Abstract
Many spectrin mutations that destabilize tetramer formation and lead to hereditary hemolytic anemias are located at the N-terminal region of α-spectrin, with the Arg28 position considered to be a mutation hot spot. We have introduced mutations at positions 28 and 45 into a model peptide, Spα1-156, consisting of the first 156 residues in the N-terminal region of α-spectrin (αN). The association of these α-spectrin peptides that have single amino acid replacements with a β-spectrin model peptide, consisting of the C-terminal region of β-spectrin (βC), was determined, and structural changes due to amino acid replacements were monitored by nuclear magnetic resonance (NMR). We found evidence for similar and very localized structural changes in Spα1-156Arg45Thr and Spα1-156Arg45Ser, although these 2 mutant peptides associated with β-spectrin peptide with significantly differing affinities. The Spα1-156Arg28Ser peptide showed an affinity for the β-spectrin peptide comparable to that of Spα1-156Arg45Ser, but it exhibited substantial and widespread spectral changes. Our results suggest that both Arg45 replacements induce only minor structural perturbations in the first helix of Spα1-156, but the Arg28Ser replacement affects both the first helix and the following structural domain. Our results also indicate that the mechanism for reduced spectrin tetramerization is through mutation-induced changes in molecular recognition at the αβ-tetramerization site, rather than through conformational disruption, as has been suggested in prior literature.
Introduction
The α and β subunits of human erythrocyte spectrin both consist of multiple homologous sequence motifs, with each motif presumably folding into 3 helices, which bundle to form a structural domain similar to the structures determined forDrosophila spectrin1 and chicken brain spectrin.2,3 Both α- and β-spectrin associate at the N-terminal end of the β-subunit and the C-terminal end of the α-subunit (dimer nucleation site) with high affinity (nM dissociation constant [Kd] values) to give αβ heterodimers.4,5 It has been suggested that spectrin dimers then associate to form spectrin tetramers, with an association site at the other end of the dimers, involving 2 sets of identical, low-affinity (μM Kd) interactions between the N-terminal region of the α-subunit (αN) of one αβ dimer and the C-terminal region of the β-subunit (βC) in another αβ dimer to give an (αβ)2 tetramer.6,7 Sequence homology studies predict that about 30 residues in this αN region, prior to the first structural domain, fold into a helical conformation; likewise, about 60 residues in the βC region, following the last structural domain, are predicted to fold into 2 helices.8,9 These one- and 2-helix regions are termed partial domains for α- and β-spectrin, respectively. It is assumed that the dimer to tetramer formation involves association of these partial domains to form a triple helical bundle similar to the structural domains.7,8 Spectrin tetramers in the human erythrocyte play a critical role in maintaining the architecture, and therefore the integrity, of the red cell membrane. Many hereditary hemolytic anemias involve spectrin mutations that destabilize tetramer formation (produce low levels of spectrin tetramers and high levels of dimers).10,11 In fact, nearly all cases of hereditary elliptocytosis (HE) or its aggravated form, hereditary pyropoikilocytosis (HPP), can now be related to specific mutations in spectrin. Many lie in or near the tetramerization sites in either α- or β-spectrin.10 Recently, spectrin oligomerization has been suggested to be cooperatively coupled to membrane assembly and is a linkage targeted by many hereditary hemolytic anemias.12
Early studies showed that these spectrin mutants exhibited abnormal proteolysis patterns.13 Partial trypsin digestion of α-spectrin yields a major fragment, the αI domain,13with a molecular mass of 80 kd (αI80). This fragment undergoes a secondary cleavage after residue Arg45 and/or Lys48 in the αI domain to give another fragment of 74 kd (αI74, a fragment lacking the first 45 to 48 amino acids in the αI domain).10 For some spectrin mutants, the level of αI74 fragment increases at the expense of the αI80 fragment in the trypsin digestion pattern, and they are referred to as αI74 abnormalities.10,14It has generally been assumed that the αI74 abnormality is due to a conformational change in the first helix in α-spectrin (helix 3 as named by Delaunay and Dhermy10 or Helix C′ in our work).15 Many mutations of this type lead to HE or HPP. However, clinical expression is more complicated than simple mutations in spectrin molecules because mutations may occur in a simple heterozygous state, in a compound heterozygous state, or in conjunction with a low-expression allele (αLELLY). It has been widely documented that each of these combinations exhibits dramatically differing clinical, morphologic, and biochemical properties.10 Systematic studies of different factors contributing to HE and HPP abnormalities are needed to better understand the diseases at the molecular level.
The mutations at positions 45 and 28 are of particular interest in this work. The Arg to Thr mutation at position 45 (Arg45Thr) induces a variety of clinical symptoms, ranging from asymptomatic to mild elliptocytosis with compensating hemolysis.11 However, the Arg45Ser mutation has been reported to induce more severe symptoms, with the range of symptoms correlated with the extent of mutant spectrin expression.10,16 Mutations at position 28 from Arg to His, Leu, Ser, or Cys all induce severe clinical symptoms.10,17 All of these mutations apparently reduce αβ tetramerization.10
In this study, we focus on the effects of specific amino acid replacements on α- and β-spectrin association at the tetramerization site. Amino acid replacements at positions 28 and 45 of α-spectrin were introduced into an extensively studied model peptide, Spα1-156,15,18,19 to give Spα1-156Arg28Ser (Arg28Ser), Spα1-156Arg45Ser (Arg45Ser), and Spα1-156Arg45Thr (Arg45Thr). The association of these α-spectrin mutant peptides with a model peptide of β-spectrin, Spβ1898-2083 (Spβ),20 was determined, and structural changes due to mutations were monitored by nuclear magnetic resonance (NMR). These 2 α- and β-peptides were chosen to mimic the N-terminal end of α-spectrin and the C-terminal end of β-spectrin, respectively. The specific sequences of the peptides were used to reflect properly phased fragments of spectrin to give stable and functional model peptides.20 21
Our data suggest that structural changes in Arg45Thr and Arg45Ser are similar and quite limited, although these 2 mutant peptides associate with β-spectrin peptide with differing affinities. Substantial NMR spectral changes were observed in Spα1-156Arg28Ser, although its affinity for the β-spectrin peptide was comparable to that of Arg45Ser.
Materials and methods
Recombinant spectrin peptides
Spectrin peptides Spα1-156 and Spβ1898-2083 (Spβ) were expressed and purified as before.18 19 Spα1-156, the native peptide without any amino acid replacement at positions 28 and 45 (Arg28/Arg45), served as the parent peptide for single amino acid replacement to Ser at position 28 to give Spα1-156Arg28Ser (Arg28Ser) and at position 45 to give Spα1-156Arg45Ser (Arg45Ser), and to Thr at position 45 to give Spα1-156Arg45Thr (Arg45Thr), following the QuikChange Site-Directed Mutagenesis method (Stratagene, La Jolla, CA). Plasmids with the desired mutation were sequenced at the University of Illinois at Chicago DNA core facility, and the identities of all 3 mutations were confirmed.
αβ-Spectrin association affinity measurements
We have shown that the native Arg28/Arg45 (Spα) and Spβ peptides are good model systems to study α- and β-spectrin association at the tetramerization site.18 20 The association affinity of each mutant peptide with Spβ was assessed qualitatively by Kd of the Arg28/Arg45-Spβ (αβ) complex obtained from the results of gel-filtration measurements. Although Kd values obtained by gel-filtration methods do not provide absolute quantitative values for affinities, they are a rapid and simple means for good comparison of relative affinities. All samples were prepared in 5 mM phosphate buffer with 150 mM NaCl at pH 6.5 (NMR sample buffer). Three different concentrations (7, 12, and 30 μM) of α- and β-peptides were used. The α- and β-peptides at each concentration were incubated at 4°C overnight to ensure complete association. Incubation for longer periods (eg, 36 hours) was also tested, but no difference was observed. Each mixture was then loaded onto a Superdex 75 (Pharmacia, Piscataway, NJ) size exclusion column running on a BioLogic HR (BioRad, Hercules, CA) system, using the NMR sample buffer as the eluting buffer at 4°C to separate the αβ complex from free α- and β-peptides. Each fraction collected was analyzed by denaturing sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Overlapping peaks in elution profiles (Figure1) were deconvoluted with peak-fitting routines in PeakFit V.4.0 (SPSS Science, Chicago, IL), using the positions of the individual α and β peaks as fixed components. In all cases, the peak fitting gave R2 > 0.999. Peak areas for the αβ complex and the free α- and β-peptides were obtained. The relative concentrations of each species were then calculated by dividing each peak area by the corresponding molar extinction coefficient. The molar extinction coefficients used were 16 500 M−1 cm−1 for all Spα1-156 peptides, 31 010 M−1 cm−1 for the Spβ1898-2083 peptide, and 47 510 M−1 cm−1 for αβ complex. The proportionality constant between peak area and concentration was obtained from the initial concentration values. Kd values and uncertainties were obtained from the equilibrium concentrations and calculated from the free energy expression. Values obtained from different initial concentrations were averaged and are reported as mean values of Kd.
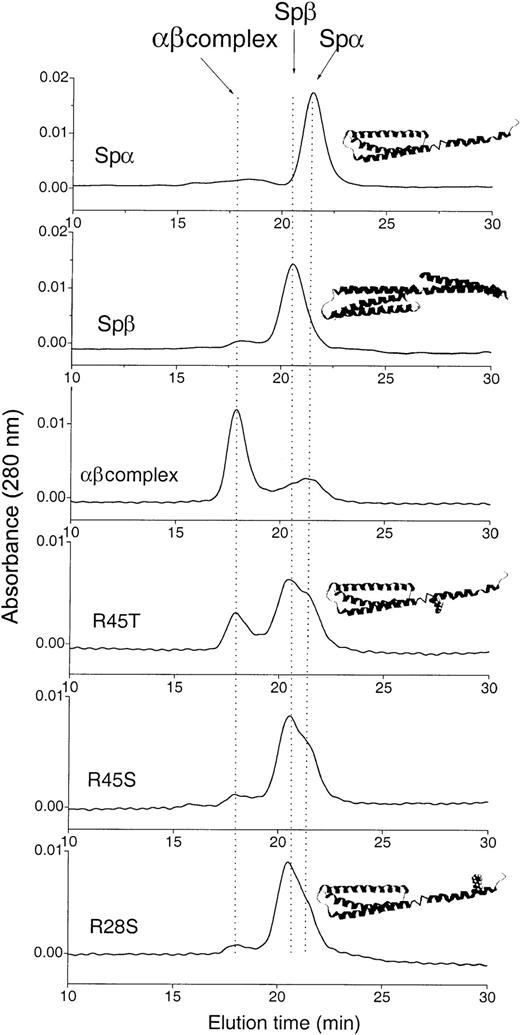
Gel filtration of mixtures of Spβ1898-2083 and SpαArg28/Arg45, Arg28Ser, Arg45Ser, or Arg45Thr at 7 μM.
The top 2 chromatograms of the individual sample are shown as references; additional measurements were done at 12 and 30 μM (data not shown). The position of each species is indicated by an arrow on the top chromatogram. We used 5 mM phosphate buffer with 150 mM NaCl at pH 6.5 (NMR sample buffer) for sample incubation and as eluting buffer. Schematic representations of the native Spα1-156 and Spβ1898-2083 structures are shown as insets above the corresponding chromatograms. The positions of the Arg28 and Arg45 mutations are shown by addition of the side chains at their sequence positions in insets above the corresponding chromatograms.
Gel filtration of mixtures of Spβ1898-2083 and SpαArg28/Arg45, Arg28Ser, Arg45Ser, or Arg45Thr at 7 μM.
The top 2 chromatograms of the individual sample are shown as references; additional measurements were done at 12 and 30 μM (data not shown). The position of each species is indicated by an arrow on the top chromatogram. We used 5 mM phosphate buffer with 150 mM NaCl at pH 6.5 (NMR sample buffer) for sample incubation and as eluting buffer. Schematic representations of the native Spα1-156 and Spβ1898-2083 structures are shown as insets above the corresponding chromatograms. The positions of the Arg28 and Arg45 mutations are shown by addition of the side chains at their sequence positions in insets above the corresponding chromatograms.
NMR experiments and analysis
All α-spectrin peptide samples (Arg28/Arg45, Arg28Ser, Arg45Ser, and Arg45Thr) for NMR studies were prepared and labeled with15N as described previously.15 Two-dimensional heteronuclear single quantum coherence (2D HSQC) spectra of15N-labeled samples were then obtained.15 19Solution characteristics for the 3 peptides with single amino acid replacements were quite similar to those of the native peptide.
The extent of the chemical-shift changes in spectra for Arg28Ser, Arg45Ser, and Arg45Thr in comparison with that of the native Arg28/Arg45 peptide is expressed as normalized chemical-shift changes: Δδ, in parts per million, calculated by the methods described previously22 using the equation: Δδ = [Δδ + 0.17Δδ]1/2, where ΔδHN and Δδ15N are the chemical-shift changes for the amide protons and the amide nitrogens, respectively, between the parent (Arg28/Arg45) and mutated (Arg28Ser, Arg45Ser, and Arg45Thr) peptides.
Results
Association of Arg28Ser, Arg45Ser, and Arg45Thr with Spβ
As shown in the column elution profile in Figure 1, the samples containing the native Arg28/Arg45 (7 μM) and Spβ (7 μM) existed mostly as an αβ complex. However, for samples containing Spβ and either Arg45Ser or Arg28Ser, the elution profile shows only low levels of αβ complex. In contrast, a significant amount of complex formation was observed in the sample containing Spβ and Arg45Thr. The Kd values shown in Table 1indicate that, in comparison with the native Arg28/Arg45 peptide, Arg45Thr exhibited about a 7-fold reduction in its affinity with Spβ, whereas Arg45Ser and Arg28Ser exhibited about a 50- to 60-fold reduction in their affinity with Spβ. The value for Arg28/Arg45 (Table 1) is qualitatively similar to those reported previously, in the range of approximately 10−6 to 10−7M.6,23,24 Using a solid-state assay, we previously obtained the concentration for 50% inhibition (IC50) of 0.14 μM for binding of Arg28/Arg45 to Spβ20 and 0.3 μM for Arg28/Arg45 and intact β-spectrin.18 Thus, the values in Table 1 are about 3 to 7 times higher than those obtained by a solid-state assay. However, the relative affinities among these peptides are clearly defined, with the native Arg28/Arg45 peptide having the highest affinity, Arg45Ser and Arg28Ser having the lowest affinities (and being statistically equivalent to each other), and Arg45Thr having an intermediate affinity, between the Arg45Ser and Arg28Ser substituted peptides and the native Arg28/Arg45 peptide. It is interesting to note that patients with the Arg28Ser spectrin mutation suffer severe symptoms10,17 and some patients with the Arg45Ser spectrin mutation also suffer severe symptoms,10,16 whereas patients with the Arg45Thr mutation show only mild symptoms.11 Therefore, our αβ model-peptide association affinities appear to be correlated with the severity of disease symptoms.
Dissociation constants for Spα:Spβ peptide complexes in samples containing native and substituted Spα 1-156 peptides and native Spβ1898-2083 peptides
| Peptide . | Kd(μM) . | Standard error . |
|---|---|---|
| Arg28/Arg45 | 0.93 | + 1.25 |
| − 0.53 | ||
| Arg45Thr | 6.5 | + 9.0 |
| − 3.8 | ||
| Arg45Ser | 44 | + 20 |
| − 14 | ||
| Arg28Ser | 59 | + 30 |
| − 20 |
| Peptide . | Kd(μM) . | Standard error . |
|---|---|---|
| Arg28/Arg45 | 0.93 | + 1.25 |
| − 0.53 | ||
| Arg45Thr | 6.5 | + 9.0 |
| − 3.8 | ||
| Arg45Ser | 44 | + 20 |
| − 14 | ||
| Arg28Ser | 59 | + 30 |
| − 20 |
Kd values were calculated as described in the “Materials and methods” section, using data at 7, 12, and 30 μM; uncertainties are asymmetric because Kd cannot be less than zero.
Structural information from NMR studies
We have determined the solution secondary structure of the native Arg28/Arg45 peptide by NMR methods and have found a total of 4 helices in Arg28/Arg45.15,25 The first 20 residues are in a random coil conformation, followed by a helix of 25 residues (residues 21-45, referred to as helix 3 by some authors, or generally referred to as Helix C′15), which is linked to the next helix by a random coil of 7 residues. The second, third, and fourth helices are bundled, whereas the first helix appears to be a lone helix.
From the resonance of the Arg28/Arg45 native peptide (Figure2A), chemical shifts in the 2D HSQC spectra of Arg45Thr, Arg45Ser, and Arg28Ser (Figure 2B-D) were used to evaluate environmental changes for residues 21 to 45 in particular, along with the rest of the molecule. Chemical-shift differences are interpreted as environmental changes because chemical shifts are very sensitive to their local environments.
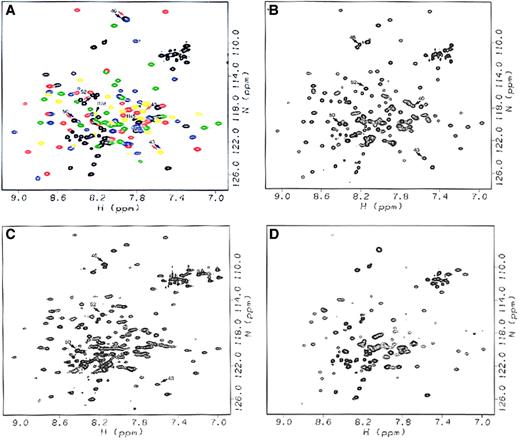
HSQC spectra of native Arg28/Arg45, Arg45Thr, Arg45Ser, and Arg28Ser peptides.
(A) Native Arg28/Arg45 peptide, with resonances corresponding to the initial unstructured sequence 1-20 and the loop regions between helices 46-52, 82-87, and 119-122 shown in black; resonances corresponding to the initial Helix C′ shown in yellow; and resonances corresponding to Helices A1, B1, and C1 of the triple helical bundle shown in red, green, and blue, respectively. Arrows denote resonances affected by the mutations, with black arrows noting the mutated residues (Arg45 and Arg28), red arrows noting resonances affected by the Arg45Thr mutation, and blue arrows noting resonances affected by the Arg45Ser mutation. (B) Arg45Thr peptide, with arrows noting resonances whose chemical shifts are affected by the mutation. (C) Arg45Ser peptide, with arrows noting resonances affected by the mutation. (D) Arg28Ser peptide, for which missing resonances can be seen by comparison with (A). All spectra were acquired for the samples in NMR sample buffer at 20°C.
HSQC spectra of native Arg28/Arg45, Arg45Thr, Arg45Ser, and Arg28Ser peptides.
(A) Native Arg28/Arg45 peptide, with resonances corresponding to the initial unstructured sequence 1-20 and the loop regions between helices 46-52, 82-87, and 119-122 shown in black; resonances corresponding to the initial Helix C′ shown in yellow; and resonances corresponding to Helices A1, B1, and C1 of the triple helical bundle shown in red, green, and blue, respectively. Arrows denote resonances affected by the mutations, with black arrows noting the mutated residues (Arg45 and Arg28), red arrows noting resonances affected by the Arg45Thr mutation, and blue arrows noting resonances affected by the Arg45Ser mutation. (B) Arg45Thr peptide, with arrows noting resonances whose chemical shifts are affected by the mutation. (C) Arg45Ser peptide, with arrows noting resonances affected by the mutation. (D) Arg28Ser peptide, for which missing resonances can be seen by comparison with (A). All spectra were acquired for the samples in NMR sample buffer at 20°C.
To our surprise, the spectra of Arg45Thr and Arg45Ser were quite similar to that of Arg28/Arg45, indicating that the amino acid replacements at position 45 caused only minor local conformational changes regardless of amino acid side-chain differences (Ser versus Thr). The chemical shifts for Gly46, the residue adjacent to the replaced residue at position 45, in the spectra for both Arg45Ser and Arg45Thr were very large, but were easily identified because the Gly resonances typically appear in a characteristic and well-isolated region in the spectra. However, the replaced residues, Ser in Arg45Ser and Thr in Arg45Thr, could not be identified with confidence and were not considered further.
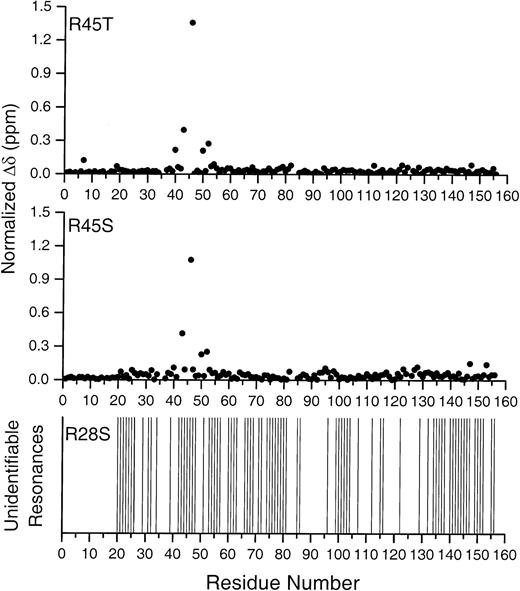
To obtain more specific information on the differences in the Arg45Ser and Arg45Thr peptides, we plotted the normalized chemical-shift changes relative to Arg28/Arg45 against the residue number (Figure 3). Chemical shifts for most of the residues in Arg45Ser and in Arg45Thr were within 0.1 ppm of those in Arg28/Arg45. For both Arg45Ser and Arg45Thr, Δδ for residue 43 was about 0.4 ppm, and the largest Δδ value was that of residue 46 (larger than 1 ppm). The changes for both Arg45Ser and Arg45Thr were quite similar. These data strongly suggest that the conformational changes in Arg45Ser and Arg45Thr are similar and remain limited to only small regions in the vicinity of residue 45.
Plot of the backbone amide chemical-shift changes (Δδ) against residue numbers in Spα1-156Arg45Ser and Arg45Thr relative to Arg28/Arg45, and missing resonances in Arg45Ser.
Sample identity is indicated at the top of each plot. Normalized chemical-shift changes in the top 2 plots were calculated as described in “Materials and methods.” Residues that could not be identified because of changes in the mutant spectra (residues 44, 45 in Arg45Thr; and 42, 45 in Arg45Ser) are left blank. The bottom plot notes resonances missing in the Arg28Ser HSQC spectrum, as compared with that of the native peptide.
Plot of the backbone amide chemical-shift changes (Δδ) against residue numbers in Spα1-156Arg45Ser and Arg45Thr relative to Arg28/Arg45, and missing resonances in Arg45Ser.
Sample identity is indicated at the top of each plot. Normalized chemical-shift changes in the top 2 plots were calculated as described in “Materials and methods.” Residues that could not be identified because of changes in the mutant spectra (residues 44, 45 in Arg45Thr; and 42, 45 in Arg45Ser) are left blank. The bottom plot notes resonances missing in the Arg28Ser HSQC spectrum, as compared with that of the native peptide.
Discussion
Structural effects
The behavior of the Arg28Ser mutant is perhaps the most intriguing. A large number of resonances are missing, but rather than being localized to a region close to the Arg28Ser mutation site, the missing resonances are distributed throughout the full structure of the peptide, including the triple helical bundle, as shown in the bottom plot of Figure 3. Similarly, many resonances throughout the structure, including both the initial Helix C′ and the triple helical bundle, are equivalent in chemical shift and line widths to those of the native peptide. There are basically 3 possibilities that could give rise to the substantially altered spectrum seen: (1) The peptide is substantially unfolded or denatured into a “random coil” conformation, but remains monomeric in solution; (2) the peptide forms stable dimers or higher oligomeric states; or (3) the peptide exhibits transient conformational association or transient peptide–peptide association, giving rise to exchange effects in the NMR spectrum.
The first possibility, substantial unfolding or denaturation, could be consistent with the Arg28Ser mutation destabilizing Helix C′. However, the Arg28Ser spectral changes are distributed throughout the full 156-residue sequence, rather than being confined to the Helix C′ that contains the mutated residue (Figure 3; lower plot). Unfolding or denaturation should also generate intense new signals in the center of the HSQC spectrum, characteristic of unstructured peptide, similar to those we see from the first 20 residues prior to the start of Helix C′, but no such signals were observed. Similarly, about 30% to 40% of the resonances distributed throughout the structure are essentially equivalent to those of the native peptide, both in chemical-shift position and in approximate line width. There appears to be no plausible unfolding or denaturation mechanism that could produce such a pattern. Furthermore, the Arg28Ser NMR solution characteristics (particularly solubility and temperature at which aggregation is induced) are similar to those of the native and the Arg45Ser and Arg45Thr substituted peptides, and its affinity for the Spβ peptide is statistically equivalent to that of the Arg45Ser peptide, also suggesting that the Arg28Ser peptide maintains its conformational integrity.
The second possibility, that of forming stable dimers or higher oligomeric states, is inconsistent with the observed spectral characteristics. An Arg28Ser dimer would be approximately 37.4 kd in molecular mass and should generate very broad lines compared with the native peptide, which we have previously shown to be in a monomeric state under NMR conditions.15 Again, there appears to be no plausible mechanism by which oligomerization could selectively eliminate some resonances along the helices, but leave others in neighboring residues unaffected in both chemical shift and line width.
The third possibility, namely conformational exchange that is at an intermediate time scale, either by transient association of the Helix C′ with the triple helical bundle within one peptide or by transient association of one Arg28Ser peptide with another Arg28Ser peptide, should selectively broaden resonances for residues involved in the association, but would leave other resonances essentially unaffected. This is, in fact, the pattern observed. A detailed examination of the NMR data also revealed that the spectral disturbances were distributed nonuniformly throughout the structure. Classifying resonances as either similar to those of the native peptide or missing or unidentifiable in Arg28Ser, we found that more than 80% of the Helix C′ resonances were affected and that nearly 80% of Helices A1 and C1 in the first structural domain were affected, whereas less than 50% of Helix B1resonances were affected. It is possible that the Lys79 residue, and probably the Lys150 residue, located in Helices A1 and C1, respectively,25 provide electrostatic repulsion between the exterior AC face of the triple helical bundle and the Helix C′ Arg28 residue, thus inhibiting interaction between the Helix C′ and the triple helical bundle in the native peptide. However, the Arg28Ser substitution deletes the cationic surface side chain, and thus may also permit transient association between the Helix C′ and the exterior AC face of the triple helical bundle, inducing the exchange characteristics that appear suggestive in the Arg28Ser HSQC spectrum. This association could occur within individual peptides or could be through transient peptide–peptide association involving the specific faces noted above. Thus, even the Arg28Ser Helix C′ is not necessarily drastically altered, but may simply exhibit a very weak transient association with the triple helical bundle or allow weak peptide–peptide association.
Mechanism of reduced affinity for αβ association in Arg28Ser, Arg45Ser, and Arg45Thr
It is crucial to determine whether the impaired αβ association in Arg28Ser, Arg45Ser, and Arg45Thr associated with a single amino acid replacement is due to a disruption of helix 3 (Helix C′) conformation, as has widely been suggested,10,11,16,17 26 or is simply a disruption of molecular interactions involved in the αβ molecular recognition and association, without significant change in Helix C′ conformation in the α-peptide.
Perrotta et al11 used a predictive method that integrates 6 different methods of analysis to predict secondary structure topology of Arg45Ser and Arg45Thr and suggested that the structure of a helix including residues 36 to 51 should be significantly perturbed by the single amino acid replacement. When residue 45 is replaced with Thr, the C-terminal segment of this helix is predicted to be perturbed, showing a lower consensus score, suggesting a less-well-formed helix.11 The Ser replacement at position 45 has been predicted to produce a more profound effect, disrupting the C-terminal part of this helix to an even larger extent.11 These predicted differential disruptions of Helix C′ were then hypothesized to be the basis of differences in the clinical expression of the similar phenotypic mutations at the Arg45 site (with HE giving rise to increased αI78).11
However, our results suggest that the clinical differences between Arg45Ser and Arg45Thr mutations are most probably due to differences in the association interaction itself, caused by respective amino acid substitutions. NMR results showed that the amino acid replacement from Arg to Ser or Thr at position 45 induced very localized chemical changes with similar patterns around position 45, suggesting minor conformational change, with residue 45 as the last residue in helix 3 (Helix C′).15
Arg45Thr may be more favored in the association with Spβ because of its additional methyl group when compared with Arg45Ser. Residue 45 is in a “d” position in the heptad (abcdefg) repeat pattern in this helix,15 which is presumably involved in hydrophobic interaction between helices in helical bundles. (Residue 28 is an “a” position in the heptad repeat pattern in this helix.15) The ratio of Kd values that we obtained for Arg45Ser and Arg45Thr is around 5 to 6, which corresponds to a ΔG difference of about 1.1 kcal/mol. This value agrees with the well-established ΔΔG values for a cavity-creating mutation of one methylene group, which is approximately 1.0 to 1.2 kcal/mol.27-29 Thus, the additional methyl group in Thr might play a role in enhancing the association of Arg45Thr with β-spectrin, as compared with Arg45Ser and β-spectrin. Pascual et al2 suggested that when Arg is replaced by Thr or Ser, a loss of electrostatic interaction between the Arg45 residue and a residue in β-spectrin causes impaired binding of α- and β-spectrin at the tetramerization sites. However, this explanation does not provide a rationale for the affinity differences between Arg45Ser and Arg45Thr because Thr and Ser both lack the ionizable group needed to give positive charges for the suggested interactions.
The differences in Kd values obtained in this study suggest differences in free energy between Arg28/Arg45 and Arg45Ser (ΔΔGArg45Ser) of about 2.1 kcal/mol, and of about 1.1 kcal/mol between Arg28/Arg45 and Arg45Thr (ΔΔGArg45Thr). Thus, both Arg45Thr and Arg45Ser may have lost an electrostatic contribution of about 1 kcal/mol in the bundling interaction, as suggested by Pascual et al.2Arg45Ser alone may have lost an additional 1 kcal/mol, which can be attributed to hydrophobic and/or van der Waals contact interactions. This interaction might come from the aliphatic part of the side chains in the Arg residue, which is, of course, almost entirely lost when Arg is replaced by Ser, but to a lesser extent when substituted by Thr, with its additional methyl group. This involvement of the aliphatic portion of Arg residues in hydrophobic interaction was also noted in the structure of chicken brain spectrin peptide3 and ofDrosophila spectrin.1 The Arg28 residue has been predicted to form a salt bridge to Glu2069 in the β-spectrin partial domain,20 and loss of that salt bridge should also substantially reduce binding affinity.
In summary, the replacement of Arg with Ser at position 28 induces substantial spectral changes that are difficult to delineate further. We suggest that the replacement of Arg with Ser may remove the repulsive interaction between Helix C′ and the hydrophilic exterior surface of Helices A and C in Arg28Ser. Thus, our results do not require a drastically altered Helix C′ conformation in Arg28Ser, but simply suggest that Helix C′ may transiently associate with the first structural domain or that there is transient Arg28Ser peptide–peptide association. This interpretation is consistent with the similar affinities of Arg28Ser and Arg45Ser, in that such a transient association for a small fraction of the time should not significantly reduce the accessibility of the Helix C′ binding site. The results for the Arg28Ser, Arg45Ser, and Arg45Thr substitutions at this time demonstrate that the effects of each single amino acid replacement are unique to the substitution, and need to be carefully evaluated to understand the effect of replacement on local conformation as well as on overall conformation and behavior of the protein. It is possible that the αI74 fragment observed in some spectrin mutants, as discussed above, may not be due to a conformational change in the first helix in α-spectrin, but may be due to disruption of helix bundling with β-spectrin consequent to disrupted local molecular interactions. The first helix (Helix C′ or helix 3), when not bundled with the helices in β-spectrin, may exhibit a more flexible or “looser” helical conformation25 that may be more susceptible to proteolysis than the tightly packed triple helical bundle. Our studies also suggest that for therapeutic development, it may not be necessary to use an approach to correct a drastically altered helix 3 (Helix C′) structure, but instead only a subtle manipulation of a specific molecular interaction/recognition may be needed to return to the normal helical bundling that occurs in spectrin tetramerization.
We thank Shahila Mehboob and Bing-Hao Luo from Loyola University of Chicago for providing the Spβ peptide.
Supported in part by grants from the United States National Science Foundation (NSF) MCB9801870 (L.W.-M.F.), the United States National Institutes of Health (NIH) HL57604 and American Heart Association Midwest Affiliate 0051630Z (M.E.J.), and an American Heart Association Midwest Affiliate predoctoral fellowship 9910169Z (S.P.). This research made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant RR02301 from the Biomedical Research Technology Program, National Center for Research Resources. Equipment in the facility was purchased with funds from the University of Wisconsin, NSF (DMB-8415048 and BIR-9214394), NIH (RR02301, RR02781, and RR08438), and the US Department of Agriculture.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
L. W-M. Fung, Department of Chemistry, Loyola University of Chicago, 6525 N Sheridan Rd, Chicago, IL 60626; e-mail:lfung@luc.edu; or Michael E. Johnson, Center for Pharmaceutical Biotechnology, University of Illinois at Chicago, 900 S Ashland, Chicago, IL 60607; e-mail: mjohnson@uic.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal