Abstract
For many years, human hemoglobin (Hb) isolated from erythrocytes has been investigated as a potential oxygen delivery therapeutic. Advantages with respect to the need for blood typing were balanced with various undesirable properties of cell-free Hb, including cost, overall oxygen affinity, alterations in cooperativity, and ready dissociation into toxic dimeric species. The use of total gene synthesis has resulted in very high levels of functional human Hb expression inEscherichia coli, but there remains a desire for effecting the crosslinking of the hemoglobin tetramer and providing for ready means for increasing the globular molecular weight. In this communication, we report a novel method for linking alpha chains. By circularly permuting one alpha sequence, the second alpha chain in the Hb tetramer can be linked with glycine residues to form 2 bridges across the central cavity. The second alpha chain thus presents its amino and carboxyl termini on a solvent exposed surface, providing for additional polymerization of oxygen-carrying subunits or attachment of any other peptide-based therapeutic.
Introduction
A mechanistic and predictive understanding of how a primary sequence can fold to a single tertiary structure remains undefined. Since a thorough understanding of this process will enable the rational design of unique protein structures, this remains an area of active research. One theory suggests that short continuous regions within a protein develop local interactions early in the folding process to minimize the number of accessible structures and catalyze the formation of a functional protein. Radical perturbations of protein structure by circular permutation is one technique being used in an attempt to explore this issue. In addition, utilization of circular permutation itself can be exploited in the design of new protein structures.
A circularly permuted protein is created in 2 steps. First, the original termini are linked to form a circular polypeptide. New termini are then created by cleavage of a peptide bond at a location distant from the original termini. Goldenberg and Creighton engineered the first circularly permuted sequence by chemically condensing the termini of bovine pancreatic trypsin inhibitor (BPTI) and generating new termini by limited proteolysis.1 This circularly permuted variant was shown to refold, in vitro, to a native functional conformation. This seminal experiment suggested that the location of the termini has little effect on the final 3-dimensional structure of the protein. Luger and coworkers genetically circularly permuted phosphoribosyl anthranilate isomerase.2 TheEscherichia coli translated variant was structurally and functionally similar to the wild-type protein. Several other monomeric proteins have since been circularly permuted through genetic manipulations, all of which maintain a nativelike function.3-11 Single subunits of multimeric proteins can also withstand circularly permuted sequences. For example, circularly permuted catalytic chains of aspartate transcarbamoylase combined in vitro with regulatory chains produced a folded and functional multimeric protein.12 However, all of these polypeptides are single-domain cooperatively folding units.
More recently, circularly permuted variants of multidomain proteins have been accomplished. Circular permutation within the NAD binding domain of glyceraldehyde-3-phosphate dehydrogenase produced a nativelike functional protein.13 The termini of the T4 lysozyme were also moved from one domain to another without significant effects on the stability or function of the protein.14These results suggest that local sequence continuity within defined structural domains is not required for proper protein folding.
The remarkable similarity between circularly permuted isomers suggests that the linear organization of secondary structures has little effect on the final 3-dimensional structures. In fact, Viguera et al have addressed this question more directly by exhaustively permuting the SH3 domain of α spectrin, demonstrating that any loop region could be redefined as the new termini without effecting the folded structure.15 However, these circular permutations have relied on the fact that the termini of many proteins are found close together in their respective 3-dimensional structures. The termini could thus be linked with the smallest polypeptide required to span the distance between them and lock them in a geometry consistent with the wild-type protein structure. The fact that some proteins have been shown to fold from 2 independent polypeptide fragments suggests that this tight connectivity of the termini is not an absolute requirement for successful circular permutations.16-20 However, only recently has one explored the ability of circularly permuted sequences to fold with long linking sequences separating the original termini or additional structural domains designed within the linking region itself. Baird et al inserted calmodulin into a circularly permuted green fluorescent protein.21 To further explore the flexibility allowed within the linear sequence and spatial organization of amino acids to define a tertiary structure, we have constructed a circularly permuted variant which links 2 subunits within the human hemoglobin (Hb) tetramer.
Human hemoglobin is a heterotetrameric protein which consists of 2 α globin subunits and 2 β globin subunits. Each α globin associates with a β globin to form a dimer and 2 dimers associate to form the hemoglobin tetramer. We effected the circular permutation of one α globin by linking its termini with the sequence of another α globin within the tetramer. The oligomeric nature of hemoglobin provides an ideal system for characterizing a circularly permuted sequence whose termini are linked with another structural domain, since each of the subunits naturally associate to form the functional tetramer.
Furthermore, there is a significant amount of commercial interest in stabilizing the hemoglobin tetramer with the introduction of covalent linkages across the dimeric interface. Cell-free hemoglobin solutions are being explored for use in surgical and traumatic blood loss. However, hemoglobin dissociates into dimers within the vasculature and the dimers are rapidly filtered by the kidneys. Numerous chemical methods of crosslinking the dimers have been investigated. Only one genetic fusion of globin subunits has been reported wherein 2 α globin genes are fused in tandem to generate a di-α globin. The circular permution we describe is a unique method of genetically stabilizing the hemoglobin tetramer. We will also discuss the immplications of this protein for the future design of enhanced oxygen-carrying therapeutics.
Materials and methods
Genetic constructions
The E coli strain DH5α was used in all the genetic engineering and protein expression. Plasmids pHS471 and pWHS486, used in the genetic constructions, were generated previously by Hernan et al.45 These vectors encode the desVal variants of α and β globin in which the native amino terminal valines have been replaced with initiator methionine residues.
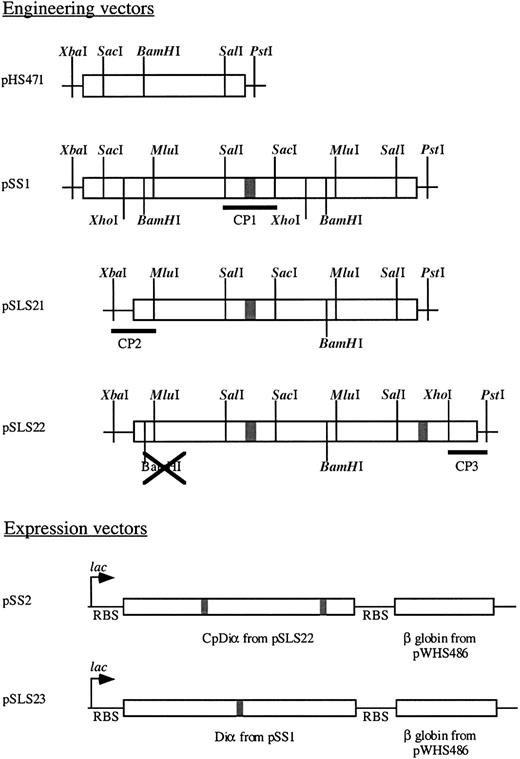
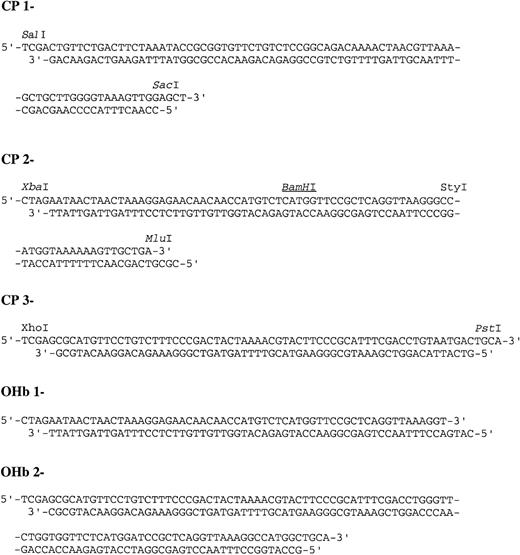
Several steps were required in the construction of the circularly permuted hemoglobin (CpHb). Each vector utilized in the construction is schematically represented in Figure 1. First, a tandem fusion of 2 α globin genes was created utilizing the α desVal gene (pHS471). pHS471 was digested with SalI andPstI to yield a vector containing the majority of the first α globin. Then an α globin gene fragment was generated from the digestion of pHS471 with SacI and PstI. This fragment was then ligated into the vector along with a linking cassette, CP1 (Figure 2), which codes for the last portion of the first α globin gene beginning with theSalI restriction site, a glycine codon, and the first portion of the second α globin gene through the SacI restriction site to generate pSS1, a Diα globin gene. Then, cassette CP2 (Figure 2) and a 400-bp fragment generated from anMluI/BamHI digest of pSS1 was ligated into pHS471 digested with XbaI and BamHI to generate pSLS21. Finally, cassette CP3 (Figure 2) and a 400-bp fragment generated from an XhoI/BamHI digest of pSS1 was ligated into pSLS21 digested with BamHI and PstI to generate pSLS22 (Figure 1). The coding region of pSLS22 encodes a circularly permuted α globin in which the termini are linked by a -gly-α globin-gly- sequence.
Vectors utilized in engineering and expression of CpHb and DiαHb.
Boxed areas are coding sequences, where shaded box is a glycine linker. Vector names are at the left with restriction sites and cassettes used denoted.
Vectors utilized in engineering and expression of CpHb and DiαHb.
Boxed areas are coding sequences, where shaded box is a glycine linker. Vector names are at the left with restriction sites and cassettes used denoted.
Expression operons were then constructed consisting of the α globin constructs and the β DesVal globin gene under the control of a singlelac promoter. The tandemly fused diα globin gene (pSS1) and the circularly permuted Diα globin gene (pSLS22), isolated by digestion of pSS1 and pSLS22 with XbaI and PstI, and the β globin gene, isolated by digestion of pWHS486 withPstI and HindIII, were ligated into pUC18 digested with XbaI and HindIII to generate pSS2 and pSLS23, respectively. Each of these operons are schematically represented in Figure 1. DNA sequencing of each construct was performed at the University of Illinois DNA Sequencing Facility using a Perkin/Elmer DNA Sequencer (Wellesley, MA) with polymerase chain reaction (PCR) amplification. Sequencing reactions were stopped with fluorescently labeled dideoxy nucleotides. Universal and reverse primers for pUC sequencing initiated the reactions.
Protein expression and purification
Native human hemoglobin, HbAo, was isolated and purified as described previously.46 The DiαHb and CpHb were expressed in E coli DH5α cells. The cultures were grown in 1 L of 2XYT media (16 g tryptone, 10 g yeast extract, and 5 g NaCl/L) containing 200 μg/mL ampicillin and 0.5 mM δ-aminolevulinic acid in 6 L shake flasks at 37°C. δ-aminolevulinic acid is a heme precursor after the committed step in the biosynthesis and has been shown to increase hemoglobin production in E coli.47 After 36 to 48 hours, the cells were harvested by centrifugation at 8000g for 5 minutes and the cell paste was removed and stored frozen at −70°C.
CpHb was purified by the following methods. Cell paste was allowed to thaw over a stream of carbon monoxide (CO), and all buffers used during the lysis and purification procedure were saturated with CO. Thawed cells were resuspended in 5 times (wt/vol) 10 mM NaH2PO4, pH 6.0, 1 mM ethylenediaminetetraacetic acid (EDTA). Cells were lysed by 3 to 4 passes through a Stansted AO-116 cell disrupter (Stansted Fluid, Stansted, United Kingdom). After cell lysis, 80 units/mL DNase and 8 units/mL RNase were added to the mixture and allowed to incubate at room temperature for 1 hour. The mixture was then centrifuged at 100 000g in a Beckman L8-M ultracentrifuge (Beckman Instruments Incorporated, Fullerton, CA) for 30 minutes. The supernatant was retained and the pH adjusted to 6.0 with 20 mM NaH2PO4. The supernatant was then loaded onto a carboxy methyl cellulose column (Whatman, Clifton, NJ) equilibrated with 10 mM NaH2PO4, pH 6.0, 1 mM EDTA. The column was then washed with 4 column volumes of 10 mM NaH2PO4, pH 6.0, 1 mM EDTA, and finally eluted with a step gradient to 20 mM Tris-HCl, pH 7.0, 1 mM EDTA. Further purification was accomplished by high-pressure liquid chromatography. The protein samples were first exchanged into Millipore water or 10 mM NaH2PO4, pH 6.0, 1 mM EDTA using a Sephadex G-25 column (Pharmacia Biotech, Peapack, NJ). The protein was then loaded onto a HiLoad SP Sepharose column (Pharmacia Biotech) equilibrated with 10 mM NaH2PO4, pH 6.0, 1 mM EDTA. The column was washed with one column volume of the equilibration buffer. The circularly permuted hemoglobin was eluted with a linear gradient to 20 mM NaH2PO4, pH 7.0, 1 mM EDTA. The same method was used for DiαHb purification with the exception of equilibration of the loading and washing buffers to pH 6.5 and the elution buffers to pH 7.5.
Protein characterization
Electrospray mass spectometry was performed on the protein samples at the University of Illinois Mass Spectrometry Facility using a Quattro-70 mass spectrometer (Micromass, Manchester, United Kingdom). Purified protein samples were exchanged into water using a Sephadex G-25 column. The samples were diluted to a concentration of 10 pmol/μL into a 50:50 acetonitrile:water solution containing 0.2% formic acid for the experiments.
N-terminal sequencing was performed at the University of Illinois Genetic Engineering Facility. One nanomole of protein was sequenced by automated edman degradation48 using a 470A gas phase sequencer (Applied Biosystems, Foster City, CA) with an online PTH amino-acid analyzer.
Polyacrylamide gel electrophoresis (PAGE) was performed on the purified protein samples. Sodium dodecyl sulfate (SDS)–PAGE was performed after boiling in 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 2.5% SDS, 5% β-mercaptoethanol, and 0.001% bromophenol-blue. Native-PAGE was performed in 10 mM Tris-HCl, pH 8.0, 1 mM EDTA. Both the denaturing and nondenaturing gels were run on a 10% to 15% preformed gradient gel using standard protocols on the Phast System (Pharmacia Biotech). The gels were stained with Coomassie.
Heme–globin stoichiometry
To assess the heme-to-globin ratio in each construct, the protein content and heme content were measured independently. Protein samples were diluted to an approximate [heme] of 10 μM. Heme content was assessed by a pyridine hemochromogen assay.49One mL of protein was added to 3 mL of 4.1 M pyridine, 0.1 M NaOH and allowed to incubate at room temperature for 1 hour. A few grains of dithionite were added to the sample; the sample was mixed and the absorbance at 556 nm was determined. The heme content was calculated using ε556 = 0.032 μM−1cm−1for the reduced pyridine heme complex. The globin content was assessed with the absorbance at 280 nm. The 280-nm absorbance is attributable to aromatic side chains (phenylalanine, tyrosine, and tryptophan). Since each globin domain contained an equal complement of these residues, the absorbance at 280 is a good indicator of proportional globin content. The ratios of these independent measures were normalized to a ratio of 4:1 for HbAo.
Spectroscopy methods
Protein samples were reduced by the addition of a few grains of dithionite and exchanged into air-saturated water over a Sephadex G-25 column. The oxygen-bound spectrum was taken in a U-3300 spectrophotometer (Hitachi, Tokyo, Japan). Then a few grains of dithionite were added to the sample and a deoxy spectrum was taken in a septa-sealed cuvette. Finally, the CO bound form was generated by gently bubbling the sample with CO for 15 seconds.
Circular dichroism spectra were recorded using a J-720 spectropolarimeter (Jasco, Victoria, British Columbia, Canada) using a 0.1 cm pathlength quartz cell. The CD signal of the cuvette and buffer was recorded and subtracted from the experimental samples. Protein solutions were adjusted to a heme concentration of 10 mM in 10 mM KPO4, pH 7.4, and CO saturated. Helical contents were estimated using extinction coefficients at 222 nm as reported by Greenfield and Fasman for α helix and random coil with the assumption that no β-pleated sheet structure was present in the proteins.50
Oxygen equilibrium
Oxygen equilibrium analysis was performed using a modified Imai cell on an apparatus made in-house. The sample cell was described previously.44 White light from a tungsten source was chopped at a frequency of about 630 Hz by passing through an Oriel chopper (Oriel, Stratford, CT). Fiber bundles directed the light through the sample cell mounted over a homemade magnetic stirrer and into an Oriel 1/4 meter monochrometer where 560 nm or 430 nm light was isolated, depending on sample concentration. Signal detection was accomplished with a photomultiplier powered by a PS310 high-voltage power supply and amplified with a SR530 lock-in amplifier (all from Stanford Research Systems, Sunnyvale, CA). The cell was thermostated with a circulating water bath at 25°C. Gas exchange was accomplished by venting air or nitrogen over the sample surface after passing through a flow meter and a gas wash bottle thermostated in the circulating water bath. Gas flow was maintained at a rate to exchange ligands over a 45-minute period (about 20-30 mL/min). Oxygen concentrations were monitored with a YSI 5331 Clark electrode (Yellow Springs Instruments, Yellow Springs, OH). The polarization voltage of 0.8 volts for the electrode and current to voltage conversion were supplied by a Bioanalytic Systems CV-27 voltamograph (West Layfayette, IN). The voltage outputs for both the oxygen electrode and optical signals were recorded on Hewlett Packard 3840 multimeters (Hewlett-Packard, Palo Alto, CA), then transferred to a PC.
The experiments were performed in 50 mM Tris-HCl, pH 7.4, 0.1 M [Cl−], 25°C. Protein concentrations for HbAo and the recombinant hemoglobins were 60 μM and 5 μM in heme, respectively. Since there was no concentration dependence on the degree of cooperativity or oxygen affinity for the crosslinked variants, 5 μM concentrations of heme were sufficient to measure accurate oxygen binding parameters. An enzymatic reduction system was also added to the samples to minimize the amount of oxidized hemoglobin.51As a measure of the degree of cooperativity of the proteins, Hill coefficients were determined from each isotherm. Each protein's response to allosteric effectors was determined by the addition of 0.1 mM inositol hexaphosphate (IHP). Finally, isotherms for 3 different pH values were measured to establish a Bohr effect. The 3 buffers used were as follows: 50 mM BisTris, pH 6.5, 0.1 M [Cl−]; 50 mM Tris-HCl, pH 7.4, 0.1 M [Cl−]; 50 mM Tris-HCl, pH 8.5, 0.1 M [Cl−].
Results
Design of the circularly permuted hemoglobin
Figure 3 is a schematic of the linear amino acid sequence and folded structure of the circularly permuted hemoglobin (CpHb). An open-reading frame coding for the circularly permuted Diα globin was constructed. The coded polypeptide consisted of the following: (1) an initiator methionine residue, (2) residue 49-141 of an α globin sequence (the first portion of a circularly permuted α globin), (3) a glycine residue, (4) a wild-type α globin sequence (the linking sequence), (5) a second glycine residue, and (6) residue 1-48 of an α globin sequence (the second portion of a circularly permuted α globin). The completed gene construct was coexpressed with a wild-type β globin under the control of a single promoter. As a control, the DiαHb described by Looker et al22 was also constructed. This protein consists of the tandem fusion of 2 α globins with a single linking glycine residue.
Schematic of the circularly permuted hemoglobin.
The linear polypeptide of the circularly permuted Diα globin can fold into 2 distinct 3-dimensional structures. However, only one is capable of assembling with β globins, as shown. The fact that CpHb consists of both α and β globin components suggests that the schematic shown above accurately depicts the structure of CpHb.
Schematic of the circularly permuted hemoglobin.
The linear polypeptide of the circularly permuted Diα globin can fold into 2 distinct 3-dimensional structures. However, only one is capable of assembling with β globins, as shown. The fact that CpHb consists of both α and β globin components suggests that the schematic shown above accurately depicts the structure of CpHb.
Large structural transitions occur during the oxygenation of the hemoglobin tetramer. High-resolution crystal structures of both the oxy (R state) and deoxy (T state) states of hemoglobin were used in the design of the circularly permuted Diα globin.23,24 In both states the termini of the α globins are highly ordered. The α carbons of the amino and carboxy termini of complementary α globins are situated about .85 nm (8.5 Å) away from one another in both the R and T states. Since the maximum distance between 2 α carbons of a polypeptide is .38 nm (3.8 Å), the addition of one residue to link the termini will span the majority (.76 nm [7.6 Å]) of the distance. A single glycine residue was utilized as the linking segments to allow for maximal conformational freedom and minimize steric bulk in the protein. Furthermore, Looker et al have previously described the use of a single glycine residue in a tandem fusion of α globins. The refined crystal structure of this variant was shown to cause minimal perturbation to the protein structure.22 25 Molecular symmetry in hemoglobin enables the extension of this linkage scheme to both sets of termini.
The selection of a permutation site within the α globin sequence was more difficult. The α globin is a very efficiently folded domain with the majority of the polypeptide involved in secondary structure, intersubunit interactions, or heme binding. This significantly limited the choice in permutation sites. Secondary structure is generally regarded as important to the stability of proteins, whereas loop regions connecting the secondary structure are less important.26 Moreover, the SH3 domain of α-spectrin was exhaustively permuted in each of the loop regions with little structural effect, suggesting that loop regions are more appropriate targets for cleavage.15 However, several loop regions are involved in the stabilization of the oligomeric structure and transitions between R and T states.27 For example, scissoring of the EF corner is critical to the intersubunit communication of a ligand binding event.28 Furthermore, the ability of the globin to bind to the prosthetic heme group relies on the formation of a hydrophobic cavity which is partially defined by loop regions. Elimination of these critical regions in the protein left the loop between helix C and D as the optimal site for the new termini of the circularly permuted α globin. Serine 49, within the CD corner, was chosen as the new amino termini for the circularly permuted variant.
Biochemical characterization
Both DiαHb and CpHb were expressed to a high level inE coli. The proteins were purified to homogeneity and Edman degradation revealed that the amino terminal sequence of the circularly permuted Diα globin was consistent with cleavage of the initiator methionine.29 This was expected from the rules described previously for N-terminal processing in E coli.30 Electrospray mass spectrometry, performed on each variant, reconfirmed methionine cleavage of the CpHb α globin segments. Furthermore, each variant contained α globin and β globin subunits of the expected molecular weight confirming that no posttranslational modification of the subunits occurred in vivo (data not reported).
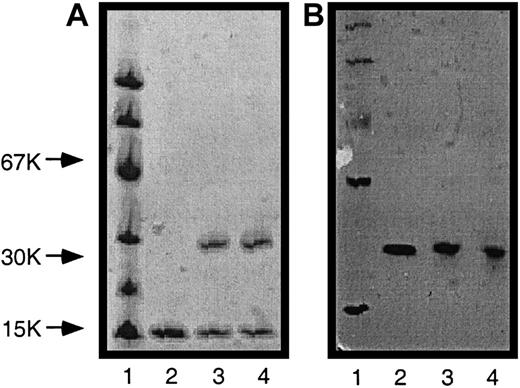
SDS-PAGE also revealed that each variant consisted of 2 components, β globins and α globins (Figure 4A). The native-PAGE then demonstrates that each variant assembles into an oligomeric structure consistent in molecular weight with 1 tetramer (Figure 4B). In addition, Coomassie staining of wild-type β globins and α globins is known to be equal in intensity. Since the α globins contain an amino acid content identical to 2 α globins, the equal staining intensity of the β globin and α globin bands suggests that each recombinant variant contains a globin ratio consistent with the expected oligomeric assembly, as depicted in Figure3.
SDS-PAGE and native-PAGE of recombinant hemoglobins.
(A) SDS-PAGE; (B) native-PAGE. Lane 1: MW standards; lane 2: HbAo; lane 3: DiaHb; lane 4: CpHb.
SDS-PAGE and native-PAGE of recombinant hemoglobins.
(A) SDS-PAGE; (B) native-PAGE. Lane 1: MW standards; lane 2: HbAo; lane 3: DiaHb; lane 4: CpHb.
The variant's capacity to bind heme was clear from the whole-cell CO difference spectrum obtained for each protein. However, this did not preclude the possibility that only a partial complement of heme was binding to each protein. To assess the ratio of heme bound to globin domains, the heme content and protein content were measured independently as described in “Materials and methods.” Each protein was shown to contain a full complement of 4 hemes per tetrameric unit (Table 1).
Ratio of heme to globin in each construct
| . | [heme] (μM) . | A280 . | [heme]:A280 . | [heme]:globin* . |
|---|---|---|---|---|
| HbAo | 2.21 | .3401 | 6.50 | 4.0 |
| DiαHb | 2.28 | .344 | 6.63 | 4.1 |
| CpHb | 2.08 | .340 | 6.12 | 3.8 |
| . | [heme] (μM) . | A280 . | [heme]:A280 . | [heme]:globin* . |
|---|---|---|---|---|
| HbAo | 2.21 | .3401 | 6.50 | 4.0 |
| DiαHb | 2.28 | .344 | 6.63 | 4.1 |
| CpHb | 2.08 | .340 | 6.12 | 3.8 |
Obtained from normalization of the data to a value of 4 for HbAo.
Spectroscopic characterization
The electronic environments around the heme were investigated by the optical spectra of the oxygen-bound, carbon monoxide–bound, and deoxygenated species. The optical spectrum of each ligation state of each variant was nearly identical to HbA. The absorbance maximum for each variant and ligation state were all within 1 nm of HbA. Furthermore, the different ligation states were reversible since the carbon monoxide–bound state was competed off with oxygen and light, the deoxygenated state was created with the reduction of free oxygen by dithionite, and the carbon monoxide or oxygen-bound forms could be reformed with bubbling of the appropriate gas through a deoxygenated solution.
The secondary structure of the proteins was assessed with far UV circular dichroism spectroscopy. Helical contents were estimated to be 74%, 75%, and 78% for HbAo, DiαHb, and CpHb, respectively. These values are in good agreement with the 72% estimated from analysis of the crystal structure.31
Oxygen equilibrium
Oxygen equilibrium measurements were taken to assess the ligand affinity and response to allosteric effectors. These measurements are exquisitely sensitive to the quaternary transitions and functional integrity of hemoglobin. Although DiαHb exhibited an oxygen affinity similar to HbA, CpHb displayed an oxygen affinity that was 5-fold greater as evidenced by the partial pressure (atmospheres) for 50% saturation (p50) of these proteins listed in Table 2.
Oxygen equilibrium parameters for recombinant hemoglobins
| . | HbAo . | DiαHb . | CpHb . |
|---|---|---|---|
| p50(mmHg) | 5.0 ± 0.2 | 4.5 | 0.9 ± 0.1 |
| nmax | 3.0 | 2.1 | 2.0 |
| IHP effect | .9 | NA | .7 |
| Bohr effect | − .55 | NA | − .37 |
| . | HbAo . | DiαHb . | CpHb . |
|---|---|---|---|
| p50(mmHg) | 5.0 ± 0.2 | 4.5 | 0.9 ± 0.1 |
| nmax | 3.0 | 2.1 | 2.0 |
| IHP effect | .9 | NA | .7 |
| Bohr effect | − .55 | NA | − .37 |
IHP indicates inositol hexaphosphate; NA, not applicable.
The cooperativity of the variants were described by the mathematical model developed by Archibald Hill described previously.32The Hill coefficients are reported as nmax in Table 2. The ability of known allosteric regulators of hemoglobin (IHP and protons) to shift the oxygen affinity was also investigated with the circularly permuted variant. Both IHP and protons preferentially bind to and stabilize the deoxy state of hemoglobin causing a right shift in the oxygen-binding curve and increasing the overall p50. The IHP effect is reported as the change in p50 with the addition of 0.1 mM IHP, Table 2. The Bohr effect, or increase in oxygen affinity with a decrease in pH was determined by the independent measurement of oxygen affinity at 3 different pH values. The slope of this curve, ∂log(p50)/∂pH, at a pH of 7.5 defines the alkaline Bohr effect. CpHb exhibits a Bohr effect of about 60% that of HbA.
Discussion
A variety of structural and functional techniques have been utilized in the investigation of the crosslinked and circularly permuted hemoglobin variants. Although there are minor differences, the recombinant proteins are remarkably similar to HbAo. The proteins assemble to the expected oligomeric structure and incorporate a full complement of heme. The identity of the optical spectra of 3 different ligation states suggests that the electronic environments of the heme cavities are similar. Furthermore, each protein folds to a structure with about 75% helical content consistent with HbAo. The only perturbations to these proteins were observed in the more sensitive functional assays.
The decrease in oxygen-binding cooperativity is not surprising. The DiαHb described herein has previously been characterized and exhibited a similarly reduced cooperativity as reported here.22 Although the α globin termini are in close proximity in both the R state and T state, there is no information on their proximity in intermediate ligation states. It is possible that during the quaternary transition these termini separate. A covalent linkage would hinder this transition, resulting in diminished cooperativity. The fact that DiαHb and CpHb exhibit similar reductions in cooperativity suggests that this reduction could be attributed to the linkage of the termini and not cleavage of the CD loop region in CpHb. Nichols et al demonstrated that this loop region is a highly conserved rigid structural element between the oxy and deoxy conformations.33 The identification of this rigid body implies that motion of this loop as a unit may be involved in intersubunit communication and required for hemoglobin cooperativity.33 Since the disruption of this loop in CpHb does not effect the cooperativity of hemoglobin, the loop must be stabilized by tertiary or quaternary interactions rather than the peptide backbone, which is discontinuous in the circularly permuted variant.
Allosteric regulators of oxygen affinity play an important physiologic role. These effectors bind to sites distinct from the heme cavity and preferentially stabilize the deoxy state of hemoglobin. The ability of the circularly permuted protein to respond to allosteric regulators suggests that the protein is structurally similar to HbAo in these regions distant from the heme cavity. IHP is a potent allosteric regulator which binds with a significantly higher affinity to the same cavity as 2,3 diphosphoglycerate.34-36 This cavity is defined by several positively charged residues at the β1β2 interface. CPHb exhibits only a slight decrease in the observed IHP effect, indicating that the protein environment around the cavity remains similar to HbAo.
Protons also preferentially bind to the T state of hemoglobin. In the acidic environment of respiring tissues, the oxygen affinity of hemoglobin is reduced to increase the release of oxygen. This results from a change in acid-based equilibrium constant (pKa), between the T and R states, of several titratable amino acids.37-39 Perutz et al38proposed a mechanism for the Bohr effect in which the N-terminal amino group of the α globins form a salt bridge with β146His in the deoxy conformation, increasing their proton affinity. Carbamylation of this amino group results in a diminished Bohr effect.40In a similar fashion, the linkage of both α globin termini and incorporation of the amino groups into the peptide backbone in the circular permutations is probably responsible for the observed decrease in Bohr effect. Furthermore, the Bohr effect is reduced by about 40% compared with HbAo. This is similar to the 20% to 30% Bohr effect contribution estimated for the N-terminal amino groups, suggesting that the remaining regions responsible for the Bohr effect are still intact.40-42
Implications on protein folding and design
The number of successfully engineered circular permutations and the identification of naturally occurring circular permutations is tribute to the remarkable insignificance of the termini location in the folding or stability of many proteins.43 This demonstrates that a vectorial folding mechanism, in which the protein folds in a linear fashion as it is translated, is not a requirement for a functional protein in vivo. However, it does not eliminate this as a possible folding pathway. It is likely that local regions of the protein fold to initiate more distant tertiary interactions. At least in some proteins the spatial uncoupling of protein regions will not prevent the protein from folding. In fact, peptide fragments have been shown to initiate protein folding independently then associate to form a complete protein.16-20 Work presented here extends this belief by demonstrating that the spatial separation of the circularly permuted globin fragments with a long linking domain (an α globin) will assemble to a functional protein.
The repeating nature of structural motifs in the CpDiα globin presents a particular problem in the folding of this protein. CpDiα globin can be thought of in 2 forms: (1) a circularly permuted α globin with termini fused by a second α globin, as discussed throughout this paper, or (2) a tandem fusion of 2 circularly permuted α globins. Each of these possibilities represents a distinct 3-dimensional structure schematically depicted in Figure 3. The difference is that only the first form is in a geometry capable of assembling with β globins to form the functional tetramer. The ability of the CpDiα globin to assemble with β globins and express nativelike function clearly eliminates the second form as a significant product of bacterial expression. In this light, a vectorial folding mechanism is unlikely, as this would favor the first form. However, it is not clear what factors prevent the second form and favor the first form. For example, it is possible that the second form is initially generated then reassembled into the first form in the presence of β globins. This is consistent with the theory that β globins act as a scaffold for the folding and assembly of α globins.44 In CpHb the presence of β globins may be energetically driving the folding of the CpDiα globin into the first form, by stabilizing this form relative to the second form.
Summary
The successful construction of this multiglobin circular permutation opens up many possibilities in the future design of supramolecular protein structures, by suggesting that protein domains can be engineered within the circular permutation site of another protein. These new multidomain or multiprotein stuctures can be spatially oriented as desired by altering the circular permutation site of the primary protein, enabling the rational design of unique multidomain proteins. It could also enable the fixed approximation of proteins with physiologic protein–protein interactions as well as the pharmacuetical design of complexes that require distinct protein functionalities in close proximity.
In the field of oxygen-carrying therapeutics this protein provides a building block for the design of an enhanced oxygen delivery vehicle. By linking the native α globin termini and generating new surface-exposed termini we have generated a stabilized tetramer with the ability to genetically link other functional proteins. Since the original α globin termini are within the central cavity of the tetramer, the relocation of these termini will enable the fusion of additional proteins with minimal perturbation to the tetramer's function. The design of hemoglobin polymers by tandem fusions of multiple circularly permuted hemoglobins is already underway. Utilization of this system to produce hemoglobin polymers would provide a significant advantage over chemically polymerized hemoglobins. First, the highly efficient and exact nature of protein production in E coli ensures a monodisperse solution rather than the polydisperse distributions observed with chemical polymerization methods. Second, elimination of chemical modification methods will improve yields and streamline processing. Finally, its recombinant nature enables functional optimization via mutagenesis for a defined application as our understanding of oxygen delivery from the extracellular environment improves.
The exposed termini of CpHb could also be utilized for the fusion of other proteins. Fusions with superoxide dismutase (SOD) (Sigma, St Louis), catalase, or other radical scavenging enzymes have the potential to minimize known toxic side effects of current oxygen-carrying therapeutics. Fusion with an erythropoeitin consensus sequence could be designed to induce endogenous red blood cell formation coincident with the in vivo degradation of the oxygen-carrying therapeutic. Alternatively, the defined vascular stability of human hemoglobin in plasma could be exploited as a pharmaceutically active peptide carrier for nearly any peptide-based product.
Thanks to Mark McLean and Aretta Weber for expert editorial assistance.
Supported by National Institutes of Health grants GM31756 and GM33775.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stephen G. Sligar, University of Illinois, 405 North Mathews, Urbana, IL 61801; e-mail:s-sligar@uiuc.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal