Abstract
Survivin, a member of the inhibitors-of-apoptosis gene family, is expressed in a cell-cycle–dependent manner in all the most common cancers but not in normal differentiated adult tissues.Survivin expression and regulation were examined in acute myeloid leukemia (AML). Survivin was detected by Western blot analysis in all myeloid leukemia cell lines and in 16 of 18 primary AML samples tested. In contrast, normal CD34+ cells and normal peripheral blood mononuclear cells expressed no or very low levels of survivin. Cytokine stimulation increasedsurvivin expression in leukemic cell lines and in primary AML samples. In cultured primary samples, single-cytokine stimulation substantially increased survivin expression in comparison with control cells, and the combination of G-CSF, GM-CSF, and SCF increased survivin levels even further. Conversely, all-trans retinoic acid significantly decreased survivinprotein levels in HL-60, OCI-AML3, and NB-4 cells within 96 hours, parallel to the induction of myelomonocytic differentiation. Using selective pharmacologic inhibitors, the differential involvement of mitogen-activated protein kinase kinase (MEK) and phosphatidylinositol-3 kinase (PI3K) pathways were demonstrated in the regulation of survivin expression. The MEK inhibitor PD98059 down-regulated survivin expression in both resting and GM-CSF–stimulated OCI-AML3 cells, whereas the PI3K inhibitor LY294002 inhibited survivin expression only on GM-CSF stimulation. In conclusion, these results demonstrate thatsurvivin is highly expressed and cytokine-regulated in myeloid leukemias and suggest that hematopoietic cytokines exert their antiapoptotic and mitogenic effects, at least in part, by increasing survivin levels.
Introduction
Survivin, a member of the inhibitors-of-apoptosis (IAP) family of proteins, is present during fetal development but is undetectable in terminally differentiated adult tissues. However, survivin is prominently expressed in transformed cell lines, in all the most common human cancers, and in approximately 50% of high-grade non-Hodgkin lymphomas.1-4,Survivin suppresses apoptosis induced by Fas, Bax, caspases, and anticancer drugs.5 Conversely, the down-regulation ofsurvivin by antisense oligonucleotides induces apoptosis in vitro.6,7 Although survivin protein lacks the ability to directly inhibit caspase-3,8 it binds quantitatively to a new IAP-inhibiting protein, Smac/Diablo,9 10 raising the possibility that it might suppress caspases indirectly by freeing other IAP family members from the constraints of this protein. Taken together, these studies support the notion that survivin exerts an antiapoptotic effect.
Survivin expression is cell-cycle–dependent. In proliferating cells, survivin is expressed at high levels in the G2/M phase and is rapidly down-regulated after cell-cycle arrest.11 Recent studies suggest thatsurvivin also plays a role in cell cytokinesis, and the same function has been observed for the survivin-homologous ancient baculovirus IAP repeat (BIR)–family proteins inCaenorhabditis elegans and yeast.12-14 The role of survivin in cell division control is thought to involve caspase-dependent loss of p21 and deregulation of mitotic transition.12 Moreover, BIR-family proteins are required for the targeting of members of the Aurora family of kinases to metaphase chromosomes, thereby controlling chromosome segregation and cytokinesis.15,16 On the basis of these collective findings, therefore, survivin is considered to play a pivotal role in linking cell death and cell proliferation.17 18
Survival and growth of hematopoietic cells exquisitely depend on the presence of appropriate cytokines that can be provided through either autocrine production or paracrine secretion by stromal cells in the bone marrow micro-environment.19 Cytokines contribute to the regulation of the apoptotic threshold of normal and leukemic cells by modulating the expression and function of different families of pro- and antiapoptotic proteins.20,21 In particular, granulocyte macrophage–colony-stimulating factor (GM-CSF) exerts its biologic activity by binding its receptor, which in turn activates multiple intracellular signal transduction pathways through the common β subunit.22 Among these pathways, both the mitogen-activated protein kinase kinase/extracellular-signal regulated kinase (MEK/ERK) and the phosphatidylinositol-3 kinase (PI3K) pathways have been linked to the induction of resistance to apoptosis and the ability of hematopoietic cells to grow autonomously.20,23Although both pathways regulate the expression and function of several Bcl-2 family members, such as Mcl-1,24,25 the downstream events linking GM-CSF–initiated biochemical events to either proliferation or survival of hematopoietic cells are incompletely elucidated. Growth factor–mediated regulation of IAP expression has recently been demonstrated in endothelial cells.26,27However, the ability of hematopoietic cytokines to affectsurvivin expression in myeloid cells has not been studied. Moreover, unlike other IAPs, survivin gene expression is not influenced by NF-κB signaling,17 28 and little is known about other potentially involved signal transduction pathways.
In the study reported here, we examined the regulation ofsurvivin expression in acute myeloid leukemia (AML). Our results demonstrate that survivin is expressed in AML cell lines and in primary AML samples and that expression is up-regulated by hematopoietic cytokines and inhibited by all-trans retinoic acid (ATRA). We further demonstrate that survivin expression is regulated through MEK/ERK and PI3K pathways and can be modulated by selective signal transduction inhibitors.
Materials and methods
Cell lines and primary samples
Human leukemia cell lines were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal calf serum, 1 mM L-glutamine, and 50 μg/mL penicillin–streptomycin. For Mo7e cells, medium was supplemented with 100 U/mL GM-CSF (Immunex, Seattle, WA). Bone marrow and peripheral blood were obtained from patients with AML and normal donors after informed consent was obtained according to institutional guidelines. Mononuclear cells were purified by Ficoll-Hypaque (Sigma Chemical, St Louis, MO) density-gradient centrifugation and cultured in AIM-V medium (Gibco-BRL, Gaithersburg, MD) supplemented with cytokines (Amgen, Thousand Oaks, CA). Blast percentages and other characteristics of primary AML samples are listed in Table1.
Cell culture studies
OCI-AML3 or HL-60 cells (0.5 × 106 cells/mL) and mononuclear cells purified from the bone marrow of patients with AML (1 × 106 cells/mL) were treated with cytokines for 48 hours in serum-free RPMI 1640 or AIM-V medium, respectively. To block the MEK/ERK and the PI3K pathways, OCI-AML3 cells were washed twice with serum-free RPMI 1640 medium, resuspended at 0.2 × 106 cells/mL in the presence of PD98059 (2′-amino-3′-methoxyflavone29,30; CalBiochem, La Jolla, CA), LY294002 (2-[4-morpholinyl]-8-phenyl-[4H]-1-benzopyran-4-one;31Sigma), or the appropriate concentration of vehicle (dimethyl sulfoxide [DMSO]) for 2 hours at 37°C before the addition of GM-CSF (100 U/mL). In other experiments, HL-60, OCI-AML3 cells (0.3 × 106 cells/mL), and NB-4 cells (0.1 × 106 cells/mL) were cultured in the presence of ATRA (1 μM) for up to 96 hours. Cells were harvested at different times; live cells were counted by trypan blue exclusion, and the morphologic characteristics were evaluated under a light microscope after staining with HEMA quick stain solution (Biochemical Sciences, Swedesboro, NJ).
Western blot analysis
Cells were washed twice with phosphate-buffered saline (PBS) buffer and lysed at 4 × 104 cells/μL in cell lysis buffer (20 mM HEPES, pH 7.4, 0.25% NP-40 containing protease inhibitor cocktail; Boehringer Mannheim, Indianapolis, IN) for 10 minutes on ice. Equal amounts of lysate (equivalent to 5 × 105 cells) were subjected to SDS-PAGE to 12% polyacrylamide gels. Proteins were transferred to Hybond-P (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) membranes and reacted with polyclonal antibody againstsurvivin for 2 hours at room temperature. After they were washed, membranes were probed with a horseradish peroxidase–conjugated secondary antibody and reacted with ECL reagent (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). Anti–β-actin blot was made in parallel as a loading control. Signals were detected by a PhosphorImager (Storm 860, version 4.0; Molecular Dynamics, Sunnyvale, CA) and quantified by Scion Image software (Scion, Frederick, MD). Results were expressed as survivin/β-actin ratios.
Reverse transcription–polymerase chain reaction
OCI-AML3 cells were treated with either PD98059 or LY294002, as described above, and RNA was isolated with STAT-60 solution (Tel-Test, Friendswood, TX). One microgram total RNA was reverse-transcribed withsurvivin reverse primer (5′TCTCCTTTCCTAAGACATT3′) by AMV reverse transcriptase (Boehringer Mannheim) at 42°C for 1 hour. Polymerase chain reaction (PCR) amplification reaction mixtures (25 μL) contained cDNA, survivin forward primer (5′CACCACTTCCAGGGTTTA3′), the reverse primer, survivin probe (5′TGGTGCCACCAGCCTTCCTGTG3′), and TaqMan Universal PCR master mix (PE Applied Biosystems, Foster City, CA). Thermal cycle conditions included holding the reactions at 50°C for 2 minutes and at 95°C for 10 minutes and cycling for 40 cycles between 95°C for 15 seconds and 60°C for 1 minute. Results were collected and analyzed by an ABI Prism 7700 Sequence Detection System (PE Applied Biosystems).
Cell-cycle analysis
OCI-AML3 and HL-60 cells (0.5 × 106), cultured under various conditions, were harvested at different times, washed twice with cold PBS, and fixed with 2 mL ice-cold ethanol (70% vol/vol in water) for 1 hour at 4°C. After centrifugation, fixed cells were exposed to 500 μL propidium iodide (PI) staining solution (25 μg/mL PI, 180 U/mL RNase, 0.1% Triton X-100, and 30 mg/mL polyethylene glycol in 4 mM citrate buffer, pH 7.8; all from Sigma) for 1 hour at 4°C and analyzed using a FACScan flow cytometer (Becton Dickinson, San Jose, CA). Cell-cycle distribution was then analyzed using the ModFit LT software (Verity Software House, Topsham, ME).
Results
Expression of survivin protein in leukemic cell lines and primary AMLs
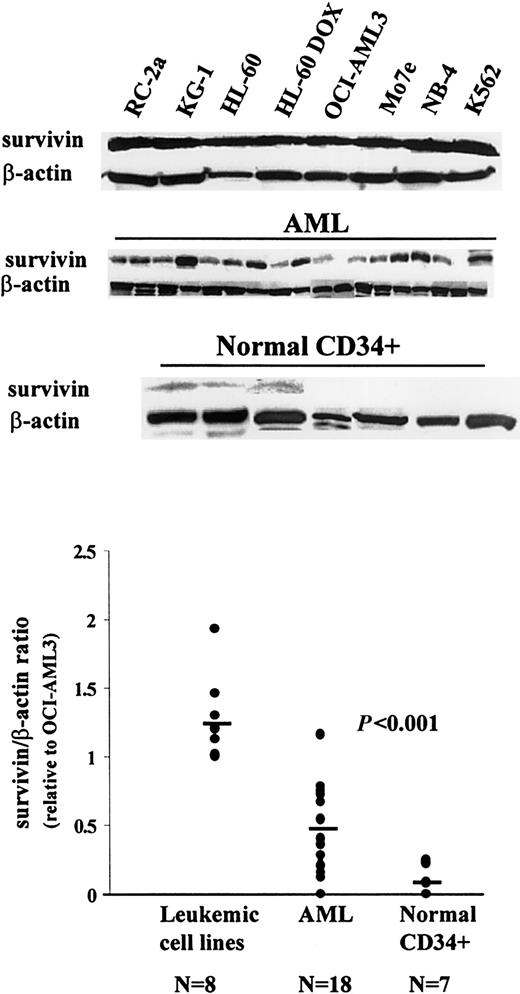
Survivin expression in myeloid leukemias has not yet been studied, though preliminary evidence that survivin mRNA is present in leukemic cell lines and in primary AML samples was reported from our group.32 Here we examinedsurvivin protein expression by Western blot analysis. As shown in Figure 1, all leukemic cell lines tested (RC-2a, KG-1, HL-60, HL-60 DOX, OCI-AML3, Mo7e, NB-4, and K562) expressed survivin protein at comparable levels (1.26 ± 0.32, survivin/β-actin ratio; survivin/β-actin ratio for OCI-AML3 = 1). Sixteen of 18 AML bone marrow samples showed variable levels of survivin (Figure 1). The meansurvivin/β-actin ratio related to OCI-AML3 in these samples (± SD) was 0.44 ± 0.31 (range, 0 to 1.17; Figure 1, Table 1). Hence, as in other malignancies, survivin protein is widely expressed in leukemic cell lines and primary AML blasts. We also examined survivin protein expression in normal CD34+ cells obtained by magnetic-bead sorting of 4 bone marrows from normal donors and 3 peripheral blood mononuclear cell samples obtained after G-CSF mobilization. Three bone marrow samples were weakly positive, and all others were negative forsurvivin expression (0.08 ± 0.11; Figure 1, Table 1). Likewise, survivin protein expression was not detectable in 3 unseparated peripheral blood mononuclear cell samples (data not shown).
Western blot analysis of survivin expression in leukemic cell lines, primary AML samples, and normal CD34+ cells.
Cell lysates equivalent to 0.5 × 106 cells were loaded on each lane. The experimental conditions are described in “Materials and methods.”
Western blot analysis of survivin expression in leukemic cell lines, primary AML samples, and normal CD34+ cells.
Cell lysates equivalent to 0.5 × 106 cells were loaded on each lane. The experimental conditions are described in “Materials and methods.”
Induction of survivin protein expression by cytokines
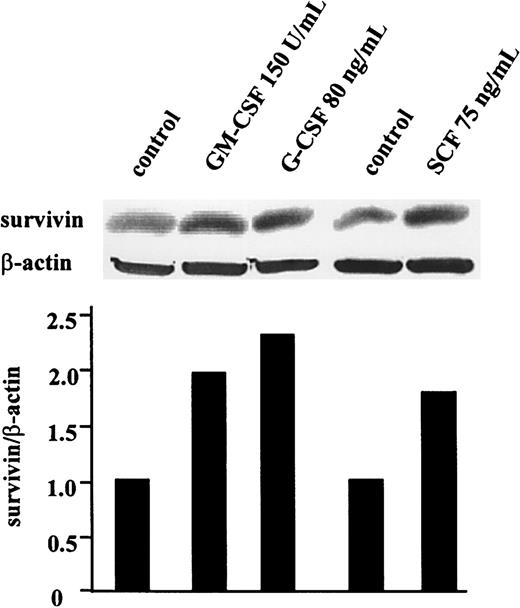
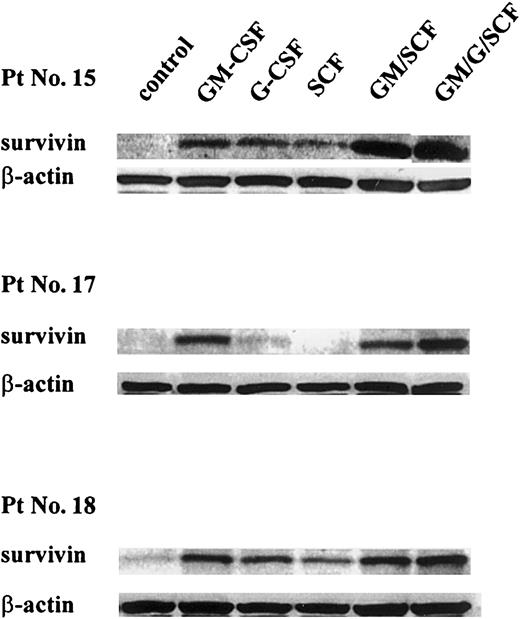
Because survival and growth of leukemic cells largely depend on the presence of appropriate cytokines, we also tested the effect of different hematopoietic cytokines on survivin expression in human leukemic cell lines and primary AML blasts. As shown in Figure2, survivin protein levels were 2-fold in GM-CSF (150 U/mL), 2.3-fold in G-CSF (80 ng/mL), and 1.8-fold in stem cell factor (SCF; 75 ng/mL)–treated OCI-AML3 cells compared to cells cultured in cytokine- and serum-free medium for 48 hours. Similar results were obtained in HL-60 cells (data not shown). In 3 AML bone marrow samples studied (patients 15, 17, 18; Table 1)survivin protein level decreased dramatically after 48-hour culture in cytokine- and serum-free medium (Figure3). Treatment with GM-CSF (100 U/mL) and, to a lesser extent, G-CSF (50 ng/mL) and SCF (100 ng/mL) substantially increased survivin protein levels compared to control cells, and combinations of these cytokines resulted in the induction of highersurvivin levels (Figure 3).
Cytokines stimulate survivin protein expression in OCI-AML3 cells.
Cells were cultured in serum-free RPMI 1640 medium with GM-CSF (150 U/mL), G-CSF (80 ng/mL), or SCF (75 ng/mL). After 48 hours, cells were lysed, and survivin levels were compared with those in untreated cells by Western blot analysis.
Cytokines stimulate survivin protein expression in OCI-AML3 cells.
Cells were cultured in serum-free RPMI 1640 medium with GM-CSF (150 U/mL), G-CSF (80 ng/mL), or SCF (75 ng/mL). After 48 hours, cells were lysed, and survivin levels were compared with those in untreated cells by Western blot analysis.
Effect of cytokines on survivin expression in primary AML blast cells.
Cells were cultured in serum-free AIM-V medium with GM-CSF (100 U/mL), G-CSF (50 ng/mL), SCF (100 ng/mL), or various combinations of these cytokines for 48 hours. Then they were lysed, and survivinprotein levels were assessed by Western blot analysis. Results were quantitated by PhosphorImager. GM, GM-CSF; G, G-CSF; and Pt No., patient number as shown in Table 1.
Effect of cytokines on survivin expression in primary AML blast cells.
Cells were cultured in serum-free AIM-V medium with GM-CSF (100 U/mL), G-CSF (50 ng/mL), SCF (100 ng/mL), or various combinations of these cytokines for 48 hours. Then they were lysed, and survivinprotein levels were assessed by Western blot analysis. Results were quantitated by PhosphorImager. GM, GM-CSF; G, G-CSF; and Pt No., patient number as shown in Table 1.
ATRA-induced differentiation and inhibition of survivinprotein expression
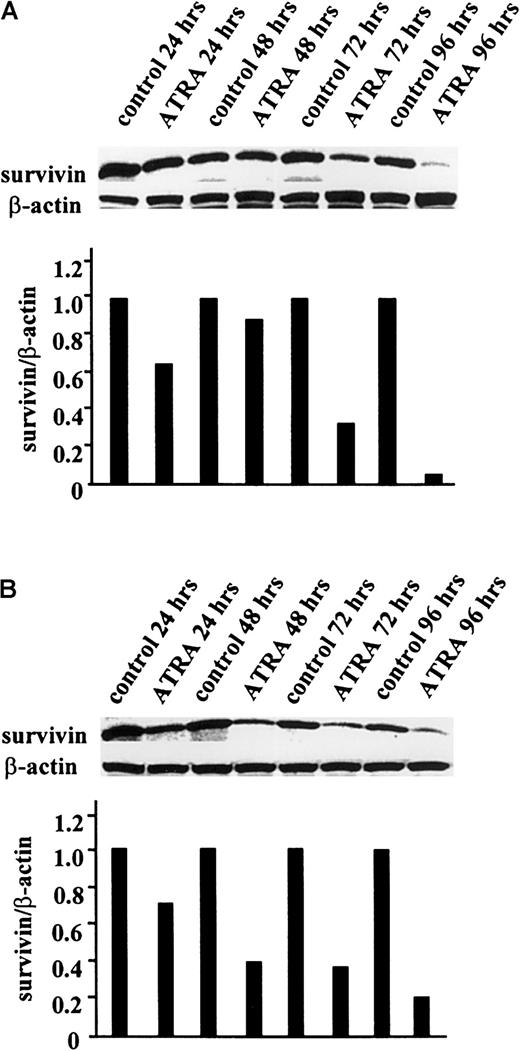
Like other antiapoptotic proteins, such as Bcl-2 and Mcl-1,33 34 survivin expression is likely to be differentially regulated during differentiation. We therefore examined the effect of ATRA-induced leukemia cell differentiation onsurvivin expression. HL-60 and OCI-AML3 cells were treated with ATRA (1 μM) for up to 96 hours, and survivin protein levels, cell-cycle status, and differentiation were determined.Survivin protein levels were significantly decreased in the ATRA-treated leukemic cells studied (Figure4). In HL-60 cells, no significant effect was observed at 48 hours; however, at 72 hours, survivinlevels were decreased by 67% and at 96 hours they were decreased by 96% compared to the levels in untreated control cells (Figure 4A). In OCI-AML3 cells, survivin levels decreased by 80% at 96 hours (Figure 4B). Concomitant with survivindown-regulation, ATRA-treated OCI-AML3 and HL-60 cells showed inhibition of cell- cycle progression and morphologic features of myelomonocytic differentiation (data not shown). Similarly, ATRA decreased survivin protein expression and induced differentiation in NB-4 cells. After 96 hours of culture in 1 μM ATRA, survivin protein levels were decreased by 65% compared to untreated cells (data not shown).
ATRA-induced down-regulation of survivinprotein expression in HL-60 and OCI-AML3 cells.
HL-60 (A) and OCI-AML3 (B) cells were treated with 1 μM ATRA for up to 96 hours, as described in “Materials and methods.” Cells were lysed, and survivin protein levels were determined at 24, 48, 72, and 96 hours by Western blot analysis. The experiment was performed 3 times, and the results shown here are representative.
ATRA-induced down-regulation of survivinprotein expression in HL-60 and OCI-AML3 cells.
HL-60 (A) and OCI-AML3 (B) cells were treated with 1 μM ATRA for up to 96 hours, as described in “Materials and methods.” Cells were lysed, and survivin protein levels were determined at 24, 48, 72, and 96 hours by Western blot analysis. The experiment was performed 3 times, and the results shown here are representative.
Regulation of survivin expression by the MEK/ERK and the PI3K signal transduction pathways
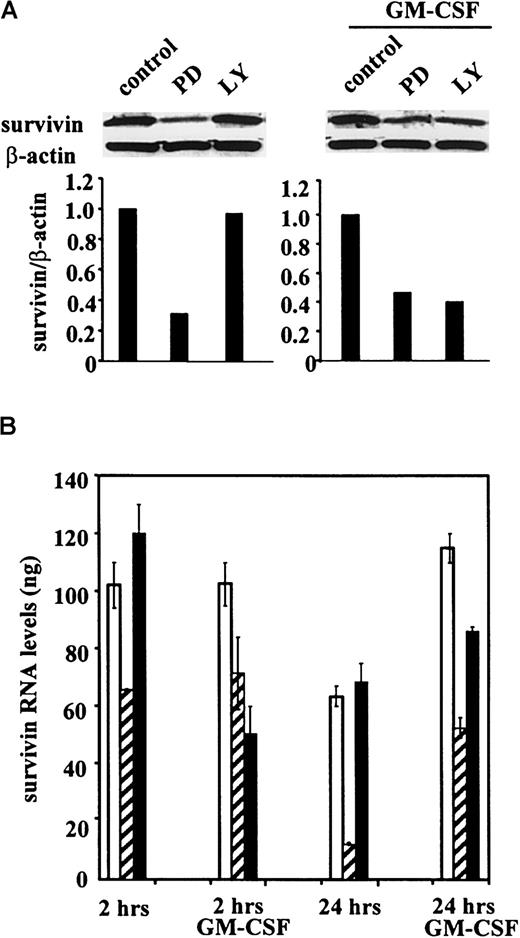
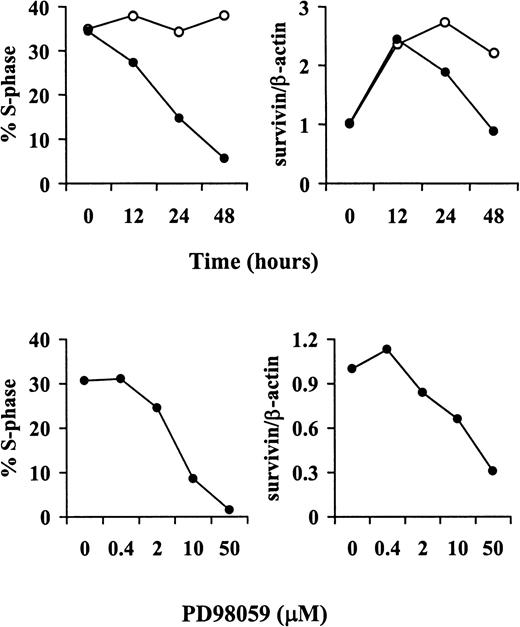
We next examined the role of the MEK/ERK and the PI3K signal transduction pathways in the regulation of basal and cytokine-stimulated survivin expression in OCI-AML3 cells using their respective pharmacologic inhibitors PD98059 and LY294002.29-31 Treatment with PD98059, but not with LY294002, significantly decreased survivin levels in the absence of cytokine stimulation (Figure5A). However, on stimulation with GM-CSF (100 U/mL), both PD98059 and LY294002 partially inhibitedsurvivin expression (Figure 5A), suggesting that both pathways are involved in GM-CSF–mediated regulation ofsurvivin. To examine whether survivin expression is regulated at the transcriptional level, RNAs from untreated and GM-CSF–stimulated OCI-AML3 cells, cultured in the presence or absence of PD98059 or LY294002, were analyzed by quantitative real-time RT-PCR with survivin-specific primers. Consistent with the protein expression data, survivin mRNA levels were reduced only by the MEK inhibitor PD98059 under basal conditions and by both the MEK and the PI3K inhibitors on GM-CSF stimulation (Figure 5B).Survivin expression in OCI-AML3 cells was also increased by treatment with the phosphatase inhibitor sodium orthovanadate (25 to 50 μM for 48 hours), and this increase was abrogated by pretreatment with either PD98059 or LY294002 (data not shown), further supporting a role for the MEK/ERK and the PI3K pathways in the regulation ofsurvivin expression. We also assessed the effect of signal transduction inhibitors on the cell-cycle distribution of unstimulated and GM-CSF–stimulated OCI-AML3 cells. PD98059 profoundly inhibited the G1/S transition in both unstimulated and GM-CSF–stimulated cells (78% and 79% reduction in S phase, respectively), whereas treatment with LY294002 only slightly affected cell-cycle distribution in either culture condition (33% and 36% reduction in S phase, respectively) (data not shown). PD98059-induced cell- cycle arrest was time- and dose-dependent, and its kinetics and dose-response curve paralleled those of survivin expression (Figure6). Taken together, our data indicate that survivin expression is differentially regulated by the MEK/ERK and the PI3K signal transduction pathways under basal and GM-CSF–stimulated conditions and suggest that the effect of MEK/ERK blockade may be mediated, at least in part, by the inhibition of cell-cycle progression.
Regulation of survivin expression in OCI-AML3 cells by MEK and PI3K inhibitors.
(A) Western blot shows survivin protein expression in cells treated with 20 μM PD98059 (PD) or 10 μM LY294002 (LY) for 48 hours without or with GM-CSF (100 U/mL). Results of 1 of 3 independent experiments are shown. (B) Quantitative RT-PCR demonstrates the regulation of survivin mRNA expression in response to the inhibitors at 2 and 24 hours (Taqman PCR; see “Materials and methods”). ■ indicates control; ▨, PD; and ▪, LY.
Regulation of survivin expression in OCI-AML3 cells by MEK and PI3K inhibitors.
(A) Western blot shows survivin protein expression in cells treated with 20 μM PD98059 (PD) or 10 μM LY294002 (LY) for 48 hours without or with GM-CSF (100 U/mL). Results of 1 of 3 independent experiments are shown. (B) Quantitative RT-PCR demonstrates the regulation of survivin mRNA expression in response to the inhibitors at 2 and 24 hours (Taqman PCR; see “Materials and methods”). ■ indicates control; ▨, PD; and ▪, LY.
Kinetics and dose-response of PD98059-induced cell-cycle arrest and down-regulation of survivin protein expression.
OCI-AML3 cells were treated with either vehicle (control) or PD98059, harvested and stained with PI to determine DNA content (left panels) or analyzed by Western blot with a survivin-specific antiserum (right panels). Top panels show the kinetics of cell-cycle arrest and down-regulation of survivin expression in response to 20 μM PD98059. Bottom panels show the dose-response curves for PD98059 at 48 hours. Results are expressed as the percentage of cells in S phase (calculated using the ModFit LT software, left panels) andsurvivin/β-actin ratios (as quantitated by a PhosphorImager, right panels). Results of 1 of 3 independent experiments are shown. ○ indicates control; ●, PD98059 (20 μM).
Kinetics and dose-response of PD98059-induced cell-cycle arrest and down-regulation of survivin protein expression.
OCI-AML3 cells were treated with either vehicle (control) or PD98059, harvested and stained with PI to determine DNA content (left panels) or analyzed by Western blot with a survivin-specific antiserum (right panels). Top panels show the kinetics of cell-cycle arrest and down-regulation of survivin expression in response to 20 μM PD98059. Bottom panels show the dose-response curves for PD98059 at 48 hours. Results are expressed as the percentage of cells in S phase (calculated using the ModFit LT software, left panels) andsurvivin/β-actin ratios (as quantitated by a PhosphorImager, right panels). Results of 1 of 3 independent experiments are shown. ○ indicates control; ●, PD98059 (20 μM).
Discussion
In this study, we provide evidence that the recently described IAP family member survivin is widely expressed in myeloid leukemia cell lines and in almost all primary AML samples tested. Furthermore, we demonstrate that survivin expression in leukemic cells is regulated by cytokines and differentiation-inducing agents and that the modulation of major signal transduction pathways, such as the MEK/ERK and the PI3K pathways, can contribute to the regulation of survivin expression at the mRNA and protein levels.
The induction of programmed cell death is the common outcome of successful cytotoxic therapy for many different types of cancer, including AML.35-40 Multiple genetic alterations that result in the disruption of the physiological regulation of apoptosis are thought to account for the ability of leukemic cells to grow autonomously and for their clinical resistance to therapy.41-44 Recently, a new family of downstream inhibitors of caspases, the IAP family, has emerged as a potential key player in the regulation of apoptosis in cancer,45,46 and we have already demonstrated that XIAP is expressed and has prognostic relevance in AML.47 Our present results demonstrate that another IAP family member, survivin, is constitutively expressed in both myeloid leukemia cell lines and in primary AML blasts and at significantly lower levels (P < .001) in normal CD34+ cells from bone marrow or G-CSF–stimulated peripheral blood. The latter finding extends to the hematopoietic progenitors the previous report of lack of survivinexpression in normal bone marrow.17 Becausesurvivin has been demonstrated to efficiently inhibit apoptosis induced by a variety of stimuli in vitro5 and its presence has been correlated in vivo with reduced apoptotic indices and poor prognosis in solid tumors,3,4 48-50 we are currently investigating its functional and prognostic relevance in AML.
Acute myeloid leukemia is a heterogeneous disease characterized by the accumulation of leukemic blasts arrested at various stages of granulocytic and monocytic differentiation. Transcriptional modulation aimed at restoring the ability of AML cells to regulate the expression of genes resulting in differentiation is, therefore, an attractive therapeutic strategy that has proved effective in the treatment of patients with acute promyelocytic leukemia (APL).51 Our study shows that ATRA significantly down-regulates survivinexpression in AML (HL-60, OCI-AML3) and APL (NB-4) cell lines, concomitant with the induction of cell differentiation. Whether other differentiation inducers such as DMSO, hexamethylene bisacetamide, vitamin D, and butyrate are also able to down-regulatesurvivin expression is unknown. Further studies are required to elucidate whether ATRA directly inhibits survivintranscription or whether it affects survivin expression primarily because of the cell-cycle arrest that accompanies differentiation. Regardless, together with previous evidence indicating that ATRA transcriptionally down-regulates Bcl-2 and Bcl-XLexpression in leukemias,52 our findings suggest that ATRA may lower the apoptotic threshold by modulating multiple pathways, eventually rendering AML cells more susceptible to cytotoxic agents.
The present study also provides unequivocal evidence that hematopoietic cytokines such as GM-CSF, G-CSF, and SCF, alone or in combination, strongly increase survivin expression in myeloid leukemia cell lines and, most important, in primary AML samples. Further evidence that IAP family members may function as growth factor–inducible antiapoptotic genes comes from the recent observation that survivin and XIAP expression are increased in endothelial cells in response to mitogenic growth factors, resulting in a decreased sensitivity to apoptotic stimuli.26,27Previous studies from our group have also shown that quiescent, but not proliferating, leukemic progenitors overexpress Bcl-2 and Bcl-XL.53 This observation, together with the present finding of increased survivin expression in response to cytokines, raises the intriguing possibility that Bcl-2 andsurvivin may represent complementary survival pathways that are differentially regulated by the cell-cycle status of leukemic progenitors. Quiescent progenitors are protected from apoptosis and are restrained from entering the cell cycle by the expression of Bcl-2 (and possibly Bcl-XL). However, once recruited into the cell cycle, proliferating cells could switch to asurvivin-mediated survival pathway that enables them to successfully complete mitosis and avoid a “default” induction of apoptosis at cell division.17 Consistent with this hypothesis, preliminary data indicate that AML cells that survive Bcl-2 antisense treatment in vitro express high levels of survivin(B.Z.C., unpublished results, December 1999). Interestingly, in the IL-3–dependent cell line BaF3, cytokine withdrawal-induced apoptosis was inhibited by the forced expression of either Bcl-2 orsurvivin,1 suggesting that, though they act at different levels, these 2 survival pathways may indeed function in concert to prevent cell death. Cytokine-mediated up-regulation of asurvivin-dependent survival pathway might also explain the conflicting clinical results reported for cytokine “priming” strategies for the therapy of AML.54
The binding of GM-CSF to its receptor activates multiple signaling pathways, which in turn lead to the proliferation, differentiation, and survival of various hematopoietic cells.19,20,22 Here we provide the first evidence that, in addition to modulating the expression and function of Bcl-2 family members, GM-CSF–mediated activation of both the MEK/ERK and the PI3K signal transduction pathways regulates the expression ofsurvivin at both the mRNA and the protein level. This finding is consistent with the notion that GM-CSF antiapoptotic activity relies on multiple and, in part, redundant pathways.55 Interestingly, we found that disruption of the MEK/ERK, but not of the PI3K, pathway also inhibited the constitutive expression of survivin (Figure 5), suggesting that the MEK/ERK pathway might be constitutively active in the cytokine-independent OCI-AML3 cell line. Consistent with this hypothesis, recent data from our group demonstrate that active ERK species are indeed detectable in OCI-AML3 cells in the absence of cytokine or serum stimulation (M.M., manuscript in preparation, May 2000).
Regulation of mouse survivin expression requires integration of typical CDE/CHR G1 repressor elements and basal transcriptional activity by Sp1 sites, which results in a cell-cycle–regulated expression in the G2/M phase.56 Our data indicate that pharmacologic disruption of the MEK/ERK kinase module in unstimulated and GM-CSF–stimulated cells profoundly inhibits the G1/S transition, suggesting that the observed down-regulation of survivin expression may be, at least in part, secondary to the inhibition of cell-cycle progression. However, the early inhibition of survivin mRNA transcription (at 2 hours) in the absence of any detectable cell-cycle changes suggests that a direct transcriptional effect might also take place. Likewise, a cell-cycle–independent transcriptional effect is the most likely explanation for the inhibition of GM-CSF–stimulatedsurvivin expression observed in response to PI3K inhibition. Support for this hypothesis comes further from the recent observation that, in endothelial cell lines, treatment with angiopoietin-1 up-regulates survivin expression in a PI3K/AKT-dependent fashion in the absence of any effect on cell proliferation.57 Further studies are under way to elucidate the potential involvement of PI3K- and MEK/ERK-dependent transcription factors in survivin gene expression. The results reported here—the effects of cytokines, ATRA, and signal transduction inhibitors on survivin expression—may be of help in the development of novel strategies for the treatment of leukemia and other cancers by targeting antiapoptoticsurvivin.
We thank Rosemarie Lauzon for assisting in the manuscript preparation and Teresa McQueen for magnetic-bead sorting normal CD34+ cells.
Supported in part by grants from the National Institutes of Health (PO1 CA55164, PO1 CA49639, CA78810, and HL54131); by the Keck Foundation; and by an American Cancer Society International Fellowship for Beginning Investigators (M.M.). M.A. holds the Stringer Professorship for Cancer Treatment and Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michael Andreeff, Section of Molecular Hematology and Therapy, Dept of Blood and Marrow Transplantation, University of Texas M. D. Anderson Cancer Center, Box 448, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: mandreef@notes.mdacc.tmc.edu.