Abstract
The regulation of stem cell proliferation is a poorly understood process balancing rapid, massive blood cell production in times of stress with maintenance of a multipotent stem cell pool over decades of life. Transforming growth factor β1 (TGF-β1) has pleiotropic effects on hematopoietic cells, including the inhibition of primitive cell proliferation. It was recently demonstrated that the cyclin-dependent kinase inhibitors, p21Cip1/Waf1 (p21) and p27Kip1 (p27), can inhibit the proliferation of hematopoietic stem cells and progenitor cells, respectively. The relation of TGF-β1 stimulation to p21 and p27 was examined using a fine-mapping approach to gene expression in individual cells. Abundant TGF-β1 expression and p21 expression were documented in quiescent, cytokine-resistant hematopoietic stem cells and in terminally differentiated mature blood cells, but not in proliferating progenitor cell populations. TGF-β1 receptor (TβR II) was expressed ubiquitously without apparent modulation. Cell- cycle–synchronized 32D cells exposed to TGF-β1 demonstrated a marked antiproliferative effect of TGF-β1, yet neither the level of p21 mRNA nor the protein level of either p21 or p27 was altered. To corroborate these observations in primary cells, bone marrow mononuclear cells derived from mice engineered to be deficient in p21 or p27 were assessed. Progenitor and primitive cell function was inhibited by TGF-β1 equivalently in −/− and +/+ littermate controls. These data indicate that TGF-β1 exerts its inhibition on cell cycling independent of p21 and p27 in hematopoietic cells. TGF-β1 and p21 or p27 participate in independent pathways of stem cell regulation, suggesting that targeting each may provide complementary strategies for enhancing stem or progenitor cell expansion and gene transduction.
Introduction
Modulation between brisk expansion and differentiation or quiescence and self-renewal defines essential processes of stem cells. Understanding the mechanisms of stem cell cycle regulation may guide methods for altering their cell-cycle kinetics for purposes of stem cell expansion and genetic manipulation. A subset of low-molecular–weight molecules that interact with cyclin-dependent kinases (CDKs) to impair progression through G1 phase of the cell cycle has been defined. These cyclin-dependent kinase inhibitors (CDKIs) can be divided into 2 families: the Cip/Kip family including p21Cip1/Waf1 (p21), p27Kip1 (p27), and p57Kip2 (p57), which may interact with many CDKs, and the INK4 family, p16INK4A (p16), p15INK4B(p15), p18INK4C (p18), and p19INK4D (p19), which specifically inhibit Cdk4 and Cdk6.1 Both have been shown to have essential roles in arresting cell-cycle progression in a number of model systems.2 3
A component of the quaternary complex, p21 is composed of cyclin D, CDK4, and proliferating cell nuclear antigen.4 It is an effector for p53-mediated cell-cycle arrest5 and is transcriptionally regulated. We have demonstrated that restriction into entry of the cell cycle in stem cells is governed by p21 in whose absence the stem cell pool is larger, more actively cycling, and more sensitive to exhaustion. Dominant inhibitory control over stem cell cycling exerted by p21 dictates the size and kinetics of the stem cell compartment, and preserving the stem cell population is dependent on restricting access to the cell cycle.6
P27 is molecularly distinct from p21 in its carboxyl terminus; it interacts with similar, though not identical, cyclin-CDK and lacks p53 regulated expression.1,2,7 In addition, unlike p21, p27 is controlled by translational and posttranslational mechanisms.8,9 A role for p27 in hematopoiesis is supported by direct flow cytometric evidence for expression in primitive cells10 and expression in more mature progenitors,11,12 and indirectly it is supported by improved retroviral transduction in the context of antisense p27.13 We used mice engineered to be deficient in p27 to define that p27 does not affect stem cell number, cell cycling, or self-renewal but that it markedly alters progenitor proliferation and pool size.14 Therefore, distinct CDKIs govern the highly divergent stem and progenitor cell populations.
Transforming growth factor-β1 (TGF-β1) has varied effects on hematopoietic cells, enhancing the proliferation of granulocytes in response to granulocyte-macrophage colony-stimulating factor15 or inhibiting the responsiveness of progenitors to other growth-promoting cytokines.16 It has been extensively characterized as a dominant-negative regulator of the proliferation of hematopoietic cells, including the inhibition of primitive progenitor cells.17-20 TGF-β1–induced cell-cycle arrest has been shown to be mediated through p21 or p27 in multiple cell lines or cell types, among them human epithelial cell lines,21,22 fibroblast cells,23 colon cancer cell lines,24 and ovary cancer cell lines.25Recently, it has been proposed that p21 and p27 are key downstream mediators for TGF-β in hematopoietic cells,20 26 yet there is little direct evidence for this in primitive hematopoietic populations. Therefore, we sought to determine whether p21 or p27 serves as a proximate mediator for TGF-β1–induced cell cycle exit in these cell types. If p21, p27, and TGF-β1 represent independent pathways of regulating cell quiescence, targeting each may represent complementary methods of enhancing primitive hematopoietic cell cycling.
Materials and methods
Human bone marrow sampling and cell sorting
Bone marrow aspirates and peripheral blood were collected from healthy adult donors after informed consent was obtained according to the guidelines established by the Human Investigation Committee of the Massachusetts General Hospital. Low-density cells obtained by Ficoll-Hypaque (Pharmacia, Piscataway, NJ) density centrifugation were further fractionated into different subsets (CD34+CD33− and CD34+CD33+ cells from bone marrow and CD11b+ and CD19+ from peripheral blood) by magnetic bead immunoselection (Miltenyi Biotec, Auburn, CA, and Dynal, Oslo, Norway) and cell sorting (FACS Vantage; Becton Dickinson, San Jose, CA). G0 or “cytokine-unresponsive” stem cells27 were derived from CD34+ cells cultured in Iscoves modified Dulbecco medium supplemented with 200 μg/mL 5-fluorouracil (Pharmacia, Kalamazo, MI), 100 ng/mL kit ligand (stem cell factor [SCF]; R&D Systems, Minneapolis, MN), 100 ng/mL interleukin-3 (IL-3) (R&D Systems), and 10% fetal calf serum (Sigma Chemical, St Louis, MO) for 7 days and subsequently were sorted by flow cytometry (Becton Dickinson) based on absence of staining for Annexin V (Caltac Laboratories, San Francisco, CA) and propidium iodide (Sigma) or 7-amino actinomycin D (Calbiochem, La Jolla, CA) staining.
Murine bone marrow sampling and colony assay
p21 +/− and p27 +/− mice (kindly provided by Drs Tyler Jacks [MIT, Boston, MA] and Adrew Koff [Sloan Kettering Cancer Center, New York, NY] respectively) were maintained and bred at the MGH animal facility. All the homozygous and wild-type mice were generated from heterozygotes. Mice were determined to be +/+, +/− or −/− based on PCR analysis of tail clip–derived DNA. To generate the single-cell suspensions, 8- to 10-week-old mice were killed, and femora and tibiae were flushed. Bone marrow cells were cultured in 0.8% methylcellulose, 30% fetal bovine serum, 1% bovine serum albumin (BSA), 0.1 mM 2-mercaptoethanol, 2.0 mM L-glutamine of α-MEM semisolid matrix culture medium (Stem Cell Technologies, Vancouver, BC, Canada) with cytokine combinations of 50 ng/mL mu-SCF (R&D System), 20 ng/mL m-IL-3 (R&D Systems), 20 ng/mL hu-IL-6 (R&D Systems), and 2 U/mL hu-erythropoietin (Amgen, Thousand Oaks, CA). Cells were plated at 20 000/200 μL/well into 48-well plates and were placed at 37°C, 5% CO2. At day 10, myeloid and erythroid colonies were scored, totaled, and reported as colony-forming cells (CFCs) representing committed progenitor cells.
32D cell line culture and synchronization
The 32D cell line28 was maintained in 10% fetal calf serum supplemented with 10% WEHI-conditioned medium (used as the source for m-IL-3), and the medium was replaced twice weekly. WEHI-conditioned medium was withdrawn from the culture for 12 to 16 hours, and 70% to 80% cells were synchronized at G0/G1 phase as documented by propidium iodide analysis.29
3H-thymidine incorporation and cell-cycle analysis
One microcurie 3H-thymidine (DuPont-New England Nuclear, Boston, MA) was added to 20 000 cells (200 μL) in 96-well plates, and cell mixture was incubated for 8 hours at 37°C. Thymidine-incorporated cells were harvested, filtered onto 96-well microfilter plates (Packard, Meriden, CT), and evaluated by liquid scintillation beta counter (Packard). Cell-cycle analysis was performed by standard propidium iodide staining and flow cytometry (Becton Dickinson) assessment of DNA content.
Single-cell reverse transcription–polymerase chain reaction
Single-cell reverse transcription–polymerase chain reaction (RT-PCR) was performed according to a minor modification of the method of Brady et al.30 31 Briefly, cells were lysed in 5.0 μL buffer (100 mM Tris HCl, pH 8.3, 150 mM KCl, 6 mM MgCl2, 4 μM each dNTP [Pharmacia], 100 ng/mL oligo (dT)24, 3360 U/mL RNA guard [Pharmacia], 100 U/mL Prime inhibitor [5 Prime-3 Prime, Boulder, CO], 20 mM dithiothreitol [DTT], and 0.5% NP-40). Samples were heated to 65°C for 1 minute, cooled to 22°C for 3 minutes, and put on ice. One hundred units of murine Moloney reverse transcriptase (Gibco-BRL) and 2 U Avian reverse transcriptase (Boehringer Mannheim) were added, and volume was made up to 10 μL with diethyl pyrocarbonate–treated H2O. Samples were incubated at 37°C for 20 minutes before heat inactivation at 65°C for 10 minutes. Resultant cDNA was then subjected to polyadenylate tailing in 20 μL total volume (50 mM Tris HCl, pH 8.3, 0.2 M potassium cacodylate, 5 mM CoCl2, 5 mM DTT, 250 μ/mL BSA, 0.2 mM dATP, and 10 U terminal transferase [Boehringer Mannheim]) at 37°C for 15 minutes, then heated at 65°C for 10 minutes and placed on ice. PCR was performed in a final volume of 50 μL (30 mM Tris HCl, pH 8.3, 65 mM KCl, 3.1 mM MgCl2, 1 mM each dNTP, 100 μ/mL BSA, 0.05% Triton X-100, 10 μM (dT) 24-primer, and 5 U AmpliTaq polymerase [Perkin-Elmer]). Cell-free and reverse transcriptase–free samples were used as negative controls. Samples were initially amplified for 25 cycles of 30 seconds at 94°C, 1 minute at 42°C, and 3 minutes (plus 5-second extension cycle) at 72°C in a thermal cycler (9600 model; Perkin-Elmer Cetus). An additional 5 U Taq polymerase was then added, and another 25 cycles were performed.
Southern blotting
The PCR product was electrophoresed through 1.2% agarose, stained with ethidium bromide, photographed, transferred to a nylon membrane (Micron Separations) with vacuum blotter (Pharmacia), immobilized by UV (Stratagene), and hybridized to the indicated32P-radiolabeled probes (1 × 107 cpm/mL) with the use of 40 bp 3′-oligodeoxyribonucleotides derived from the following human cDNA sequences: p21Cip1/Waf1, p27Kip1, TGF-β1, and TGF-β1 receptor (type II) (hereafter referred to TβR II) in the ExpressHybridization Solution (Clontech). The hybridization temperature for the oligo probes was 37°C, and final wash conditions were 0.1 × SSC and 0.1% sodium dodecyl sulfate (SDS) at 37°C. Hybridized blots were analyzed by PhosphorImager (Molecular Dynamics) quantitation and autoradiography.
RNA isolation and Northern blotting
Total RNA was isolated from the cells using the guanidine isothiocyanate procedure coupled with ultra-centrifugation through the CsCl2 cushion. Equal amounts (10 μg) total RNA were analyzed on formaldehyde gels and blotted onto nylon membrane (Micron Separations) using a commercial UV cross-linker (Invitrogen). The cDNA probes were labeled by 32P using the random primer extension system (DuPont-New England Nuclear) with 68°C hybridization temperature for the cDNA probes in the Express Hybridization Solution (Clontech). Final wash conditions were 0.1 × SSC and 0.1% SDS at 55°C. Hybridized blots were analyzed by PhosphorImager (Molecular Dynamics) quantitation and autoradiography. Mouse p21 and p27 cDNA plasmids were kindly provided by Dr David Beach (Cold Spring Harbor Laboratory).
Western blotting
32D cells were lysed in lysis buffer (250 mM NaCl, 50 mM HEPES, pH 7.0, 0.1% NP-40, 50 mM NaF, 5 mM EDTA, pH 8.0, 100 mM sodium orthovanadate, 1 mg/mL leupeptin, 1 mg/mL aprotinin, 1 M DTT, and 50 mM phenylmethylsulfonyl fluoride), and equal amounts (30 μg) protein lysates were fractionated on a 12% SDS-polyacrylamide gel. Fractionated proteins were electrotransferred onto the nitrocellulose membrane (Millipore). The blocked protein blot was incubated with the rabbit anti–mouse p21 or p27 (Pharmingen) at the 1 μg/mL final concentration in Tris-buffered saline (TBS/T) (1 M Tris, pH 8.0, 5 M NaCl, 0.05% Tween 20) with 0.5% milk for more than 1 hour, rinsed with TBS/T buffer, and incubated with goat anti–rabbit IgG conjugated with horseradish peroxidase in the TBS/T. After washing, enhanced chemiluminescence (DuPont-New England Nuclear) was performed according to instructions from the manufacturer.
Results
The hypothesis that TGF-β1 transduces its effect on cell cycle through the modulation of p21 or p27 was initially evaluated in primary human hematopoietic cells. We previously reported that the genetic program expressed by cells undergoing specific lines of differentiation can be finely mapped during the differentiation process by semiquantitative RT-PCR in individual cells isolated by micromanipulation from clonal populations of maturing progenitor cells.30 This method has a dynamic range of 250 to 250 000 mRNA copies/cell and linearity of input quantity to output signal, with r = 0.862. It is capable of defining gene expression more precisely than can be accomplished with bulk populations of cells, and it can assess transcription levels in cell types in which obtaining large numbers of cells is not feasible. This method was applied to primary stem cells and differentiating cells in the experiments below.
P21 up-regulation during the terminal differentiation of primary hematopoietic cells
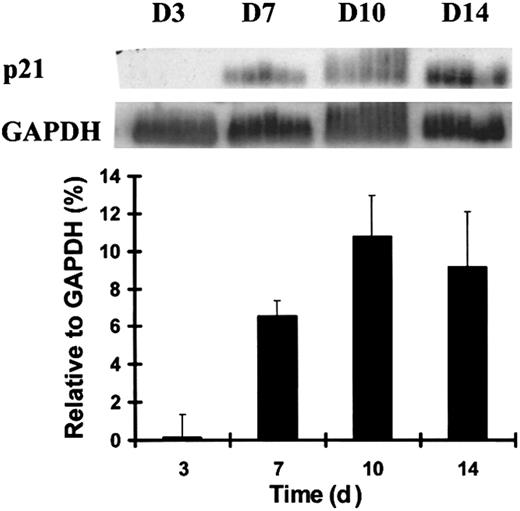
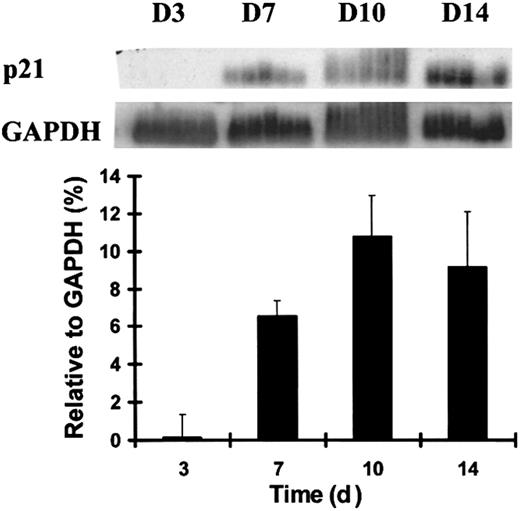
P21 is primarily regulated at the transcription level,3,32-35 in contrast to other CDKIs, which are largely controlled by translational and posttranslational mechanisms.2,8,9 We evaluated the temporal expression pattern of p21 in the terminal differentiation of human primary hematopoiesis using RT-PCR analysis during the differentiation of the myeloid progenitor cell, macrophage–monocyte colony-forming unit (CFU-M). CD34+CD38− cells were plated at limiting dilution in the medium for CFU-M development, and single daughter cells from the same original progenitor were removed by micromanipulation for subsequent analysis. During CFU-M development, p21was almost undetectable at day 3 and was up-regulated after day 7. When normalized for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, the up-regulation of p21 mRNA level was highly time dependent (Figure 1). P21 expression achieved maximal levels by day 10, when the cells achieved fully mature morphologic features and the characteristic gene expression of a terminally differentiated monocyte previously defined.30
Up-regulation of p21 mRNA during the terminal differentiation of CFU-M.
Individual human adult bone marrow cells were sorted by flow cytometry and cultured under the stated conditions. Five single cells isolated by micromanipulation and harvested at each time point from the emerging CFU-M colonies were analyzed using RT-PCR and Southern blotting. Quantitative analysis of p21 normalized to GAPDH signals presented in the graph was performed using PhosphorImager-generated quantitation of signal intensity when the poly-dT–primed RT-PCR product was probed with radiolabeled p21 or GAPDH. Data shown are from 1 of 3 similar experiments with similar results.
Up-regulation of p21 mRNA during the terminal differentiation of CFU-M.
Individual human adult bone marrow cells were sorted by flow cytometry and cultured under the stated conditions. Five single cells isolated by micromanipulation and harvested at each time point from the emerging CFU-M colonies were analyzed using RT-PCR and Southern blotting. Quantitative analysis of p21 normalized to GAPDH signals presented in the graph was performed using PhosphorImager-generated quantitation of signal intensity when the poly-dT–primed RT-PCR product was probed with radiolabeled p21 or GAPDH. Data shown are from 1 of 3 similar experiments with similar results.
Distinct expression profiles of p21 and p27 in hematopoietic cells
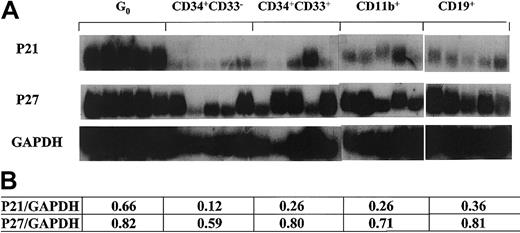
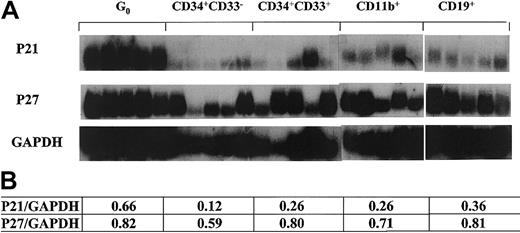
Noting the up-regulation of p21 in terminally differentiated cells, we next evaluated stem cells at the opposite end of the differentiation schema that were also quiescent. We stimulated CD34+ cells with SCF–IL-3 while maintaining high concentrations of the S-phase–specific toxin 5-flurouracil and collected the remaining viable cells at day 7 by flow cytometry.27 Cells isolated using this technique have low staining for the mitochondrial staining dye pyronin Y and the DNA staining dye Hoechst 33342, consistent with the G0 phase of the cell cycle. We and others have shown that these cells have stem cell–like characteristics, as indicated by LTC-IC assays and hu-SCID repopulating experiments.27,36 37 Individual G0 cells were sorted as were other hematopoietic cells at different stages of development, characterized by immunophenotypic profiles including CD34+CD33− and CD34+CD33+ from bone marrow and CD11b+ and CD19+ from peripheral blood. Using semiquantitative single-cell RT-PCR, we found that G0 cells expressed high levels of p21 (Figure 2). In contrast, p27 was expressed relatively evenly in all the populations tested without a pattern of altered transcript levels (Figure 2). The expression patterns of the genes tested in G0 were more uniform than others, perhaps reflecting the relative homogeneity of G0 cells. In the CD34+CD33− subset that represents early myeloid progenitors with high proliferative status, there was a very low or undetectable level of p21. In other subsets of maturing (CD34+CD33+) and fully matured (CD11b+) cells of myeloid lineage, there were elevated levels of p21 (more than 2-fold by densitometry quantitation) compared to CD34+CD33−. Heterogeneity in expression patterns within the CD34+ CD33+ and CD11b+ populations likely reflects intrinsic variability in those populations. Data in these 3 subsets were consistent with the result of CFU-M as shown above (Figure 1) and support the association of p21 transcription levels, with the state of differentiation up-regulated in quiescent primitive cells or fully mature cells. Because p27 is mainly regulated at the posttranscriptional level, data on p27 mRNA level are less informative than for p21.
P21 and p27 expression in different subsets of single human hematopoietic cells.
Flow cytometry–sorted cells were individually selected by micromanipulator and analyzed using poly-dT–primed RT-PCR with radiolabeled gene-specific probes applied to the PCR product. Panels of 5 cells of each type were used to avoid individual cell artifacts or cell heterogeneity. The lower table states the ratios derived from quantitative analysis with densitometry software (NIHimager) of the tested genes normalized to GAPDH.
P21 and p27 expression in different subsets of single human hematopoietic cells.
Flow cytometry–sorted cells were individually selected by micromanipulator and analyzed using poly-dT–primed RT-PCR with radiolabeled gene-specific probes applied to the PCR product. Panels of 5 cells of each type were used to avoid individual cell artifacts or cell heterogeneity. The lower table states the ratios derived from quantitative analysis with densitometry software (NIHimager) of the tested genes normalized to GAPDH.
Expression of TGF-β1 and TβR II in hematopoietic cells
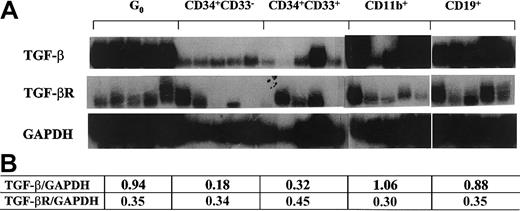
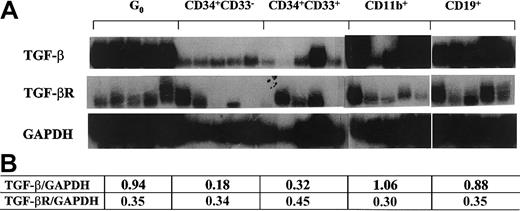
Several lines of evidence support that TGF-β1 participates in the quiescence of primitive hematopoietic cells.17,20,38,39 We evaluated the mRNA levels of TGF-β1 and its receptor in G0 stem cells and the subsets of differentiation stage-specific cells described above. TβR II was expressed throughout the cell types tested with a relatively stable expression pattern (Figure 3). In contrast, TGF-β1 was expressed at high levels at the stage of G0 stem cells and in fully mature cells. Therefore, TGF-β1 had an expression pattern similar to p21, consistent with coordinate regulation (Figure 3). This supported the possibility that p21 may be a downstream mediator of TGF-β1's effects on cell cycling, as has been documented in nonhematopoietic and transformed cell systems.23-25 40
TGF-β1 and TβR II expression in different subsets of single human hematopoietic cells.
Flow cytometry–sorted cells were individually isolated by micromanipulator and analyzed using poly-dT–primed RT-PCR probed with radiolabeled gene-specific cDNA. Panels of 5 cells of each type were used. Quantitative analysis of the tested genes normalized to GAPDH signals with densitometry software (NIHimager) are presented in the table.
TGF-β1 and TβR II expression in different subsets of single human hematopoietic cells.
Flow cytometry–sorted cells were individually isolated by micromanipulator and analyzed using poly-dT–primed RT-PCR probed with radiolabeled gene-specific cDNA. Panels of 5 cells of each type were used. Quantitative analysis of the tested genes normalized to GAPDH signals with densitometry software (NIHimager) are presented in the table.
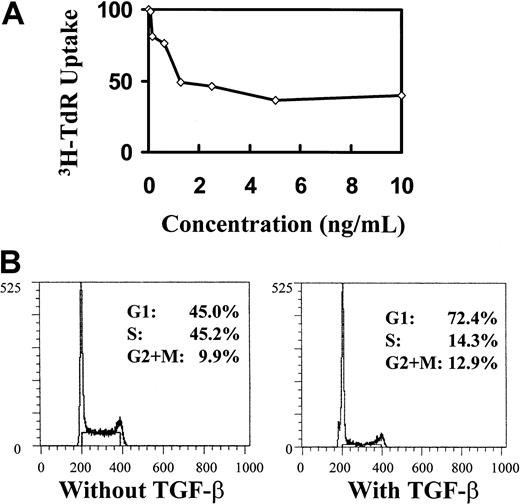
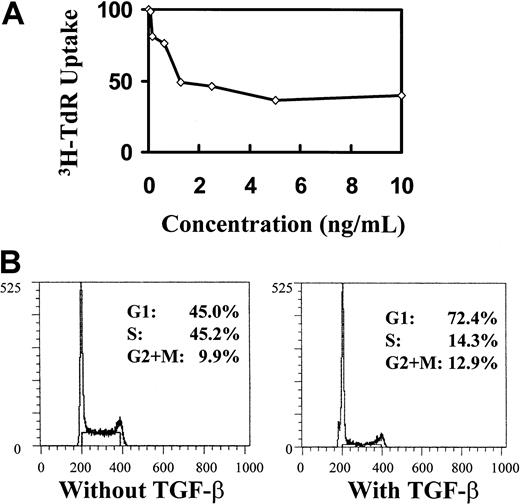
The induction of cell-cycle arrest by TGF-β1 is not associated with p21 or p27 up-regulation. To further pursue the link between p21 or p27 and TGF-β1, we asked whether p21 or p27 can be up-regulated by TGF-β1 stimulation in hematopoietic cells. Because of the limited availability of primary primitive hematopoietic cells for direct sequential measurement of RNA and protein levels, we chose the IL-3–dependent, nontransformed, multipotential cell line 32D28,29 41 as a model for studying the induction of p21 and p27 by TGF-β1. 32D cells were responsive to the inhibitory effect of TGF-β1, as shown by declining 3H-thymidine incorporation quantification of cell division and cell-cycle analysis (Figure 4). However, no elevation of p21 mRNA was detected by 72 hours, when the cells were treated with 20 ng/mL TGF-β1 (Figure 5). When the cells were synchronized in G0 by IL-3 starvation and then triggered into cell cycle with IL-3 in the presence or absence of TGF-β1, neither p21 mRNA (Figure 6) nor protein levels of p21 and p27 (Figure 7) were affected by TGF-β1 during progression through a single cell cycle. P21 levels increased as the cells entered the cell cycle (in 3 hours); this was followed by slightly elevated mRNA levels while the protein decreased during progression within the cell cycle during the course of a single cell cycle. However, TGF-β1 had no effect on these events (Figures 6, 7).
Inhibitory effect of TGF-β1 on cell proliferation and cell cycle.
Inhibitory effect shown by 3H-TdR incorporation (A) and propidium iodide staining (B) in asychronized 32D cells. 32D cells were cultured at different indicated concentrations of TGF-β1 for 48 hours, after which 3H-TdR uptake was performed. Cells used for cell-cycle analysis were treated with 20 ng/mL TGF-β1 and collected at 48 hours.
Inhibitory effect of TGF-β1 on cell proliferation and cell cycle.
Inhibitory effect shown by 3H-TdR incorporation (A) and propidium iodide staining (B) in asychronized 32D cells. 32D cells were cultured at different indicated concentrations of TGF-β1 for 48 hours, after which 3H-TdR uptake was performed. Cells used for cell-cycle analysis were treated with 20 ng/mL TGF-β1 and collected at 48 hours.
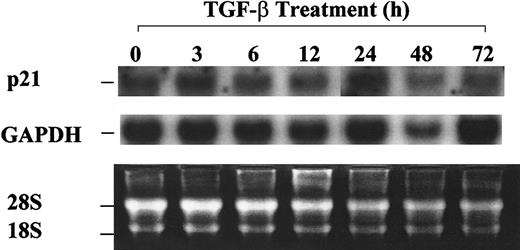
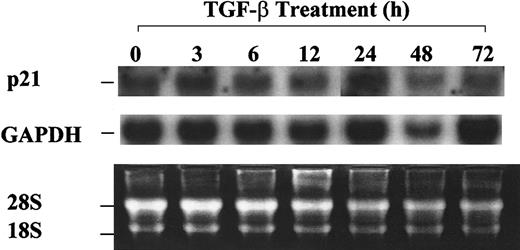
Absence of p21 up-regulation by TGF-β1 in asynchronized 32D cells.
32D cells were treated with 20 ng/mL TGF-β1 and were collected at the indicated time points. Total RNA was isolated, and Northern blot analysis was performed as described in “Materials and methods.”
Absence of p21 up-regulation by TGF-β1 in asynchronized 32D cells.
32D cells were treated with 20 ng/mL TGF-β1 and were collected at the indicated time points. Total RNA was isolated, and Northern blot analysis was performed as described in “Materials and methods.”
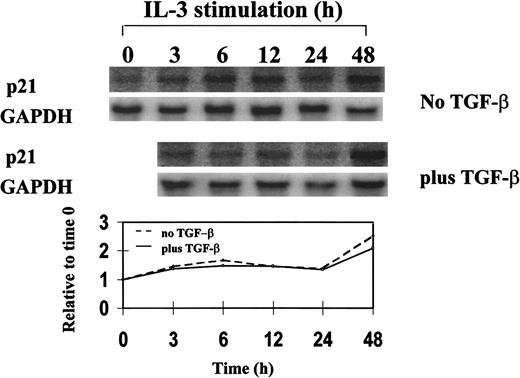
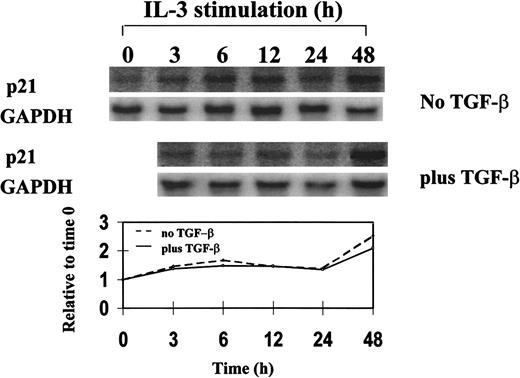
Northern blot analysis of p21 expression in synchronized 32D cells during a single cell-cycle progression in the presence or absence of TGF-β1.
32D cells were synchronized by starvation of IL-3 and restimulated with IL-3 plus or minus 20 ng/mL TGF-β1 and were analyzed for mRNA expression by Northern blotting. Single cell-cycle time was approximately 16 hours for 32D cells. The quantitative graph was made using NIHimager software comparing signal intensity at each time point with the time 0 baseline.
Northern blot analysis of p21 expression in synchronized 32D cells during a single cell-cycle progression in the presence or absence of TGF-β1.
32D cells were synchronized by starvation of IL-3 and restimulated with IL-3 plus or minus 20 ng/mL TGF-β1 and were analyzed for mRNA expression by Northern blotting. Single cell-cycle time was approximately 16 hours for 32D cells. The quantitative graph was made using NIHimager software comparing signal intensity at each time point with the time 0 baseline.
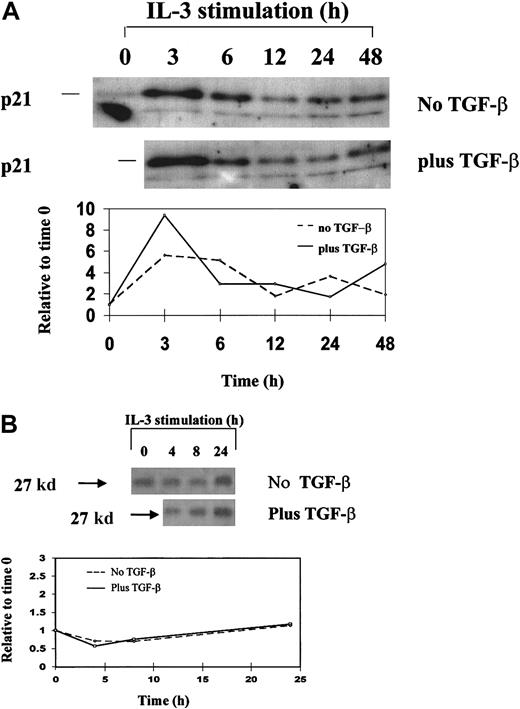
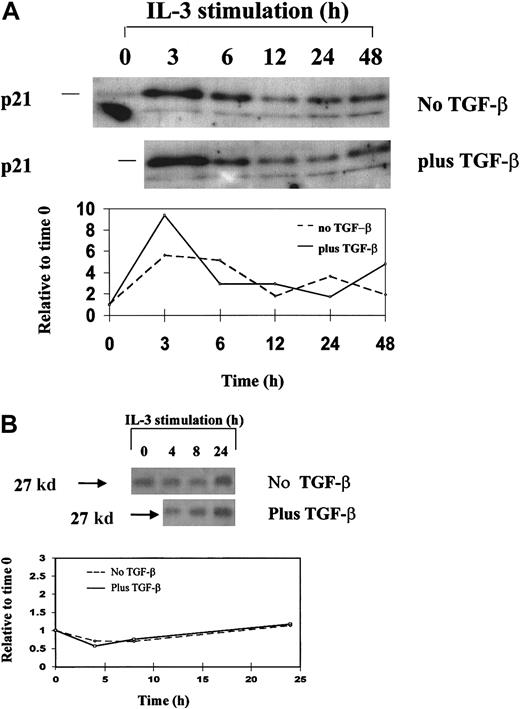
Western blot analysis.
Western blot analysis of p21 (A) and p27 (B) expression in synchronized 32D cells during a single cell-cycle progression in the presence and absence of TGF-β1. 32D cells were synchronized by starvation of IL-3 and restimulated with IL-3 plus or minus 20 ng/mL TGF-β1 and were analyzed for protein expression by Western blotting. Single cell-cycle time was approximately 16 hours for 32D cells. The quantitative graph was made using NIHimager software comparing signal intensity at each time point with time 0 baseline.
Western blot analysis.
Western blot analysis of p21 (A) and p27 (B) expression in synchronized 32D cells during a single cell-cycle progression in the presence and absence of TGF-β1. 32D cells were synchronized by starvation of IL-3 and restimulated with IL-3 plus or minus 20 ng/mL TGF-β1 and were analyzed for protein expression by Western blotting. Single cell-cycle time was approximately 16 hours for 32D cells. The quantitative graph was made using NIHimager software comparing signal intensity at each time point with time 0 baseline.
The inhibitory effect of TGF-β1 on hematopoietic cell growth is independent of p21 and p27
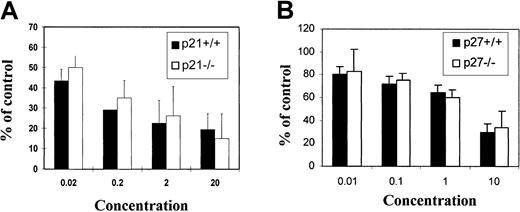
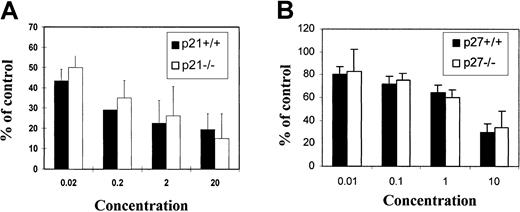
The above experiment in the 32D cell line indicated that the inhibitory effect of TGF-β1 on hematopoietic cells may not be mediated through p21 or p27. To further confirm this conclusion in primary cells, mononuclear bone marrow cells were harvested from either wild-type mice (+/+) or mice engineered to be deficient in p21 (p21−/−) or p27 (p27−/−). The 8- to 10-week-old animals were derived from the same heterozygous parents and analyzed for p21 or p27 genotype by tail clip DNA PCR. Animals from the same parent pairs were used to avoid breeding-induced artifacts. CFC assays were performed in standard methylcellulose cultures. Total CFCs were scored at day 10 to assess progenitor pool activity and long-term culture with limiting dilution (CAFC) to assess primitive cell activity. Although TGF-β1 had a dose-dependent inhibitory effect on CFCs, the effect was not different for either −/− cells or +/+ cells (Figure8). In addition, 10 ng/mL TGF-β1 was able to completely inhibit the proliferation of stem cells (CAFCs) of both genotypes (data not shown). These data demonstrate that TGF-β1 mediates antiproliferative effects independent of p21 or p27 in primitive hematopoietic cells.
Effects of TGF-β1 on colony-forming ability.
Effects on bone marrow cells from (A) p21−/− or (B) p27−/− mice in vitro. Murine bone marrow cells were plated at density of 1 × 105/mL in methylcellulose culture system supplemented with cytokine cocktails and cultured for 10 days as described in “Materials and methods.” Total CFCs were scored at day 10 by independent investigators. Data presented are mean ± SD from 4 replicates. No significant differences were observed between −/− and +/+ cells at any given dose of TGF-β1 (n = 3 independent experiments).
Effects of TGF-β1 on colony-forming ability.
Effects on bone marrow cells from (A) p21−/− or (B) p27−/− mice in vitro. Murine bone marrow cells were plated at density of 1 × 105/mL in methylcellulose culture system supplemented with cytokine cocktails and cultured for 10 days as described in “Materials and methods.” Total CFCs were scored at day 10 by independent investigators. Data presented are mean ± SD from 4 replicates. No significant differences were observed between −/− and +/+ cells at any given dose of TGF-β1 (n = 3 independent experiments).
Discussion
TGF-β1–induced alterations in cell cycle have been variably reported to signal through Smad proteins and to modulate p15, p21, or p27 to induce cell-cycle arrest in nonhematopoietic cells.7,21,42,43 In hematopoietic cells, antibody to TGF-β1 was shown to reduce p1513 and TGF-β1 stimulation to increase the fraction of p21+ cells by a qualitative in situ hybridization technique.26 However, the detailed relationship between the inhibition induced by the extracellular stimulus of TGF-β1 and its dependence on the nuclear effectors of cell-cycle inhibition in primitive hematopoietic cells, p21 and p27, has not been previously defined. Data presented here demonstrate that the action of TGF-β1 is independent of p21 and p27.
We found that quiescent primary human stem cells express high levels of p21, consistent with the functional data generated from p21−/−mice6 in which an inhibitory role of p21 was demonstrated in stem cell proliferation. Unexpectedly, we noted an increase in p21 immediately on the release of cell-cycle arrest in 32D cells, possibly reflecting the paradoxic role for p21 previously noted by others. P21 has been shown to positively affect proliferation capability after cytokine stimulation in progenitor cell pools.44,45 Mantel et al46 noted that bone marrow progenitor cells from mice proliferated poorly and formed few colonies under thymidine treatment except when transduced with a p21-encoding retroviral vector.45 Additionally, p21 and p27 were determined to be essential activators of cyclin D–dependent kinases in murine fibroblasts.47 This may be attributed to the requirement for p21 to promote the association of CDK4 with D-type cyclins. LaBaer et al48 demonstrated that low concentrations of p21 promote the assembly of active kinase complexes, and thereby entry into cycle, whereas higher concentrations are inhibitory. The stoichiometry of p21 and cyclin-CDK complexes appears crucial in determining the relative effect on transition of the cell through late G1 into S. Our observed transient rise in p21 protein immediately after the release of cell- cycle arrest in 32D cells (3-hour time point) (Figures 6, 7) is consistent with a role in cyclin-CDK complex formation to enable cycle entry.
The inhibitory role for p21 is more clearly defined and has been shown in numerous settings.49 50 When we evaluated quiescent primitive cells or terminally differentiated monocytic cells in the experiments reported here, we noted markedly elevated levels of p21 message. However, it should be noted that the association of increased p21 and terminal differentiation was cell type specific; we did not note a similar pattern in differentiating granulocytic cells (data not shown). This may also explain the insignificant change in the p21:GAPDH ratio we observed when examining CD34+ CD33+versus CD11b+ cells because the latter includes a mixture of monocytic and granulocytic cells predominated by granulocytes (Figure 2). Taken together, p21 appears to have a dual function in the hematopoietic system depending on the differentiation stage and cyclin-CDK complex status.
The coordinate expression of TGF-β1 and p21 (Figure 2) suggested a possible signaling link, which we assessed by directly measuring the expression of p21 or p27 in response to TGF-β1 inhibition. Given the heterogeneity and difficulty in cell-cycle synchronization when investigating primary human hematopoietic cells and the prohibitive constraints of cell numbers in a detailed analysis, we used the multipotent nontransformed cell line, 32D. Use of this line enabled us to more precisely assess the relationship between TGF-β1 and p21 or p27 expression at a protein and a message level. Although we recognize that immortalized cell lines offer imperfect models of primary cells, this cell line is not transformed, and therefore may offer a closer resemblance to normal cell physiology. In 32D cells, p21 or p27 was not affected by TGF-β1 despite the ability of TGF-β1 to exert an antiproliferative effect. We next validated this lack of association in a cell line by confirming the results in primary cells. Mouse strains engineered to be p21 or p27 deficient provided the clearest models for evaluating the link for TGF-β1 signaling to cell-cycle arrest mediated by these CDKIs. We reasoned that the presence or absence of the candidate downstream mediator provided a highly specific model of an in vivo effect. Bone marrow progenitor cells from −/− or +/+ mice formed colonies in methylcellulose assays, and each was inhibitable by TGF-β1. This was true for both CFC assays of mouse mature progenitors and HPP-CFC or LTC-IC assays of mouse immature primitive cells. Although compensatory changes in other cell-cycle regulators might have occurred in the knock-out animals, the absence of a definable difference in extent of inhibition in −/− compared with +/+ mice strongly argues against a role for p21 or p27 in the signaling of TGF-β1 in hematopoietic precursor cells. The absence of an altered phenotype of response to TGF-β in the absence of p21 or p27 argues against an indirect effect of TGF-β on p21 or p27 through mechanisms such as displacement from cyclin-CDK complexes.
These data demonstrate that though TGF-β1 and p21 are coordinately regulated in quiescent hematopoietic cells, there is not a causal link between TGF-β1 stimulation and p21 expression or effect. However, an earlier study by Ducos et al26 had demonstrated that the fraction of CD34+ cells expressing p21 increased after TGF-β1 stimulation, suggesting a direct link between these molecules. In situ hybridization in nonsynchronized cells was used by those investigators to define the proportion of cells expressing p21. In contrast, our studies assessed protein and mRNA levels and functional activity of TGF-β1 in the absence of p21, perhaps accounting for the differences in results. Others13 have also evaluated the relationship between TGF-β1 and p27 in primitive hematopoietic cells. Most notably, Dao et al13 demonstrated enhanced retroviral transduction when antibody to TGF-β1 and antisense p27 were used. Because neither intervention alone was sufficient to achieve this effect, these data support the notion that TGF-β1 and p27 act distinctly in the regulation of the primitive hematopoietic compartment. Indeed others51 have reported that TGF-β1 affects hematopoiesis independent of cell cycle effects entirely, inhibiting stem cell function by decreased expression of antiapoptotic genes such as Bcl-2 or increased expression of differentiative transcription factors such as GATA-1 and PU.1.
Taken together, our data and the studies noted above support the independence of the TGF-β1 effect from the CDKIs, p21 and p27. The lack of a link between TGF-β1 and these molecules has thus far only been described in hematopoietic tissue and is in contrast to the association shown in multiple other cell types.21-25 43Therefore, the involvement of p21 or p27 in mediating the TGF-β1 effect appears to be highly cell type specific. Whether other CDKIs, notably those of the INK4 family (p15, p16, p17, p18), participate in TGF-β1 effects on cell cycle remains to be explored.
Our data leave unresolved what extracellular signals may modulate p21 and p27, known to be critical for controlling hematopoietic populations. Examples of possible other signals include interferon-γ, noted to induce p21 through STAT1 signaling in A431 epidermoid carcinoma cells,52 or thrombopoietin, reported to induce p21 through STAT5 in CMK cells.53 Definition of the factors affecting p21 in stem cells or p27 in progenitor cells may provide a means of altering stem cell kinetics independent of TGF-β1. Interrupting the stimulus for p21 or p27 in conjunction with interrupting TGF-β1 may provide complementary strategies for stem cell expansion and gene transduction.
We thank Dr Tyler Jacks for providing p21+/− mice, Dr Andrew Koff for providing p27+/− mice, and Dr David Beach for providing CDKI cDNA probes.
Supported by National Institutes of Health grants HL44851, DK50234, HL55718 (D.T.S.), and KO8 DK02761 (T.C.); the Richard Saltonstall Charitable Foundation (D.T.S.); and the Deutsche Akademischer Austauschdienst (S.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David T. Scadden, Massachusetts General Hospital, Bldg 149, Rm 5212, 13th St, Charlestown, MA 02129; e-mail:scadden.david@mgh.harvard.edu.