Abstract
Natural killer T (NKT) cells are important regulators of the immune system, but their trafficking machinery, including expression of chemokine receptors, has been poorly defined. Unlike other conventional T-cell populations, we show that most NKT cells express receptors for extralymphoid tissue or inflammation-related chemokines (CCR2, CCR5, and CXCR3), while few NKT cells express lymphoid tissue–homing chemokine receptors (CCR7 and CXCR5). A population with homing potential for lymph nodes (L selectin+ CCR7+) exists only within a small subset of CD4 NKT cells. We show differential expression of chemokine receptors among NKT cell subsets: CCR4 is mainly expressed by a high cytokine (interleukin-4/interleukin-2)–producing (CD4) NKT subset, while CCR1, CCR6, and CXCR6 are preferentially expressed by the low cytokine-producing CD8 and CD4−CD8− subsets. In line with this, TARC/CCL17 (a CCR4 ligand) induces preferential chemotaxis of the CD4 NKT subset, while chemotactic activities of LARC/CCL20 (a CCR6 ligand) and MIP-1α/CCL3 (a CCR1 ligand) are focused on the CD8 and CD4−CD8− NKT cells. We conclude that, unlike conventional naive, memory, or effector T cells, the entire NKT cell population expresses nonlymphoid tissue homing chemokine receptors, yet NKT cell subsets differ considerably from each other by displaying distinct and reciprocal expression patterns of some chemokine receptors. Our results identify chemokine receptors that are potentially important for trafficking of human blood NKT cell subsets and reveal their function (cytokine production capacity)–dependent differential trafficking potentials.
Introduction
Natural killer T (NKT) cells constitute a unique class of T-lymphocyte lineage that shares some characteristics with NK cells.1,2 These cells have an extremely restricted T-cell receptor (TCR) repertoire, consisting of an invariant Vα24-JαQ chain preferentially paired with a Vβ11 chain in human beings.3,4 Although their natural TCR ligands remain to be identified, NKT cells can be generated and activated by glycolipid antigens such as α-galactosylceramide and glycosyl-phosphatidylinositol in the context of CD1d, a β2-microglobulin–associated major histocompatibility complex class I–like molecule.5-7
Human NKT cells express NK-associated NKR-P1 (CD161, the human version of mouse NK1.1) and CD45RO and are heterogeneous in expression of CD4 or CD8.8-11 NKT cells, especially the CD4+subset, are enriched in type 2 (interleukin-4+[IL-4+]) and type 0 (IL-4+interferon-γ+ [IFN-γ+]) cells.11-13 NKT cells with distinct cytokine production profiles can be generated by different types of dendritic cells or stimuli.14 Efficicient production of cytokines, particularly IL-4, is important for the regulation of the immune system by inhibition of Th1 and induction of Th2 response. NKT cells have also been implicated in tumor rejection, control of microbial infection, suppression of autoimmune diseases, and graft-versus-host disease after bone marrow transplantation.1,2 15
NKT cells are widely distributed in the body, including bone marrow, liver, thymus, lymph nodes, spleen, and lung.2 Whereas thymus and liver contain primarily CD1d-dependent NKT cells, spleen and bone marrow are enriched in CD1d-independent NKT cells, suggesting differential localization of CD1d-dependent NKT cells from other related T-cell types in mice.16 NKT cells found in different organs or tissues are heterogeneous in production of cytokines.13,16 Chemokines, differentially expressed in tissue and inflammatory microenvironments, guide homing and localization of leukocytes in the body by inducing adhesion and chemotaxis through various cell surface chemokine receptors.17-20 Thus, we postulated that chemokine receptor expression patterns of NKT cell subsets may be distinct from other T-cell subsets to allow their differential tissue localization.
We examined chemokine response and receptor expression by different human Vα24+Vβ11+ NKT cell subsets in circulation. We show that most NKT cells express tissue-homing chemokine receptors and that NKT cell subsets with different cytokine production potentials display distinctive chemokine receptor expression patterns and chemokine responses, differing from each other and from the conventional T cells. We conclude that NKT cells are specialized both in cytokine production and expression of chemokine receptors.
Materials and methods
Reagents
Antibodies to IFN-γ (4S.B3), IL-2 (MQ1-17H12), IL-5 (JES1-39D10), CD161 (DX12), CD4 (RPA-T4), CD45RA (HI100), and tumor necrosis factor-α (TNF-α) (Mab11) were purchased from Pharmingen (San Diego, CA). Antibodies to CD8 (3B5) and CD56 (MEM-188) were from Caltag (Burlingame, CA). Antibodies to CLA (HECA 452) and CD62L (DREG-200) were from our laboratory. Anti-IL4 (3010.211) was from Becton Dickinson (Mountain View, CA). Antibodies to CCR1 (53504.111) CCR2 (48607.121), CCR5 (45549.111), CCR6 (53103.111), CXCR3 (49801.111), CXCR6 (56 811.111), and CXCR5 (51505.111) were from R&D Systems (Minneapolis, MN). Antibodies to α4β7 (Act-1), CCR3 (7B11), CCR4 (1G1), CCR7 (7H12-12-2), and CCR9 (GPR96-1) were from Millennium Pharmaceuticals (Cambridge, MA). Human chemokines, thymus and activation-regulated chemokine (TARC), interferon γ inducible protein-10 (IP-10), B lymphocyte chemoattractant (BLC), I-309, EBI-1–ligand chemokine (ELC), and macrophage-inflammatory protein-1α (MIP-1α) were from R&D Systems. Human liver and activation-regulated chemokine (LARC) was from PeproTech (Rocky Hill, NJ). All antibodies to chemokine receptors have been tested on positive control cells. The anti-CCR3 used in this study binds to eosinophils.21 The anti-CCR9 used in this study binds to naive and memory T cells.22 The batches of each antibody used in the current study were shown to react with eosinophils or gut homing lymphocyte subsets as well.
NKT cell preparation
For NKT cell sources, unseparated peripheral blood mononuclear cells (for chemokine receptor analysis) (Stanford University Blood Center) or isolated Vβ11+ T cells were used (for chemotaxis and cytokine analysis). Peripheral blood mononuclear cells were prepared by density gradient centrifuge on Histopaque-1077 (Sigma, St Louis, MO). TCR Vβ11+ T cells were isolated with anti-Vβ11–biotin and streptavidin beads and sorted by a magnetic method (Vβ11+ cells > 99%, Miltenyi Biotec, Auburn, CA).
Chemokine receptor expression
Peripheral blood mononuclear cells were sequentially stained with anti-CKR, antimouse immunoglogulin G (IgG)–biotin and streptavidin-allophycocyanin plus anti-Vα24–fluorescein isothiocyanate (FITC), anti-Vβ-11–phycoerythrin, and other antibodies to various surface antigens (eg, CD4, CD8, CD56, CD161). One million cells were acquired for each fluorescence-activated cell sorter analysis. Isotype-matched mouse antibodies (IgG1, IgG2a, and IgG2b) were used to control the chemokine receptor expression. Antibodies to CCR1, CCR2, CCR5, CCR6, CCR7, CCR9, CXCR4, CXCR5, and CXCR6 are IgG2b. Antibodies to CCR4, CCR5, and CXCR3 are IgG1. Anti-CCR3 is IgG2a. All isotype (IgG1, IgG2a, and IgG2b) control antibodies did not stain NKT cells (not shown). Negative controls shown in Figures1 and 2were with IgG2b.
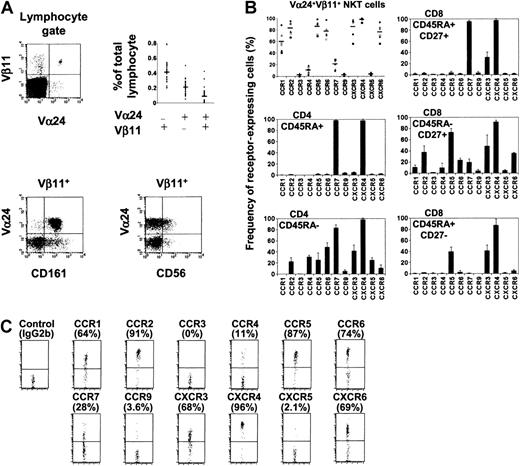
Expression of chemokine receptors by human NKT cells in circulation.
(A) Phenotype and frequency (n = 33) of human NKT cells examined in this study. (B) Chemokine receptor expression by Vα24+Vβ11+ NKT cells, CD4 (naive CD45RA+ and memory CD45RA−), and CD8 (naive CD45RA+CD27+, memory CD45RA−CD27+, and effector CD45RA+CD27−) T-cell subsets. Different symbols represent data from different donors, and horizontal bars represent averages (NKT cells, the upper left panel). Results from 6 donors (NKT cells) or averages ± SD from 4 donors (CD4 and CD8 T-cell subsets) are shown. (C) Chemokine receptor expression by Vα24+ Vβ11+ NKT cells shown as dot plots. Percentages positive are shown in parentheses.
Expression of chemokine receptors by human NKT cells in circulation.
(A) Phenotype and frequency (n = 33) of human NKT cells examined in this study. (B) Chemokine receptor expression by Vα24+Vβ11+ NKT cells, CD4 (naive CD45RA+ and memory CD45RA−), and CD8 (naive CD45RA+CD27+, memory CD45RA−CD27+, and effector CD45RA+CD27−) T-cell subsets. Different symbols represent data from different donors, and horizontal bars represent averages (NKT cells, the upper left panel). Results from 6 donors (NKT cells) or averages ± SD from 4 donors (CD4 and CD8 T-cell subsets) are shown. (C) Chemokine receptor expression by Vα24+ Vβ11+ NKT cells shown as dot plots. Percentages positive are shown in parentheses.
Distinct expression pattern of chemokine receptors among NKT cell subsets.
(A) Chemokine receptor expression by CD4, CD8, and DN NKT subsets (n = 4). (B) Dot plots of chemokine receptor expression by NKT cell subsets with percentages positive. (C) Relative frequencies of CD4, CD8, and DN NKT cell subsets in the Vα24+Vβ11+ NKT cell population (n = 11). (D) Differential cytokine production capacities of NKT cell subsets (n = 5). Isolated Vβ11+ NKT cells were stained with antibodies to Vα24, CD4, and CD8 and activated with phorbol myristate acetate and ionomycin in the presence of monensin to block internalization of surface antigens followed by intracellular cytokine analyses. Bars indicate averages of results from 4 (A), 11 (C), or 5 (D) donors. *Significant differences from the CD4 NKT subset (P < .05 [A,D]). **Significant difference from CD4 or CD8 subset (P < .02 [C]).
Distinct expression pattern of chemokine receptors among NKT cell subsets.
(A) Chemokine receptor expression by CD4, CD8, and DN NKT subsets (n = 4). (B) Dot plots of chemokine receptor expression by NKT cell subsets with percentages positive. (C) Relative frequencies of CD4, CD8, and DN NKT cell subsets in the Vα24+Vβ11+ NKT cell population (n = 11). (D) Differential cytokine production capacities of NKT cell subsets (n = 5). Isolated Vβ11+ NKT cells were stained with antibodies to Vα24, CD4, and CD8 and activated with phorbol myristate acetate and ionomycin in the presence of monensin to block internalization of surface antigens followed by intracellular cytokine analyses. Bars indicate averages of results from 4 (A), 11 (C), or 5 (D) donors. *Significant differences from the CD4 NKT subset (P < .05 [A,D]). **Significant difference from CD4 or CD8 subset (P < .02 [C]).
Chemotaxis
Chemotaxis assays were performed as previously described.23 Isolated TCR Vβ11+ T cells were used as input cells for chemotaxis. A total of 2× 105 to 5 × 105 cells were used per Transwell (Corning, Cambridge, MA) and allowed to chemotax for 3 hours. Chemokines TARC, LARC, MIP-1α, ELC, BLC, IP-10, MCP-1, MIP-1β, and I-309 were used at indicated concentrations. After chemotaxis, migrated cells in bottom wells were collected and stained with anti-Vα24–FITC, anti-CD4, and anti-CD8. Numbers of migrated cells and input cells were counted using FACSCalibur (Becton Dickinson), and the percent migrated cells of each cell subset was calculated.
Intracellular cytokine production analyses
Intracellular cytokine analyses have been performed as previously described.24 Briefly, isolated TCR Vβ11+ T cells were stained with anti-Vα24–FITC, anti-CD4–allophycocyanin, and anti-CD8–TriColor and activated for 4 hours with phorbol myristate acetate (50 ng/mL) and ionomycin (1 μM) in the presence of monensin (Sigma). Activated T cells were fixed and permeabilized with Cytofix/perm solution (Pharmingen), followed by staining with phycoerythrin-conjugated monoclonal antibodies to cytokines (IL-4, IL-15, IL-10, IL-13, IFN-γ, and TNF-α). Stained cells were analyzed with FACSCalibur and CellQuest program (Becton Dickinson). Only viable cells based upon their forward and side scatter were gated for cytokine analyses.
Statistics
Comparisons were performed by analysis of variance, and significant differences between groups were tested using a paired Student t test. Statistical significance was set atP < .05.
Results
Chemokine receptor expression by Vα24+Vβ11+ NKT cells
Vα24+Vβ11+ NKT cells represent a very small population in blood (on average 0.089% of lymphocytes, n = 33) (Figure 1A). Almost all of these circulating NKT cells express CD161, while only a subset expresses CD56 (Figure 1A). We examined Vα24+Vβ11+ NKT cells for expression of a panel of chemokine receptors. CCR1, a receptor expressed by few T cells, is expressed by about 60% of NKT cells (Figure 1B,C). Most NKT cells also express CCR2, CCR5, CCR6, CXCR3, and CXCR6. These chemokine receptors are expressed by T cells with homing potential to nonlymphoid tissues and are highly associated with inflammation.25Almost all express CXCR4, a receptor also widely expressed by T, B, and monocytes. Few NKT cells express CCR3, CCR9, and CXCR5, which are mainly expressed by eosinophils (CCR3),21 thymocytes and gut homing lymphocytes (CCR9),22 or B cells and a subset of follicle homing CD4 T cells (CXCR5).23,26 CCR7, a chemokine receptor important for T-cell homing into lymphoid tissues,27-29 is expressed by only 10% to 28% (n = 6) of NKT cells. CCR4, a receptor implicated in lymphocyte homing to skin30 and expressed on most IL-4–producing CD4 T cells,31 is expressed by about 11% of NKT cells. Although the chemokine receptor expression pattern of NKT cells is somewhat similar to that of CD45RA−CD27+ CD8 T cells, a larger percentage of NKT cells express CCR1, CCR2, CCR6, and CXCR6 compared with this CD8 T-cell subset (Figure 1B,C). Despite the distinct expression pattern of chemokine receptors by NKT cells, it is notable that there are considerable variations among donors in the fraction of NKT cells expressing each chemokine receptor (Figure1B).
Chemokine receptor expression by NKT cell subsets
Based upon expression of CD4 and/or CD8, 3 subsets of NKT cells, CD4+CD8− (CD4), CD4−CD8+ (CD8), and CD4−CD8− (DN), can be identified. The relative frequency of each subset differs considerably from person to person (Figure 2C). On average, 49% of Vα24+Vβ11+ cells are DN, 27% are CD4, and 24% are CD8 NKT cells (n = 12). The wide variation in frequency of NKT cells expressing each chemokine receptor (Figure 1B) may be related to this variation of NKT cell subset composition and prompted us to individually examine chemokine receptor expression by each NKT cell subset. CCR1 is expressed by few cells of the CD4 subset but by most CD8 and DN cells (Figure 2A,B). Slightly, but consistently, more DN (than CD8) cells express CCR1. In a similar manner, CCR6 and CXCR6 are also preferentially expressed by DN and CD8 subsets. CCR2, CCR5, and CXCR3 are expressed by most of each NKT subset with slight differences (DN ≥ CD8 ≥ CD4). In contrast, CCR4 is mainly expressed by the CD4 subset but by few cells in the CD8 or DN subset. There is no notable difference in expression of CCR7 among the NKT cell subsets. Thus, some receptors, CCR4 (CD4) and CCR1, CCR6 and CXCR6 (DN and CD8), are characteristically and differentially expressed by NKT cell subsets.
It has been reported that human CD4+ NKT cells contain many more Th0-like cells producing IL-4 and IFN-γ than CD4− NKT cells.11 Here we examined the differences in cytokine production capacity among the CD4, CD8, and DN NKT subsets that display different chemokine receptor expression patterns. As previously reported, CD4 NKT cells were far better than other subsets in producing IL-4. CD8 NKT cells are slightly but consistently better than DN cells in producing IL-4 (Figure 2D). Notably, the 3 NKT cell subsets are very different also in production of IL-2. No significant difference was observed among the NKT cell subsets in producing IFN-γ and TNF-α. Thus, the CD4 NKT cell subset contains most of the IL-2– and IL-4–producing NKT cells in circulation, while CD8 and DN NKT cells contain relatively few such cells.
Chemokine responses of NKT cell subsets
We examined the chemotactic response of NKT cell subsets to chemokines. MIP-1α (a CCR1 ligand) induced good chemotaxis of CD8 and DN NKT cells but poor chemotaxis of CD4 cells (Figure3A). LARC (a CCR6 ligand) induced robust chemotaxis of CD8 and DN NKT cells but low chemotaxis of CD4 cells. In contrast, TARC (a CCR4 ligand) induced significant CD4 NKT cell migration but did not attract DN and CD8 NKT cells well. ELC (a CCR7 ligand) induced migration of all 3 subsets at similar levels. SDF-1 (the CXCR4 ligand), MCP-1 (a CCR2 ligand), and IP-10 (a CXCR3 ligand) attracted all 3 NKT cell subsets very well (Figure 3A), while BLC (the CXCR5 ligand), thymus-expressed chemokine (TECK) (the CCR9 ligand), and I-309 (a CCR8 ligand) did not show any notable chemotactic activity for NKT cells (not shown).
Differential chemokine responses among NKT cell subsets.
(A) Chemotactic responses of CD4, CD8, and DN NKT cell subsets. Cell migration is shown as net migration after subtraction of background migration. (B) Frequency of IL-2–, IL-4–, and IFN-γ–producing NKT cells in chemotaxed cells after migration to TARC or LARC. Chemokines were used at the indicated concentration (A) or 250 ng/mL (TARC) and 1500 ng/mL (LARC) for panel B. Chemotaxed NKT cells were phenotyped by surface and/or intracellular cytokine staining followed by counting. Relative frequencies of cytokine producers were calculated by dividing numbers of cytokine-producing NKT cells after chemotaxis by those NKT cells in input (input = 1, marked by horizontal lines [B]). *Significant differences (P < .05) between CD4 and CD8 or DN NKT subsets (n = 4, [A]) or the 2 groups (n = 6 [B]).
Differential chemokine responses among NKT cell subsets.
(A) Chemotactic responses of CD4, CD8, and DN NKT cell subsets. Cell migration is shown as net migration after subtraction of background migration. (B) Frequency of IL-2–, IL-4–, and IFN-γ–producing NKT cells in chemotaxed cells after migration to TARC or LARC. Chemokines were used at the indicated concentration (A) or 250 ng/mL (TARC) and 1500 ng/mL (LARC) for panel B. Chemotaxed NKT cells were phenotyped by surface and/or intracellular cytokine staining followed by counting. Relative frequencies of cytokine producers were calculated by dividing numbers of cytokine-producing NKT cells after chemotaxis by those NKT cells in input (input = 1, marked by horizontal lines [B]). *Significant differences (P < .05) between CD4 and CD8 or DN NKT subsets (n = 4, [A]) or the 2 groups (n = 6 [B]).
To further examine whether chemokines can differentially enrich cytokine producers versus nonproducers after chemotaxis, the frequencies of IL-2–, IL-4–, and IFN-γ–producing NKT cells were compared to the input population after chemotaxis to TARC and LARC (Figure 3B). IL-2 and IL-4 producers were enriched in TARC-attracted, but not LARC-attracted, NKT cells. However, no considerable enrichment or depletion was observed for IFN-γ producers after chemotaxis to these chemokines. Thus, NKT cell subsets with distinct cytokine-producing capacities display differential responses to chemokines, and their chemokine response correlates well with the chemokine receptor expression pattern.
Expression of other homing receptors by NKT cell subsets
Along with chemokine receptors, adhesion molecules such as CD62L, CLA, and α4β7 are important for tissue-specific homing of leukocytes. CD62L is implicated in homing to peripheral lymph nodes, and it has been reported that human NKT cells are CD62Llow/−.9,10 To migrate into T-cell areas of lymph nodes, cells are thought to also coexpress CCR7.27-29 Thus, we examined the existence of CD62L+CCR7+ NKT cells in circulation (Table1). A small but distinct CD62L+CCR7+ subset is found within the NKT cell population. Most of these CD62L+CCR7+ NKT cells are CD4+, a few are DN, and virtually none are CD8+. Many NKT cells (60%-90%, n = 4) express a gut homing receptor, α4β7 (Table 1); this frequency is higher than that among conventional CD4 memory T cells (20%-30% are α4β7+). Expression of CLA, a skin homing receptor, is also detected on a subset (on average 7%-13%, n = 4) of NKT cells. Whereas CLA and α4β7 are largely mutually exclusive among conventional CD4 cells, many CLA+ NKT cells coexpress α4β7 (not shown).
Frequencies (%) of L selectin+CCR7+, CLA+, and α4β7+ cells in each NKT cell subset in circulation
| NKT cell subsets . | L selectin+CCR7+ . | CLA+ . | α4β7+ . |
|---|---|---|---|
| CD4 | 3.9 ± 2.2 | 7 ± 4.4 | 63.1 ± 9.7 |
| CD8 | 0.2 ± 0.3 | 5.6 ± 4.8 | 75.9 ± 13.2 |
| DN | 1.3 ± 0.7 | 13.4 ± 4.2 | 74.1 ± 16.1 |
| NKT cell subsets . | L selectin+CCR7+ . | CLA+ . | α4β7+ . |
|---|---|---|---|
| CD4 | 3.9 ± 2.2 | 7 ± 4.4 | 63.1 ± 9.7 |
| CD8 | 0.2 ± 0.3 | 5.6 ± 4.8 | 75.9 ± 13.2 |
| DN | 1.3 ± 0.7 | 13.4 ± 4.2 | 74.1 ± 16.1 |
Averages and SDs from 4 different donors are shown.
Discussion
Here we examined expression of chemokine receptors and homing receptors by a unique T-cell subset, Vα24+Vβ11+ “NKT” cells. The definition of NKT cells is controversial, especially in mice, because not all CD1d-restricted cells express NK1.1 and some NK1.1+ T cells are not CD1d-restricted. Surrogate markers such as CD56 and CD161 do not specifically identify human NKT cells either, because few NKT cells express CD56, and non-NKT T cells can express CD56 and CD161 (Figure1A). In human beings, identification with antibodies to TCR Vα24 and Vβ114,11 or binding with α-galactosylceramide–loaded CD1d tetramers32 33 appears to be the most specific way to identify NKT cells. Our results reveal existence not only of unique chemokine receptor expression patterns by NKT cells but also differential expression of trafficking machinery among NKT cell subsets.
Our finding of differential chemokine receptor expression patterns correlates with the wide distribution of NKT cells in tissues. Most NKT cells express CCR1, CCR2, CCR5, CXCR3, CXCR4, and CXCR6. Among these, CCR1, CCR2, CCR5, CXCR3, and CXCR6 are highly associated with “tissue” T cells found in extralymphoid tissues.24,25,34 Their chemokine ligands (eg, MIP-1α, MCP-1, MIP-1β, IP-10) are widely expressed in extralymphoid tissues and up-regulated by inflammatory signals. Furthermore, most NKT cells also express α4β7, a homing receptor to gut-associated lymphoid systems. In contrast, CCR7, a receptor required for homing into lymphoid tissues and expressed by all naive T cells and many (about 70%) memory T cells, is expressed by only about 20% of NKT cells; the follicular homing receptor, CXCR5, is expressed by few NKT cells. Thus, the chemokine receptor expression pattern suggests that, unlike naive and memory T cells, most NKT cells have the machinery to localize in extralymphoid tissues, and only a small subset of NKT cells have a homing potential to secondary lymphoid tissues. NKT cells lack the expression of CXCR5, the chemokine receptor needed to localize in the B-cell area of lymphoid tissues,35 and thus are likely to be excluded from the B-cell areas including germinal centers.
Whether blood NKT cells recirculate through peripheral lymph nodes remains unknown. In this regard, it is not clear whether the small number of NKT cells found in lymph nodes are generated in lymph nodes or migrate into the nodes via the afferent lymphs or the blood. Our results show that a small population of NKT cells in circulation expresses both CCR7 and L selectin and resides mainly within the CD4 NKT subset. L selectin and CCR7 mediate adhesive interactions of leukocytes and high endothelial venules for transmigration into lymphoid tissues, and CCR7 is required for T cells to localize in T-cell areas of lymphoid tissues in response to CCR7 ligands SLC and ELC. These chemokines are expressed by high endothelial venules (SLC, ELC) and mature DCs in T-cell areas (ELC). Therefore, the small population of NKT cells expressing CCR7 and L selectin can potentially migrate into sites of T-cell differentiation in secondary lymphoid organs.
Interestingly, NKT cell subsets differ substantially in their expression of some chemokine receptors even though NKT cell subsets share expression of many chemokine receptors as discussed above. Most notably, CD4 NKT cells (high IL-2 and IL-4 producers) express CCR4 and are highly responsive to TARC, while CD8 and DN NKT cell subsets (low IL-2 and IL-4 producers) express CCR1, CCR6, and CXCR6 and respond very well to their ligands such as MIP-1α and LARC. Thus, our results show polarization in chemokine receptor expression and chemokine response among NKT cell subsets with distinct effector functions. It has been reported that chemokine receptors are associated with the effector cytokine production capacity of CD4 or CD8 T cells.36-38CCR4 is expressed by almost all IL4-producing CD4 cells, while CXCR3 is expressed by almost all IFN-γ–producing cells.31 T cells expressing CCR2, CCR5, and CXCR6 also contain many more IFN-γ producers than total memory T cells.31 Our results suggest that NKT cells also show such differential expression pattern of chemokine receptors. In the case of NKT cells, however, the association is somewhat skewed by the unusually uniform expression of tissue homing chemokine receptors: Most NKT cells express the Th1-associated chemokine receptors (CCR2, CCR5, and CXCR3) in a less discriminatory way, while they express CCR1, CCR4, CCR6, and CXCR6 more differentially.
Tissue expression of chemokines provides insights into how NKT cells establish tissue tropisms. For example, the chemokine ligand of CCR6, a receptor expressed by the most NKT cells, is LARC, a chemokine mainly expressed in liver among various tissues.39 This is in good correlation with the enrichment of NKT cells in liver. Cytokines and inflammatory signals profoundly change the expression pattern of cytokines. Expression of LARC and β-defensin (a nonchemokine CCR6 ligand) in epithelial cells or keratinocytes is greatly enhanced by bacterial endotoxin, IL-1, and TNF-α.40,41 CCR4 ligands, TARC and MDC, are constitutively detected in thymus at high levels and in small intestine and lung at lower levels.34,42Importantly, expression of TARC and MDC is induced by Th2 cytokines IL-4 and/or IL-13 in various cell types43,44 and in inflamed skin venules,30 amplifying the recruitment of CCR4+ T cells to sites of Th2 inflammation. The CXCR6 ligand, CXCL16, is expressed in liver and lung at high levels and in spleen and lymph nodes at low levels.45 Thus, depending on the expression of 2 types of chemokines (TARC vs LARC, MIP-1α, and CXCL16) under normal and inflammatory conditions, recruitment of each NKT cell subset may be specifically controlled. Some differences in tissue localization have been observed at least in mice: There are more DN and CD8 than CD4 NKT cells in lungs, while the CD4 subset is the major NKT cell population in thymus.13 Although chemokine receptor expression and NKT cell localization in tissues could differ in human beings and mouse, our results suggest a potential mechanism to fine-control migration (or retention) of different NKT cell subsets.
In conclusion, our results show that NKT cells almost universally express inflammatory lymphocyte chemokine receptors to levels that are much higher than any other memory or effector T-cell subsets examined and yet display distinct and subset-specific expression profiles of other chemokine receptors, including CCR4 (by CD4), CXCR6, CCR1, and CCR6 (by CD8 and DN). Furthermore, expression of human immunodeficiency virus coreceptors (CCR2, CCR5, CXCR4, and CXCR6) by many CD4 NKT cells suggests that they are potential targets of human immunodeficiency virus infection. The precise NKT cell subset-specific localization in various human tissues in normal and inflammatory conditions remains unknown. Therefore, this report on expression of chemokine and homing receptors would be valuable in helping explain and predict the localization of the circulating human NKT cell subsets.
We thank Dr Lijun Wu (Millennium Pharmaceuticals) for reagents (anti-CCR4 and CCR7).
Prepublished online as Blood First Edition Paper April 17, 2002; DOI 10.1182/blood-2001- 12-0196.
Supported by a special fellowship from the Leukemia and Lymphoma Society (C.H.K.); grants AI47822, GM37734, and AI37832 from the National Institutes of Health; an award from the Department of Veterans Affairs (E.C.B.); and the FACS Core facility of the Stanford Digestive Disease Center.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Chang H. Kim, 3801 Miranda Ave, Mail code 154-B, VAMC Building 101, Room C4-111, Palo Alto, CA 94304; e-mail:chkim@stanford.edu.

![Fig. 2. Distinct expression pattern of chemokine receptors among NKT cell subsets. / (A) Chemokine receptor expression by CD4, CD8, and DN NKT subsets (n = 4). (B) Dot plots of chemokine receptor expression by NKT cell subsets with percentages positive. (C) Relative frequencies of CD4, CD8, and DN NKT cell subsets in the Vα24+Vβ11+ NKT cell population (n = 11). (D) Differential cytokine production capacities of NKT cell subsets (n = 5). Isolated Vβ11+ NKT cells were stained with antibodies to Vα24, CD4, and CD8 and activated with phorbol myristate acetate and ionomycin in the presence of monensin to block internalization of surface antigens followed by intracellular cytokine analyses. Bars indicate averages of results from 4 (A), 11 (C), or 5 (D) donors. *Significant differences from the CD4 NKT subset (P < .05 [A,D]). **Significant difference from CD4 or CD8 subset (P < .02 [C]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/1/10.1182_blood-2001-12-0196/7/m_h81322776002.jpeg?Expires=1767780004&Signature=Sp4JJ6CPFwsmcNMN7qUDm6oO-KYfDfy9FOiFFvuaQaJe2d-BbZ6b91qjQhhWumUi9~HiB2co-yKZXaF4FjWneGnRg8cezdO1VY0lg15OrGPIG5oLjmC~dQ1TrZJ6bOh9~m5OVKryFDKENE9WweTBEwREI0zDhmtL4gxCnt6jvnJ~0dUHNSEvjXxndMfl~PMHNzEfucWWlXCjPMEC1MypwGDPVH7NgTxdNG8H3nYL3XchnRTqTWF05h6BDjtwdq-fx0bI2pCkiB-Tv5aq7QDrx0YKUDoR9BiqSWsjKm1rLEbudgCxTb~aT9E~dmZiAKpAwD6nXMjSe7lZqjiHgPSp2g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Differential chemokine responses among NKT cell subsets. / (A) Chemotactic responses of CD4, CD8, and DN NKT cell subsets. Cell migration is shown as net migration after subtraction of background migration. (B) Frequency of IL-2–, IL-4–, and IFN-γ–producing NKT cells in chemotaxed cells after migration to TARC or LARC. Chemokines were used at the indicated concentration (A) or 250 ng/mL (TARC) and 1500 ng/mL (LARC) for panel B. Chemotaxed NKT cells were phenotyped by surface and/or intracellular cytokine staining followed by counting. Relative frequencies of cytokine producers were calculated by dividing numbers of cytokine-producing NKT cells after chemotaxis by those NKT cells in input (input = 1, marked by horizontal lines [B]). *Significant differences (P < .05) between CD4 and CD8 or DN NKT subsets (n = 4, [A]) or the 2 groups (n = 6 [B]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/1/10.1182_blood-2001-12-0196/7/m_h81322776003.jpeg?Expires=1767780004&Signature=U19MLDwmYrqdjrlVEH2ob2fAmFA4m~dvPHKert2HdDe5tMaSyLrK0jQIowvvuxb1CnO59niguTTODyPshFf5Tci9ttH4DW9f2rZOelQ0hjhnkgW8ZMnvwRKb8qb7SMacZqUT24DDflnoB~aTs0aqePzirfXOZcRFB8g5NS9mxSYbDj8lc0z~kjzfTFJ78S5a-N6tU0L~i6O7DIU49jNUYdUyisO6x0V-w4tTwqRnsALYGm19AXuqn8YVTSN~wGb8vFmdGHAgmRxk7rckWT32MHm0OnGJX8rypgIcdk4l3wpdU4zpZMUo0wrbrci9OZP1v8rt66kHBvkcFrBrZbLjPw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal