less active or absent in committed erythroid cells, a possibility suggested from data obtained in mouse erythroleukemic (MEL) cells.45 To this purpose, TfR mRNA stability was measured in iron-chase experiments.35 46
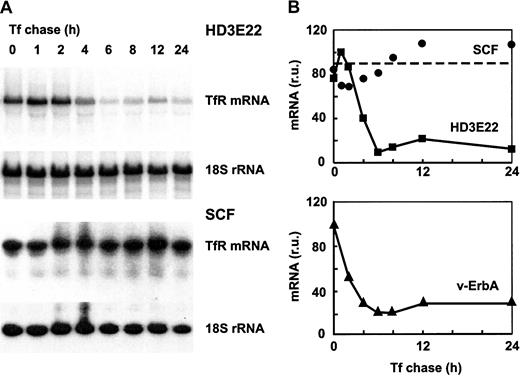
The SCF erythroblasts as well as HD3E22 cells were preincubated for 24 hours with DFO to induce iron depletion and thus a maximal level of TfR mRNA. Subsequently, the chelator was washed out and replaced by high but physiologic concentrations of Tf, which should lead to rapid turnover of TfR mRNA according to the standard model. This treatment did not induce any significant alteration of TfR mRNA stability in the primary erythroblasts (Figure 6), as expected from previous experiments (Figures 1, 3, and 5). In sharp contrast, the transformed HD3E22 cells responded with a rapid decline of TfR mRNA, with a calculated half-life of less than 2 hours, in agreement with previously published results.35 46 Under iron deprivation, SCF erythroblasts and HD3E22 cells exhibited equal levels of TfR mRNA, suggesting that the transcriptional rate of the TfR gene is similar in both cell types. Thus iron-independent stabilization of TfR mRNA apparently is an intrinsic property of primary erythroid cells, lost on leukemogenic viral transformation.
Effect of iron chase on TfR mRNA stability in primary versus transformed erythroid cells.
Transformed HD3E22 or primary SCF cells were pretreated with 50 M DFO for 24 hours to achieve maximal iron depletion. Subsequently, cells were subjected to an iron chase with 1 mg/mL Tf for the times indicated. (A) After preparation of RNA and Northern blotting, filters were sequentially hybridized with a 32P-labeled probe specific for chicken TfR and for normalization with 18S rRNA. (B) The signals from the autoradiographs were quantitated by PhosphoImage analysis. Also shown are the results from an analogous experiment performed with erythroblasts expressing the v-ErbA oncogene alone.
Effect of iron chase on TfR mRNA stability in primary versus transformed erythroid cells.
Transformed HD3E22 or primary SCF cells were pretreated with 50 M DFO for 24 hours to achieve maximal iron depletion. Subsequently, cells were subjected to an iron chase with 1 mg/mL Tf for the times indicated. (A) After preparation of RNA and Northern blotting, filters were sequentially hybridized with a 32P-labeled probe specific for chicken TfR and for normalization with 18S rRNA. (B) The signals from the autoradiographs were quantitated by PhosphoImage analysis. Also shown are the results from an analogous experiment performed with erythroblasts expressing the v-ErbA oncogene alone.
We previously showed that one of the 2 oncogenes of AEV (the transforming agent of HD3E22), v-ErbA, a constitutively active mutated version of thyroid hormone receptor,47caused a transformation-specific alteration of Fer mRNA translation.22 Thus we determined whether the same oncogene might be responsible for the leukemic phenotype of TfR mRNA regulation. Erythroblasts expressing v-ErbA29were subjected to the same experimental regime as the HD3E22 cells. Again, iron repletion reduced TfR mRNA half-life to less than 2 hours (Figure 6). Thus v-ErbA alone is indeed able to cause different aspects of a transformation-specific phenotype of iron metabolism.
Taken together, these iron-chase experiments demonstrated that the major determinant for constitutive TfR mRNA hyperexpression in primary chicken erythroblasts is a decreased and iron-independent rate of mRNA turnover. This degradation pathway is restored on transformation by AEV, or, more specifically, by one of the targets of the v-ErbA oncogene.
Distinct sensing of intracellular iron levels in primary versus transformed erythroblasts: differential localization of transferrin-bound iron
Although IRP1 activity and TfR mRNA expression correlated well in SCF and HD3E22 erythroblasts, it remained unclear why IRP activity could not be modulated by iron in the primary erythroblasts. One possible explanation was that in primary erythroblasts a major fraction of intracellular iron might be inaccessible to the cytoplasmic IRP iron sensing system by remaining “hidden” in endosomal vesicles. Given the high-level expression of TfR in primary erythroblasts and in consequence extensive internalization of Tf/TfR complexes this should lead to an expansion of the corresponding pool within the endocytic compartment.
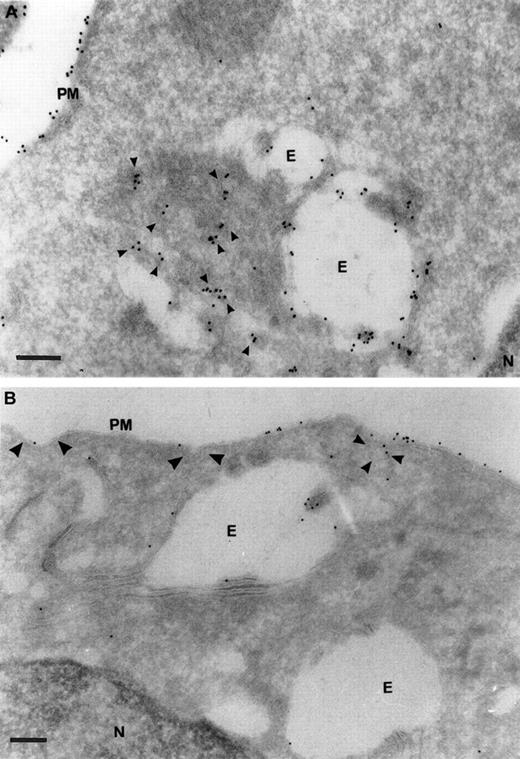
To test this hypothesis we localized TfR in primary and AEV-transformed HD3E22 erythroblasts by electron microscopy. Ultrathin cryosections were labeled with a rabbit antibody against chicken TfR (JS-8), and TfR was visualized with a rabbit antimouse Protein A/gold (10 nm) complex. High-intensity anti-TfR labeling in the primary erythroblasts was observed on the plasma membrane and in early endosomes (Figure7). In addition, the cells internalized large amounts of TfR molecules and appeared to transport TfR molecules from proximal to perinuclear compartments. This distribution pattern was completely different in the transformed HD3E22 cells, where anti-TfR gold particles were concentrated on the plasma membrane, including coated pits and vesicles and only rarely detected in early endosomes. This TfR receptor redistribution toward later endosomal compartments in primary SCF erythroblasts was previously described for HD3 erythroblasts during early stages of terminal differentiation.38
Ultrastructural localization of TfRs in SCF progenitors versus transformed HD3E22 erythroblasts.
Subcellular localization of TfRs in primary SCF progenitors (A) and AEV-transformed HD3E22 erythroblasts (B) was determined by immunoelectron microscopy of ultrathin cryosections labeled with a monoclonal murine antibody to chicken TfR and detected with Protein A/gold (10 nm) complex (see “Materials and methods”. The pictures show a section of the plasma membrane and intracellular compartments. Cisternal structures (double membranes) that enclose electron-lucent areas and electron-dense structures resembling endosome carrier vesicles are part of the early endosomal compartments. PM indicates plasma membrane; E, endosome; N, nucleus. TfR-specific labeling in coated pits (large arrowheads), coated vesicles (medium arrowheads), and small vesicular structures (small arrowheads). Note the differential amounts and distribution of the 10 nm Protein A/gold particles in the SCF progenitors versus the erythroleukemic HD3E22 cells. Magnifications: (A) × 75 000, (B) × 50 000; bars represent a length of 200 nm.
Ultrastructural localization of TfRs in SCF progenitors versus transformed HD3E22 erythroblasts.
Subcellular localization of TfRs in primary SCF progenitors (A) and AEV-transformed HD3E22 erythroblasts (B) was determined by immunoelectron microscopy of ultrathin cryosections labeled with a monoclonal murine antibody to chicken TfR and detected with Protein A/gold (10 nm) complex (see “Materials and methods”. The pictures show a section of the plasma membrane and intracellular compartments. Cisternal structures (double membranes) that enclose electron-lucent areas and electron-dense structures resembling endosome carrier vesicles are part of the early endosomal compartments. PM indicates plasma membrane; E, endosome; N, nucleus. TfR-specific labeling in coated pits (large arrowheads), coated vesicles (medium arrowheads), and small vesicular structures (small arrowheads). Note the differential amounts and distribution of the 10 nm Protein A/gold particles in the SCF progenitors versus the erythroleukemic HD3E22 cells. Magnifications: (A) × 75 000, (B) × 50 000; bars represent a length of 200 nm.
These results, together with the low levels of hemoglobin in SCF erythroblasts (L.L. et al, unpublished observations, February, 2001), suggest that in committed primary erythroblasts internalized iron bound to Tf/TfR complexes may reside in endosomes as a kind of temporary intracellular store before differentiation. With the onset of terminal differentiation, internalized iron is targeted specifically to mitochondria,10 where it becomes chelated as heme.
Discussion
In this report, we demonstrate that TfR is hyperexpressed in committed primary chicken erythroblasts (SCF erythroblasts). This hyperexpression is maintained under high levels of iron supply, contradicting the standard model of iron homeostasis, which involves iron-sensing IRPs as the prime regulators of TfR expression.7 TfR hyperexpression in erythroblasts involves an iron-independent intrinsic stabilization of TfR mRNA. Moreover, a high amount of iron-bearing Tf/TfR complexes in primary erythroblasts appears to elude the cytoplasmic IRP sensory system by residing in endosomal compartments. Transformation of erythroblasts drastically alters the regulation of iron metabolism: TfR hyperexpression is abolished, TfR responsiveness to alterations in iron levels is restored, and TfR protein relocalizes predominantly to the cell membrane.
The “standard model” of iron regulation postulates that iron homeostasis is regulated by interaction of the mRNA binding proteins IRP1 and IRP2 with IREs in mRNAs encoding proteins required for efficient iron uptake and utilization. Depending on IRE positioning, IRP binding stabilizes TfR mRNA and confers less efficient translation initiation of Fer and e-ALAS mRNAs. According to a wealth of published evidence, this model holds true for most nonerythroid cell types and, apparently, also for erythroleukemic cells.7,39,48Primary, committed erythroid progenitors and terminally differentiating erythroblasts, however, require excessive iron for hemoglobin production, thus presenting a paradox to the standard model; high levels of TfR for efficient iron uptake and high levels of e-ALAS for heme biosynthesis must be expressed simultaneously. Indeed, TfR is essential for erythropoiesis; mice lacking TfR49 have a more severe phenotype than hypotransferrinemic (hpx/hpx; mutation in the Tf gene) mice.50
There was some previous in vivo evidence for TfR hyperexpression in late-stage erythroblasts51,52 as well as embryonic erythroblasts.33 This was primarily attributed to cell type–specific transcriptional control.33 In our case, at least loss of TfR hyperexpression in AEV-transformed HD3E22 cells cannot be explained by reduced transcription. Under iron-deprivation conditions, primary SCF progenitors and leukemic HD3E22 cells could reach similarly high TfR mRNA levels, mediated by increased stabilization via IRP1 (Figures 1-3 and 5). This also ruled out that the TfR promoter is a direct target of v-ErbA/v-ErbB signaling but rather demonstrated that hyperexpression of TfR in primary erythroid progenitors must involve additional mechanisms. In primary SCF and c-Kit-erythroblasts, as well as in v-Ski erythroid progenitors (all having a finite lifespan and the potential to undergo complete erythroid maturation19,23), TfR mRNA and protein levels were not, or only slightly, down-modulated by addition of external iron. In contrast, the immortalized HD3E22 erythroblasts, representing very immature leukemic cells, responded to iron scarcity and deprivation according to the standard model, very similar to the hepatic LMH cells. Moreover, iron-dependent changes in TfR mRNA expression and apparent IRP1 activity correlated strongly in all cell types tested. Modulation of IRP1 activity could not explain the variations of absolute TfR mRNA or protein levels in the various cell types. For example, TfR mRNA levels were 40-fold lower in CEF cells than in SCF progenitors, whereas there was only a 2-fold difference in apparent IRP1 activity. A common denominator for all these apparently disparate observations can be found in the differential activity of the specific endonuclease involved in TfR mRNA turnover (Figure 6). Whereas a switch from low iron supply to high Tf induced rapid decay of TfR transcripts in HD3E22 and v-ErbA–expressing cells (half-life about 2 hours, as expected35,40), no mRNA degradation at all occurred in primary SCF erythroblasts, although the protective IRP activity was low (Figure 5). Interestingly, similar observations of increased TfR mRNA stability regardless of iron supply were reported for differentiating murine erythroleukemia cells.45 Taken together, this is a strong indication for the absence or at least massively reduced activity of the responsible regulatory RNAse in cells committed to terminal erythropoiesis. This may also explain why these effects have escaped detection so far. The majority of erythroid cell systems used are immortalized, either virally transformed or derived from leukemias, and have limited differentiation capability.
In the primary erythroblasts, IRP activity was hardly influenced by changes in external iron supply, concomitant with very little change in high-level expression of TfR. By contrast, in HD3E22 cells IRP activity was apparently the major regulator of TfR expression. Therefore, the IRP sensory system seemed to be effectively nonfunctional in primary erythroblasts. According to our data, these differences might arise from differential localization of internalized iron (Figure 7; much higher TfR abundance in endosomes of SCF erythroblasts as compared to transformed HD3E22 cells), consistent with previous data on HD3 cells, the parental cell line to HD3E22.38 Interestingly, in the latter cells, a redistribution of Tf/TfR complexes toward the endosomal network occurred on differentiation induction, during which cells started to resemble SCF erythroblasts also in other properties. In early endosomes of HD3 cells the pH is 5.8, whereas late endosomal compartments have a more acidic pH of 5.38,53 The higher pH in early endosomes may be insufficient to efficiently release iron from TfR-bound Tf. This led to the hypothesis that movement of Tf/TfR complexes to more acidic compartments during erythroid differentiation enables a more efficient uptake and utilization of iron.38In maturing red cells, the hemoglobin content increases drastically. In line with the arguments above, one of the first events of this concerted process is a massive redistribution of Tf/TfR complexes to late endosomal compartments (L.L. et al, manuscript in preparation). This leads to the intriguing hypothesis that temporary accumulation of iron-laden Tf in endosomes may be one of the prerequisites for commitment of erythroid progenitors, just prior the onset of terminal differentiation. Thereby, iron residing in endosomes would be inaccessible to the cytoplasmic IRP system. This in turn would lead to continued hyperexpression of TfRs, despite high extracellular iron supply, by increased stabilization of the TfR mRNA in addition to erythroid cell-specific transcription.
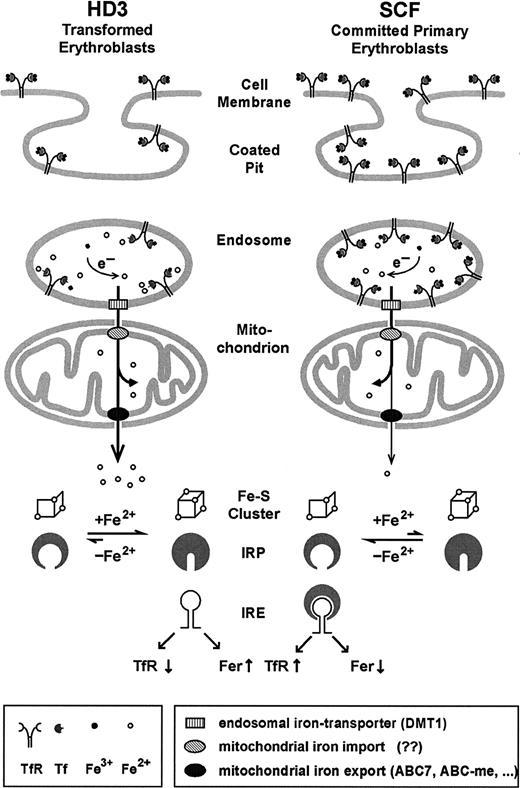
The observations described in this paper can be combined into a model for iron uptake in committed erythroid progenitors (Figure8). After endocytosis of Tf/TfR complexes, iron needs to be released from the endosomes. This apparently involves a specific iron transporter, divalent metal transporter 1 (DMT1),54-56 which colocalizes with Tf.57 DMT1 only functions at low pH49 and its mutated version causes hypochromic microcytic anemia in humans, mice, and rats. Most likely, ferric iron liberated from Tf is reduced to the more soluble ferrous form.10,58 This conclusion is strengthened by recent findings that DMT1 can transport only ferrous iron.59 Subsequently, at least in erythroid cells, iron may be specifically targeted toward mitochondria, as suggested by several independent genetic defects causing sideroblastic anemias as well as in vitro cell systems.10,60-63 For example, in differentiating erythroblasts, iron continues to flow into mitochondria even when the synthesis of protoporphyrin IX is suppressed.62 Therefore, iron released from Tf in erythroid endosomes may be shunted directly into mitochondria via protein-protein interactions until it reaches ferrochelatase, which inserts iron into protoporphyrin IX. This direct transfer of iron was recently designated as the “Kiss and Run Hypothesis.”10
Model for iron uptake and transport in transformed versus normal erythroblasts.
In committed primary erythroblasts (SCF cells), after endocytosis, a large proportion of the highly abundant iron-loaded Tf/TfR complexes resides in undissociated form in the endosomal compartment due to its relatively high pH. The fraction of ferrous iron leaving the endosomes via the action of DMT154-57 is directly shunted into mitochondria, where most of it is incorporated into heme and iron-sulfur clusters. Only minor amounts of iron-containing compounds are released into the cytosol through the action of iron transporters (candidate genes possibly involved are the ATB-binding cassette (ABC) transporter ABC760,66,67 or the erythroid-specific ABC-me protein 968) and registered by the IRP sensory system. Although transformed erythroleukemic cells (like HD3) express less TfR, their more acidic endosomes favor an enhanced flow of iron through the mitochondria (in the absence of heme synthesis). In consequence, on iron overload, IRP harbors the complete 4Fe-4S cluster, leading to the conformation that does not bind IRE elements, thus further reducing TfR expression. See “Discussion” for further details and references.
Model for iron uptake and transport in transformed versus normal erythroblasts.
In committed primary erythroblasts (SCF cells), after endocytosis, a large proportion of the highly abundant iron-loaded Tf/TfR complexes resides in undissociated form in the endosomal compartment due to its relatively high pH. The fraction of ferrous iron leaving the endosomes via the action of DMT154-57 is directly shunted into mitochondria, where most of it is incorporated into heme and iron-sulfur clusters. Only minor amounts of iron-containing compounds are released into the cytosol through the action of iron transporters (candidate genes possibly involved are the ATB-binding cassette (ABC) transporter ABC760,66,67 or the erythroid-specific ABC-me protein 968) and registered by the IRP sensory system. Although transformed erythroleukemic cells (like HD3) express less TfR, their more acidic endosomes favor an enhanced flow of iron through the mitochondria (in the absence of heme synthesis). In consequence, on iron overload, IRP harbors the complete 4Fe-4S cluster, leading to the conformation that does not bind IRE elements, thus further reducing TfR expression. See “Discussion” for further details and references.
According to this model (Figure 8), iron taken up by committed erythroid progenitors via the Tf/TfR cycle temporarily resides in hypoacidic endosomes, fails to traffic through mitochondria, and, in consequence, eludes the IRP sensory system. This, in turn, would lead to maintenance of hyperexpression of TfR in primary erythroblasts, despite physiologically high iron supply, to meet the extreme demand for iron uptake during terminal erythropoiesis. With the onset of differentiation, iron is released from endosomal Tf/TfR complexes by acidification, specifically targeted into mitochondria, and almost fully incorporated into heme. Again, traffic of iron out of the mitochondria is minimal and IRP stays in the “low iron” mRNA binding conformation. This does not necessarily interfere with the required e-ALAS translation in the primary erythroblasts, however. Because there is a high abundance of TfR mRNA (due to the absence of endonuclease), almost all the available IRP will be associated with IREs in TfR mRNA and not enough will remain to interfere with translation initiation of e-ALAS mRNA. In line with this argument, we have already previously observed a partial derepression of e-ALAS mRNA translation in committed but still undifferentiated SCF progenitors.22 The interpretation above is also consistent with the observations from transformed HD3E22 cells; due to the activity of the regulatory endonuclease there is much less TfR mRNA. So even in the presence of the same IRP activity, the molar ratio between IRP, e-ALAS mRNA, and TfR mRNA will be shifted toward an excess of IRP over the IREs in the transcripts, thus effectively blocking e-ALAS translation.
These cell type–specific mechanisms of erythroid iron metabolism may be a stringent prerequisite for committed erythroblasts, which become relaxed on leukemic transformation, where iron metabolism reverts to the “standard mode” of regulation. In primary erythroblasts, mechanisms preventing iron overload may be less important—the threat of iron toxicity may be less severe—due to the limited lifespan of committed progenitors. Transformation of erythroid cells, however, leads to continuous proliferation as well as differentiation arrest or even partial dedifferentiation. Therefore, transformed cells have to adopt an iron storage/detoxification strategy compatible with long-term proliferation.64,65 Indeed, one of the first events during establishment of the transformed state apparently is a loss of erythroid cell-specific features of iron metabolism. Transformation to the leukemic state by AEV oncogenes or other oncogenes, accompanied or not by immortalization,22 restores the ability of the cells to reduce iron uptake and to detoxify excessive intracellular iron by means of the IRP system as described by the standard model of iron homeostasis.
Supported by grants from the Herzfelder Family Foundation and the Fonds zur Förderung der Wissenschaftlichen Forschung, Austria. L.L. was the recipient of stipends from the University of Vienna and from the Hans and Blanca Moser-Foundation. The authors have reviewed the manuscript and agree with its content.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ernst W. Müllner, Institute of Medical Biochemistry, Division of Molecular Biology, Vienna Biocenter; Dr Bohr-Gasse, A-1030 Vienna, Austria; e-mail: em@mol.univie.ac.at.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal