Abstract
Children with neurofibromatosis type 1 (NF1) carry germline mutations in one allele of the NF1 gene and are predisposed to myeloid malignancies, particularly juvenile myelomonocytic leukemia (JMML). Disruption of the remaining NF1 allele can be found in malignant cells. Flow cytometric cell sorting techniques to isolate the malignant cell populations and molecular genetic methods to assay for somatic loss of the normal NF1 allele were used to study an unusual child with NF1 and JMML who subsequently had T-cell lymphoma. The data show that malignant JMML and lymphoma cells share a common loss of genetic material involving the normal NF1gene and approximately 50 Mb of flanking sequence, suggesting that the abnormal T-lymphoid and myeloid populations were derived from a common precursor cell. These data support the hypothesis that JMML can arise in a pluripotent hematopoietic cell.
Introduction
Juvenile myelomonocytic leukemia (JMML) is a relentless myeloproliferative disorder of children characterized by the monoclonal overproduction of myeloid cells.1,2 Up to 14% of cases occur in children with neurofibromatosis type 1 (NF1),3,4 an autosomal dominant disorder caused by germline inactivation of one allele of the NF1 gene on chromosome 17. JMML can involve more than the myeloid lineage5 because a malignant clonal expansion of erythroid cells has been inferred by cytogenetic,6 X chromosome inactivation7 and microsatellite polymorphic marker studies,8 and a JMML patient has been reported whose disease evolved to pre-B–cell acute lymphoblastic leukemia (ALL).9 Here we describe a boy with NF1 who was brought for treatment for JMML and in whom a T-cell lymphoma later developed. Molecular genetic and flow cytometric analyses provided strong evidence that both malignant clones derived from a common precursor with pluripotent potential, suggesting that JMML is a stem cell disorder.
Study design
Case report
A 3½-year-old boy with NF1 inherited through the maternal lineage was brought for treatment for JMML. Physical examination showed numerous café au lait spots and enlarged tonsils but an absence of hepatosplenomegaly. His white blood cell count was 96 700/μL, with 4% circulating myelocytes/metamyelocytes, a hemoglobin level of 10.1 g/dL, and a platelet count of 218 000/μL. The bone marrow showed an overwhelming myeloid predominance with less than 5% blasts and a normal karyotype, 46,XY. His peripheral blood myeloid cells formed colony-forming unit granulocyte-macrophage colonies in methylcellulose cultures without exogenous growth factors.
During the next 4 months leukocytosis persisted and was complicated by a worsening anemia and thrombocytopenia with an enlarging spleen, failure to thrive, and airway obstruction caused by hypertrophied tonsils. There was no response to isotretinoin administered at 100-200 mg/m2 per day. Adenotonsillectomy and splenectomy were performed, and histopathologic examination of the adenoids and tonsils revealed a dense infiltration with myeloperoxidase-positive cells. Similarly, the enlarged spleen showed expansion of the red pulp by immature myeloid cells. New, diffuse adenopathy and hepatomegaly developed 6 weeks later. A lymph node biopsy revealed a T-cell expansion consistent with lymphoma. He received combination high-dose chemotherapy, but respiratory distress, anasarca, and renal failure ensued, which led to his death 8 months after the diagnosis of JMML.
Flow cytometry
Monoclonal antibodies were obtained from Becton Dickinson Immunocytometry Systems (San Jose, CA), DAKO (Carpinteria, CA), and Pharmingen (San Diego, CA). Flow cytometric analysis of the lymphomatous node and bone marrow aspirate was performed as previously described.1 Using a FACS Vantage (Becton Dickinson), viable cells from an enlarged lymph node were separated into CD4+surface CD3−(CD4+sCD3−) lymphoma cells, and CD4+sCD3+ (phenotypically normal T cells) populations and viable bone marrow cells were purified as bright CD45+ with bright CD5+ (phenotypically normal T lymphocytes), bright CD45 with intermediate side scatter (monocytes), and intermediate CD45 with high side scatter (maturing granulocytes) populations. The cells were lysed immediately with DNA preparation buffer (Gentra Systems, Minneapolis, MN) and were snap-frozen in liquid nitrogen.
DNA extraction and analysis for loss of constitutional heterozygosity
DNA was isolated from unfractionated and sorted blood, bone marrow, spleen, and lymph node populations as described.11To screen for loss of heterozygosity (LOH) at NF1, 4 intragenic polymorphisms were assayed: EVI-20,12 an Alu repeat,13 a dinucleotide repeat,14 and a complex repeat.15 The extent of the chromosome 17 LOH region was determined by assay of polymorphic loci UT17212 and by 9 loci defined in the Genome Database (http://gdbwww.gdb.org/) by their identification numbers—D17S926 (GDB, 199252), D17S805 (GDB, 188452), D17S1294 (GDB, 686175), D17S1800 (GDB, 607032), D17S250 (GDB, 177030), D17S836 (GDB, 1218969), D17S1806 (GDB, 607848), D17S1830 (GDB, 1218973), and D17S928 (GDB, 1218974). Each locus was assayed using the polymerase chain reaction (PCR) to amplify DNA segments that contained a variable number of short nucleotide repeats. Thermocycle parameters and procedures for genotyping each locus have been described.12 15Radiolabeled PCR products were resolved by electrophoresis. Loss and retention of heterozygosity was determined by comparing the alleles detected in the blood of both parents with the allele(s) detected in the patient's tissues.
Results and discussion
LOH for NF1 served as a marker of somatic inactivation of the normal allele in various hematopoietic compartments and cells showing LOH at NF1 are likely to be derived from a common precursor cell. Similarly, X chromosome inactivation has been used to demonstrate the clonality of mononuclear cells in girls with JMML.7 To test whether the lymphoid and myeloid cells shared a common LOH of NF1 in this patient, subpopulations of normal and aberrant cells were identified and purified using flow cytometry and subjected to genetic analysis.
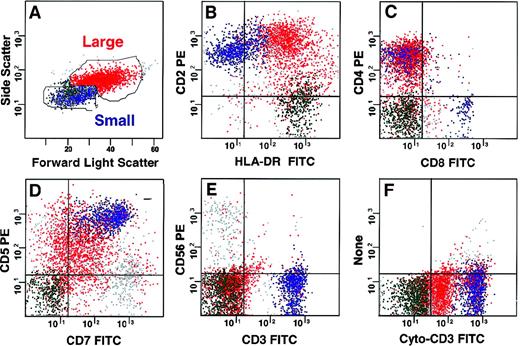
Cells from the enlarged lymph node showed 2 populations by forward and side scatter displays (Figure 1). The population of small cells revealed a mixture of phenotypically normal B, T, and natural killer lymphoid cells. The large lymphoma cells (red), which comprised 40% of viable cells, were consistent with T-cell lymphoma. Using surface (s) staining for CD4 and CD3, the T cells were sorted into an abnormal and a phenotypically normal population defined as CD4+sCD3− and CD4+sCD3+, respectively. Cells with this abnormal phenotype could not be identified in the bone marrow (lower limit of detection less than 0.5%). Bone marrow was also used to purify aberrant myeloblasts expressing CD7 and normal T lymphocytes (CD4+sCD3+), as well as monocytoid forms and maturing neutrophils.
Multidimensional flow cytometric analysis of lymph node cells.
Two populations could be defined based on forward and side light scatter (A). The larger T-cell lymphoma population (red) expressed CD2 and HLA-DR (B), CD4 without CD8 (C), and CD5 and CD7 (D). It did not express CD3 or CD56 on the surface (E), but it had CD3 in the cytoplasm (F) and were CD5dim, CD7dim, CD10dim, CD1a−, TdT−, CD13−, and CD34− and did not express B lymphoid markers (data not shown). Small cells in the lymph node contained normal B (green, 20%) and T (blue, 24%) lymphocytes and natural killer cells (gray, 7%).
Multidimensional flow cytometric analysis of lymph node cells.
Two populations could be defined based on forward and side light scatter (A). The larger T-cell lymphoma population (red) expressed CD2 and HLA-DR (B), CD4 without CD8 (C), and CD5 and CD7 (D). It did not express CD3 or CD56 on the surface (E), but it had CD3 in the cytoplasm (F) and were CD5dim, CD7dim, CD10dim, CD1a−, TdT−, CD13−, and CD34− and did not express B lymphoid markers (data not shown). Small cells in the lymph node contained normal B (green, 20%) and T (blue, 24%) lymphocytes and natural killer cells (gray, 7%).
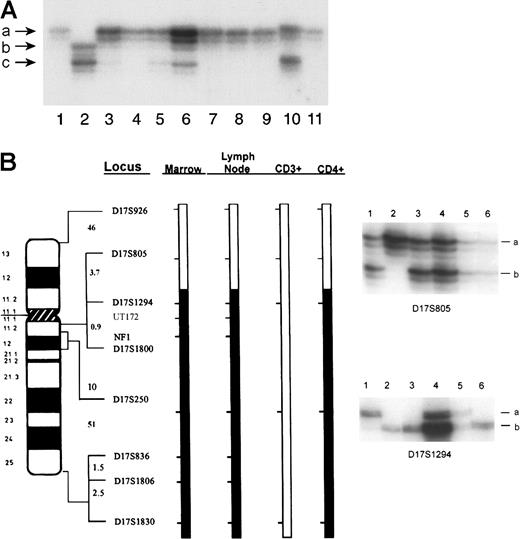
Loss of the normal NF1 allele inherited from his father was detected in the patient's blood and bone marrow specimens obtained at the initial diagnosis of JMML (Figure 2A, lanes 3 and 4, respectively). A similar loss of the normal paternalNF1 allele was identified in the lymph node and in bone marrow cells obtained with the onset of diffuse adenopathy (lanes 5 and 7, respectively), in maturing monocyte and neutrophil fractions purified from the bone marrow (lanes 8 and 9, respectively), and in the immunophenotypically aberrant CD4+sCD3−subpopulation of cells purified from the lymphomatous lymph node (lane 11). Unfractionated spleen cells (lane 6) showed a marked reduction in the signal derived from the paternal allele, a result that is consistent with an admixture of NF1−/− andNF1 +/− cells. In contrast, the intensities of the mutant maternal and normal paternal alleles were similar in the phenotypically normal CD4+CD3+ T cells isolated from the lymph node (lane 10) and from the phenotypically normal CD5+ T cells purified from bone marrow (data not shown), suggesting that these cells were not involved in the malignant process.
Analysis of tissue from the patient and of blood from his parents for LOH at
NF1. (A) DNA samples were amplified using oligonucleotides corresponding to the intragenic marker EVI-20. DNA obtained from the mother with NF1 (NF1±, lane 1) and the unaffected father (NF1+/+, lane 2). Patient peripheral blood (lane 3) and unfractionated bone marrow (lane 4) were obtained at the time of initial presentation and diagnosis of JMML. Lymph node (lane 5), spleen (lane 6), unfractionated marrow (lane 7), maturing marrow monocytes (lane 8), and marrow-derived neutrophils (lane 9) and the lymph node subpopulations—CD4+sCD3+ (lane 10) and CD4+sCD3− (lane 11)—were obtained at the time of the diagnosis of lymphoma. Different alleles are designated in size order, beginning with the letter a. (B) Identical chromosome 17 loci showed LOH in both primary leukemic and lymphoma cells. Bars represent a schematic of the patient's paternally derived chromosome 17 in bone marrow, lymph node, sCD3+ (phenotypically normal CD4+ T cells), and CD4+sCD3−(abnormal lymphoma) cells, designated as CD4+. The heterozygosity status of informative loci is indicated by the shaded bar for loci that lost heterozygosity or the open bar for loci that retained heterozygosity. Interlocus genetic distances in centiMorgans (cM) are indicated to the left of each interval as estimated from a chromosome 17 sex-averaged map (http://www.marshmed.org/genetics/). Data illustrating the retention of heterozygosity at the D17S805 locus and the loss of heterozygosity at the D17S1294 locus are shown to the right. DNA samples are as follows: lane 1, patient's unaffected father; lane 2, patient's mother affected with NF1; lane 3, patient's unfractionated bone marrow; lane 4, patient's lymph node; lane 5, CD3+ cells from the lymph node; lane 6, CD4+cells from the lymph node; positive and negative controls are not shown. For each locus, 2 alleles, designated simply as a and b, are segregating in this family. Differences in band intensity are caused by differences in the amount of DNA loaded per lane.
Analysis of tissue from the patient and of blood from his parents for LOH at
NF1. (A) DNA samples were amplified using oligonucleotides corresponding to the intragenic marker EVI-20. DNA obtained from the mother with NF1 (NF1±, lane 1) and the unaffected father (NF1+/+, lane 2). Patient peripheral blood (lane 3) and unfractionated bone marrow (lane 4) were obtained at the time of initial presentation and diagnosis of JMML. Lymph node (lane 5), spleen (lane 6), unfractionated marrow (lane 7), maturing marrow monocytes (lane 8), and marrow-derived neutrophils (lane 9) and the lymph node subpopulations—CD4+sCD3+ (lane 10) and CD4+sCD3− (lane 11)—were obtained at the time of the diagnosis of lymphoma. Different alleles are designated in size order, beginning with the letter a. (B) Identical chromosome 17 loci showed LOH in both primary leukemic and lymphoma cells. Bars represent a schematic of the patient's paternally derived chromosome 17 in bone marrow, lymph node, sCD3+ (phenotypically normal CD4+ T cells), and CD4+sCD3−(abnormal lymphoma) cells, designated as CD4+. The heterozygosity status of informative loci is indicated by the shaded bar for loci that lost heterozygosity or the open bar for loci that retained heterozygosity. Interlocus genetic distances in centiMorgans (cM) are indicated to the left of each interval as estimated from a chromosome 17 sex-averaged map (http://www.marshmed.org/genetics/). Data illustrating the retention of heterozygosity at the D17S805 locus and the loss of heterozygosity at the D17S1294 locus are shown to the right. DNA samples are as follows: lane 1, patient's unaffected father; lane 2, patient's mother affected with NF1; lane 3, patient's unfractionated bone marrow; lane 4, patient's lymph node; lane 5, CD3+ cells from the lymph node; lane 6, CD4+cells from the lymph node; positive and negative controls are not shown. For each locus, 2 alleles, designated simply as a and b, are segregating in this family. Differences in band intensity are caused by differences in the amount of DNA loaded per lane.
To further investigate whether JMML and lymphoma cells derived from a common progenitor, loci spanning the length of chromosome 17 were assayed for LOH. Although multiple loci showed LOH in JMML cells and lymphomatous lymph node cells (CD4+sCD3−), representative data for the D17S805 locus, which retained heterozygosity, and D17S1294, which lost heterozygosity, are shown in Figure 2B. The loci that lost heterozygosity were identical and spanned the long arm of the chromosome, a large region greater than 50 Mb in length (Figure 2B). In contrast, the normal CD4+sCD3+ T cells from the lymph node retained heterozygosity at all chromosome 17 loci tested. These data strongly implicate a single genetic event that resulted in loss of the normal paternal NF1 allele in a progenitor cell that gave rise to both the myeloid leukemia and the lymphoma clones.
The most likely genetic mechanism of LOH in this case is a recombination between D17S805 and D17S1294 of a maternal and a paternal chromatid during the S/G2 phase of the cell cycle of an ancestral cell. All possible recombinants would have 2 apparently normal chromosome 17 homologs, which is consistent with the results of the 2 independent cytogenetic normal analyses of bone marrow from our patient. One recombinant would carry the unaltered NF1 maternal chromosome and a paternal chromosome in which the 17q arm with theNF1+ allele had been replaced with the corresponding maternal 17q region carrying the NF1 allele. This putative progenitor cell would carry 2 maternal NF1alleles, 2 maternal alleles for the additional approximately 500 genes in the LOH region, and a single maternal and paternal allele for the remaining chromosome 17 loci. Therefore, the progenitor cell showed maternal isodisomy for nearly all loci at chromosome 17q. Isodisomy resulting from a single mitotic recombination is a common mechanism of LOH in other tumor types.17 18
Although NF1 allele loss was generally restricted to the myeloid compartment in previous studies of NF1-associated leukemia, it has been reported in B-lineage lymphoid cells, such as an Epstein-Barr virus (EBV)-transformed lymphoblastoid cell line derived from a child with JMML.6 Additional evidence that the stem cell giving rise to JMML has pluripotent potential comes from a patient with JMML, whose disease evolved to B-cell acute lymphocytic leukemia.9 Our data extend these reports by demonstrating clonal proliferation of malignant T-lymphoid lineage cells in a patient with JMML.
If the normal NF1 gene is inactivated in a pluripotent hematopoietic stem cell, it is unclear why children with NF1 are strongly predisposed to JMML but not to lymphoid malignancies. This observation is not restricted to humans; the adoptive transfer of murine Nf1-deficient fetal liver cells consistently induces a JMML-like myeloproliferative disorder without lymphoid abnormalities.19 Together, the human and murine data suggest that loss of NF1 (or Nf1) confers a proliferative advantage that is predominately expressed in the myeloid lineage. As suggested by a previous report20 showing polyclonality in the T-lymphoid lineage in a child with JMML, the emergence of a T-cell lymphoma may require the development of cooperating genetic mutations that are distinct from NF1inactivation.
NF1 encodes neurofibromin,21 and genetic and biochemical studies have shown that NF1 functions as a tumor suppressor gene in immature myeloid cells by negatively regulating Ras.22 Inactivation of NF1 is associated with the constitutive activation of Ras signaling resulting in hyperactivation in response to stem cell factor, IL-3, and granulocyte-macrophage colony-stimulating factor (GM-CSF).19 23-25 These data raise the possibility that the myeloproliferative phenotype seen in JMML is caused by deregulated Ras signaling in response to GM-CSF and other myeloid growth factors. Presumably, additional mutations within the aberrantNF1−/− clone of our patient resulted in the outgrowth of a malignant T-lymphoid precursor clinically manifested as a T-cell lymphoma.
In conclusion, analysis of this unusual patient provides insights into the clonal origins of JMML and the proliferation ofNF1-deficient hematopoietic cells, and our data support the hypothesis that at least some cases of JMML originate in a pluripotent hematopoietic stem cell.
Acknowledgment
We thank Beverley Tork-Storb for performing the CFU-GM cultures.
Supported by grants from the National Institutes of Health (CA72614 to K.M.S.; P30 HD28834 to the University of Washington Child Health Resource Center), the United States Army Medical Research and Material Command (NF960048 to K.S.), the National Cancer Institute (CA09351), and the Leukemia and Lymphoma Society of America.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Laurence J. N. Cooper, Clinical Research Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, D3-100, Seattle, WA 98109-4417; e-mail:lcooper@fhcrc.org.