Abstract
The human and the murine glycoprotein platelet IIb (GPIIb) promoters are megakaryocyte specific in human and murine cell systems, respectively. Here we show that the murine promoter is, however, highly active when transfected in K562 human cells in which the human promoter is almost inactive. A murine promoter, in which the enhancer element was replaced by the human, retrieves its megakaryocytic specificity in human cell lines. The human and murine GATA-binding sites located in the enhancer region display slight sequence divergence next to the consensus GATA core sequence. Gel shift experiments show that, although the murine and the human GATA sequences both bind GATA-1, the murine sequence alone forms an additional complex (B) not detected with the human sequence. When the murine GATA-containing region is replaced by the human in the context of the murine GPIIb promoter, megakaryocyte specificity is restored in the human cell lines. A G nucleotide 3′ to GATA appears crucial because its substitution abrogates B but not GATA-1 binding and restores megakaryocyte specificity to the murine promoter. Conversely, substitution of the human GATA-1 binding sequence by its murine homologue that binds both GATA-1 and complex B induces an abnormal activity for the human promoter in K562 cells. Altogether, our data suggest that limited changes in the GATA-containing enhancer of the GPIIb promoter can induce the recruitment of accessory proteins that could be involved in alteration of a megakaryocyte-restricted gene activation program.
Introduction
The mechanism by which a lineage-specific pattern of gene expression is progressively established during hematopoietic differentiation is still not completely understood. Recent studies have pointed to a growing number of transcription factors relevant to the developmental program leading to hematopoietic stem cell expansion and lineage determination. Gene targeting inactivation of SCL/Tal-1 and Rbtn-2 transcription factors leads to similar defects in early hematopoietic development.1-3 Other transcription factors are also important regulators later in the hematopoietic process. For example, GATA-1 is expressed in erythroid, megakaryocytic, mast, and eosinophilic cells,4-7 and a number of genes expressed in these cells contain GATA motifs in critical cis-regulatory elements.8,9 Gene targeting experiments of GATA-1 have led to a developmental arrest at the proerythroblastic stage and to apoptosis, resulting in embryonic lethality by day 11.5 of gestation.10-14 Megakaryocytes lacking GATA-1 by a lineage-selective knockout also arrest their maturation, but undergo proliferation rather than apoptosis.15 16 These results underline the critical role of GATA-1 in both megakaryocytes and erythroid lineage and show that the same transcription factor can be recruited for different cellular functions and may act at different stages in different lineages.
The combinatorial association of transcription factors may account for modulation of lineage differentiation. For example FOG, for friend of GATA, is a cofactor for GATA-1 that was shown to be crucial for normal erythroid and megakaryocytic differentiation.17 FOG inactivation revealed that FOG−/− erythroid cells displayed a blockage of maturation reminiscent of GATA-1−erythroid precursors. However, loss of FOG leads to the specific ablation of the megakaryocytic lineage at a very early stage, contrasting with the late blockage of megakaryocytic differentiation occurring with GATA-1 inactivation. This finding suggests that, although acting as an essential cofactor of GATA-1 in erythroid cells, FOG may act independently from GATA-1 in megakaryocytes.18This action illustrates that association between transcription factors can be modulated according to the cell type and to the differentiation stage, leading to specific functions in transcription regulation.
DNA cis-acting elements of gene promoters, the targets of these transcription factors, represent the other central actors in promoter activity regulation. It is probably the concerted differential engagement of cis elements according to the lineage context that regulate the fine-tuning of transcription. For example, most of the megakaryocytic gene promoters contain GATA and Ets binding sites, the association of which appears to be crucial for the control of the transcription level and cell specificity of these genes.19-22
Among them, the gene coding for glycoprotein platelet IIb (GPIIb, αIIb) is one of the most extensively studied. We had previously proposed a model explaining how GPIIb gene transcription is switched off in nonmegakaryocytic cells, whereas it is kept active all along the megakaryocyte differentiation.23 Indeed, GPIIb promoter contains an enhancer that bears GATA and Ets elements8 active in erythroid and megakaryocytic cells.19 The GPIIb promoter also contains ubiquitous positive cis-active elements. Both types of elements are smothered by the action of a potent repressor, depending on the cellular context. This repressor is also active in megakaryocytic cells, where it tunes the GPIIb gene transcriptional activity to a medium level. The transcription of GPIIb gene and its cell specificity is thus exquisitely regulated by the balance between a repressor and different positive cis-acting elements.23 This balance could be regulated during the differentiation of early hematopoietic progenitors and/or during the course of the embryonic development. GPIIb gene was for a long time thought to be a specific, although early, marker of megakaryocytes and platelets.24 However, its expression was detected on the common progenitor of erythroid and megakaryocytic cells.25 Recently, the expression of GPIIb-IIIa was detected on avian multilineage hematopoietic cells.26 This finding suggests that, although detected in early hematopoietic progenitors, GPIIb expression is turned off in the course of progenitor commitment. In parallel, GPIIb expression increases while the megakaryocytic differentiation program is engaged.24,27 28
In this paper, we show that the murine GPIIb promoter sequence and organization is very similar to that of the human. However, when placed in a heterologous context, ie, the murine promoter in human cell lines, the balance between the repressor and the positive elements is deregulated. We show that the sequences adjacent to the GATA binding site of the enhancer region bind an unknown DNA binding protein and play an important role in this imbalance. Whether the recruitment of an additional DNA-binding protein next to the GATA site is involved in the alteration of the regulation of the GPIIb promoter activity is discussed.
Materials and methods
Plasmid constructions
The basic chloramphenicol acetyl-transferase (CAT) plasmids used were pBLCAT3,29 pBLCAT2, and pRSVCAT (Promega Biotech, Madison, WI).30 A 129svj Mouse Genomic Library in the Lambda FIX®II Vector (Stratagene, La Jolla, CA) was screened with a previously described 583-base pair (bp) mouse genomic DNA probe.31 A positive clone was digested byEcoRI, yielding 2 genomic fragments that were subcloned into the pBluescriptII vector (Stratagene): a 3.1-kb fragment containing the region extending from −3500 to −320 according to the transcription start site, and a 1.3-kb fragment extending from −320 to +1000. Because the initiation translation start site is located at position +33 (the A of the ATG codon), the −320/+32 region was amplified by polymerase chain reaction (PCR), using the −320/+1000 fragment as a template and was cloned in a shuttle vector (TA-cloning, Invitrogen, San Diego, CA). The −3500/−320 EcoRI fragment was introduced in the 5′ EcoRI site of the −320/+32 fragment. The resulting −3500/+33 construct was digested byHindIII restriction enzyme, generating a −2700/+32 fragment, subcloned into the pBLCAT3 vector, upstream from the CAT gene. Further digestion of this 2700 construct produced a murine GPIIb genomic fragment, extending from −899, −538, −396 to +32, respectively. The human 813 construct was already described19 and contains the human GPIIb promoter sequences starting from position +33, relative to the transcription start site +1 to −813.32 The human −598/−406 GPIIb enhancer19 was PCR-amplified and linked to the −396/+32 region that was described above. One copy of the −598/−406 enhancer was inserted, in direct or reverse orientation. The cohesive ends of the −598/−406 and of the −396/+32 fragments were designed in a way such that the GATA and Ets elements of the human enhancer were inserted at the same position of the murine GATA (mGATA) and Ets sites.
Site-directed mutagenesis
Site-directed mutagenesis was performed on the murine −899 and human −813 GPIIb constructs, with the Transformer Site-directed Mutagenesis Kit (Clontech, Palo Alto, CA), according to the manufacturer's instructions. The mutated nucleotides are indicated in bold type, and consensus sequences for transcription factor binding sites are underlined. The oligonucleotide used to obtain the murine −456 GATA mutant of the murine promoter designated as −899 mG* was (−443)5′ AGCTGTTCTCCCC TTCTAAGACCAGAGG 3′(−470). Swapping of the mGATA sequence on the murine −899 construct by the human GATA (hGATA) sequence was obtained by using the oligonucleotide 5′ GTAAGCAAGCTGCTGCCCCCGATAAAA CCTGAGGCTGTCATCA 3′. Swapping of the hGATA sequence on the human −813 construct by the mGATA sequence was obtained by using the oligonucleotide 5′ CGGGGAAGGAGAAGGAAGCTGTTCTCCCCTGATAAGACCAG AGGCTTCTGTATC 3′.
Cell culture
HEL, K562, LIN-175, and MEL cells were grown in RPMI-1640 medium (Gibco-BRL, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal calf serum (FCS; ATGC, F), 2 mmol/L glutamine, and 100 U/mL penicillin/streptomycin (Gibco-BRL). Cells were grown at 37°C in a 5% CO2 incubator. HeLa and 3T3 cells were grown in DMEM medium (Gibco-BRL) in the same conditions. Human CD34+ progenitor cells were obtained from umbilical cord blood. Mononuclear cells were obtained by centrifugation on ficoll Lymphoprep (Nycomed Pharma, Gibco-BRL) and were then enriched for CD34+ cells by a 2-round separation procedure, using the CD34 magnetic cell isolation kit MiniMacs (Miltenyi-Biotech, Paris, France) (typically to greater than 95% purity). To obtain megakaryocytic differentiation, the CD34+ cells were cultured at a density of 3.105 cells/mL in Stemα (Tebu, France), with the following recombinant human (rH) cytokines (Preprotech Tebu, France): 50 ng/mL rH Tpo, 6 U/mL rH interleukin-3 (IL-3), 10 U/mL rH IL-6, and 4 U/mL rH IL-11, and incubated in 5% CO2 at 37°C. The erythroid differentiation of the CD34+ cells was performed as previously described.33 Megakaryocytic and erythroid cultures were harvested after 12 days. Human cell surface phenotype was determined by flow cytometry using PerCP-conjugated antihuman CD34 (Becton Dickinson, Mountain View, CA), fluorescein isothiocyanate (FITC)-conjugated antihuman glycophorin A (GPA) (Immunotech, France), and phycoerythrin (PE)-conjugated antihuman CD41 (DAKO, France) antibodies, according to the manufacturer's instructions.
Reverse transcription and PCR amplification of RNA
Total RNA was prepared from different cell lines and primary culture from CD34+ cells, using the TRIzol reagent (Gibco-BRL), according to the manufacturer's protocol.34Reverse transcription and PCR amplification was carried out as previously described.35 Oligonucleotide primers were synthesized by Eurobio (Les Ullis, France), according to the sequence information previously described for murine GPIIbM and HPRT,35 human GPIIbH,36 and GAPDH.37
DNA transfection
Plasmids were isolated by alkaline lysis and purified on anion exchange columns (Jetstar Genomed, Quantum Europe, France). Nonadherent cells were transfected by the electroporation method by using a gene pulser (BioRad Laboratories, Hercules, CA), as previously described.38 Adherent HeLa and 3T3 cells were transfected by the calcium phosphate method.39 The pRSV-luciferase plasmid (Promega Biotech) was used as an internal standard expressing firefly luciferase under the control of the Rous Sarcoma virus promoter.40
Luciferase and CAT assays
Cells were harvested 48 hours after transfection, and cell extracts were obtained by 3 cycles of freeze and thaw lysis. Luciferase activity of the extracts was measured using the Luciferase Assay system (Promega Biotech) and a luminometer (MicroLumat LB96P, EG&G Berthold) and expressed in arbitrary units. CAT assays were performed as described elsewhere.30 The amount of protein in the extract tested was normalized as a function of the luciferase activity. Acetylation of 14[C] chloramphenicol was determined by quantification of the radioactivity on thin-layer chromatography plates by phosphoImager analysis using the Image Quant software (Molecular Dynamics, Sunnyvale, CA). CAT activity corresponds to the percentage of conversion of chloramphenicol to acetylated forms. All assays were performed within the linear range of acetylation reaction.
Nuclear protein extracts
Nuclear protein extracts were prepared from the following cells: HEL, K562, LIN-175, MEL, human megakaryocytes, and erythroblasts obtained from CD34+ progenitor cells differentiation, according to the rapid method of Schreiber et al.41 They were quantified according to Bradford coloration protocol (BioRad) and stored at −80°C.
DNA probes
The synthetic oligonucleotides used in electrophoretic mobility shift assay (EMSA) experiments were synthesized by Eurobio (Les Ullis). One strand was 5′ end-labeled with T4 polynucleotide kinase and annealed with an excess of the nonlabeled complementary strand at a 1:4 ratio. All fragments were purified from unincorporated radioelements using Quick Spin Columns (Roche Molecular Biochemicals), and radioactivity incorporation was measured by scintillation counting (Beckton Dickinson LS-1800). The hGATA oligonucleotide containing the human −463 GATA site has already been described.8 Its sequence was (−451) 5′ AGCTGCTGCCCCCGATAAAACCTGAGG 3′(−477). The mGATA oligonucleotide sequence containing the murine −456 GATA site was (−443) 5′ AGCTGTTCTCCCCTGATAAGACCAGAGG 3′(−470). Consensus sequences for transcription factor binding sites are underlined. Other oligonucleotides were used to correspond mGATA and hGATA sequences with mutations (bold letters). Their corresponding sequences are reported in the figures of interest. The same oligonucleotides were used as cold competitors. The consensus Sp1-binding site from the SV40 early promoter was used as nonspecific cold competitor control.42
Electrophoretic mobility shift assays
The gel retardation assays were performed as already described,8 by a combination of the procedures of Halligan and Desiderio43 and of Singh et al.44 For competition studies, unlabeled competitors were added to the binding reaction and nuclear extracts, 5 minutes at room temperature, according to the ratio described in each experiment, prior to the addition of radioactive probes. Rat antimouse GATA-1 monoclonal and rabbit anti-human Stat6 polyclonal antibodies were purchased from Santa Cruz Biotechnology. For gel supershift assays, 2 μg of antibodies were added to the binding reaction and nuclear extracts, 2 hours at 4°C prior to the addition of radioactive probes.
Scatchard analysis and determination of dissociation constants (Kd)
EMSAs were performed with increasing known concentrations of the radiolabeled probes (0.02-6 ng), incubated with constant, nonsaturating amounts of nuclear proteins (10 μg). The amount of free and bound probes formed in binding reactions were quantified by phosphoImager (Molecular Dynamics) using the Image Quant software. Scatchard plot analyses were done as described.45 The ratio between Bound and Free DNA probe (B/F) versus specific DNA-protein binding concentration [Bound] (μmol/L) was plotted, and the straight lines were drawn by fitting the data using a linear regression, with the Scatchard equation: B/F = −(1/Kd) × [Bound] + Bmax/Kd, where Kd is the apparent equilibrium dissociation constant and Bmax is the maximal number of binding sites. The Kd value corresponds to (as the inverse of) the affinity of the probe for its protein-binding site, because it is equal to the probe concentration that yields half-maximal binding of protein. The Bmax value corresponds to the maximal number (density) of specific binding sites and thus depends on the amount of nuclear protein extract. They were calculated according to the Scatchard equation for each DNA/protein complexes formed.
Results
Murine, human, and rat GPIIb promoters display strong sequence homology and conservation of cis-acting elements
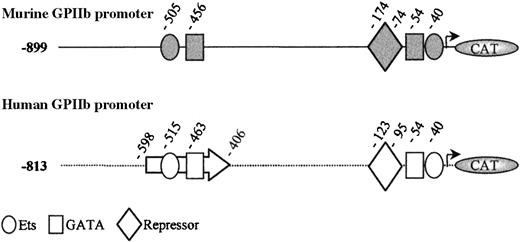
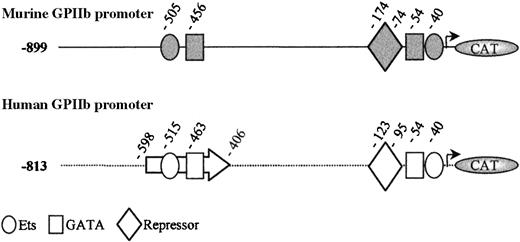
Alignment of the human −880/+3246 and murine −901/+32 GPIIb promoters yielded an overall identity score of 64% (data not shown). The previously described human −598/−406 enhancer region displays 71% homology with the murine −588/−396 region. As previously reported, the −456 GATA and −505 Ets sequences are well aligned with the −463 GATA and −515 Ets sites of the human promoter.31 We have previously demonstrated that these human sites bind GATA-1 and PU.1, respectively,8,47 and are responsible for the erythro-megakaryocytic activity of the human GPIIb enhancer.19 These sites have also been shown to be important for the rat GPIIb promoter function.28 The murine −174/−74 region is 60% homologous to the human −198/−81 region, containing a repressor element that was shown to be crucial for megakaryocyte specificity of the human promoter.23,48 A repressor element was also described on the −183/−70 region of the rat GPIIb promoter.49 The −56/+32 proximal domain of the murine promoter appeared highly conserved when compared with the human corresponding region (75% homology). This region contains the −54 GATA/−40 Ets tandem sites, which are active in the human and the rat promoters.8,49 50 Figure 1illustrates the similar organization between the murine and the human promoters.
Schematic representation of the murine (gray) and human (white) GPIIb promoter fragments extending from −899 and −813 to +32 bp, respectively.
Nucleotide position of the different sites and elements are indicated from the initiation start site. Boxes represent GATA sites; circles, Ets sites; and diamonds, the repressor elements. The human enhancer was delimited by an arrow.
Schematic representation of the murine (gray) and human (white) GPIIb promoter fragments extending from −899 and −813 to +32 bp, respectively.
Nucleotide position of the different sites and elements are indicated from the initiation start site. Boxes represent GATA sites; circles, Ets sites; and diamonds, the repressor elements. The human enhancer was delimited by an arrow.
Murine GPIIb promoter loses its megakaryocyte specificity in human cell lines
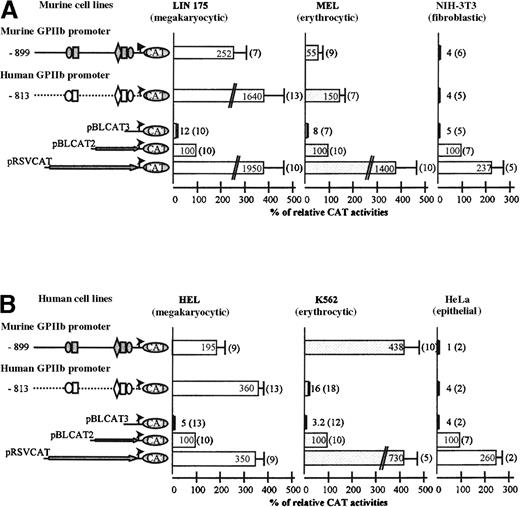
The cell-specificity of the murine −899/+33 GPIIb promoter fragment was analyzed in several cell lines (Figure2). The murine LIN-175 and human HEL cells were used as megakaryocytic cell lines that express GPIIb,51 murine MEL cells display erythroid features and do not express GPIIb,52 whereas human K562 cells display erythro-megakaryocytic features but with low GPIIb expression53 (Figure4B). Murine NIH-3T3 and human HeLa cells were used as nonhematopoietic controls. To make the results comparable between the different cell lines, the CAT activities of the promoter fragments were normalized by using the pBLCAT2 control vector activity as reference, arbitrarily taken as 100%.
Comparison of the transcriptional activities of the murine and human GPIIb promoter fragments, in murine (A) and human (B) cell lines.
Murine and human GPIIb promoters (−899/+33 and −813/+33, respectively) were cloned upstream of CAT gene in pBLCAT3 plasmid and transfected in different cell lines. The pRSVCAT plasmid containing the CAT gene driven by the RSV promoter was used as a positive control, and the promoter less pBLCAT3 plasmid was used to assess background level. In each assay, the pRSV-luciferase plasmid was cotransfected as an internal control of transfection. Cellular extracts were prepared 48 hours after transfection, and the CAT assays were normalized according to the luciferase activity of each extract. The CAT values were expressed relatively to the pBLCAT2 plasmid, containing the CAT gene driven by the ubiquitous TK promoter, which was taken as the 100% value to compare CAT activity between cell lines, and were presented in bar graph. Each value is the average of a number of independent experiments indicated in parentheses. (A) Transfection experiments in murine cell lines: LIN-175 (megakaryocytic), MEL (erythrocytic), and NIH-3T3 (fibroblastic). (B) Transfection experiments in human cell lines: HEL (megakaryocytic), K562 (erythrocytic), and HeLa (epithelial).
Comparison of the transcriptional activities of the murine and human GPIIb promoter fragments, in murine (A) and human (B) cell lines.
Murine and human GPIIb promoters (−899/+33 and −813/+33, respectively) were cloned upstream of CAT gene in pBLCAT3 plasmid and transfected in different cell lines. The pRSVCAT plasmid containing the CAT gene driven by the RSV promoter was used as a positive control, and the promoter less pBLCAT3 plasmid was used to assess background level. In each assay, the pRSV-luciferase plasmid was cotransfected as an internal control of transfection. Cellular extracts were prepared 48 hours after transfection, and the CAT assays were normalized according to the luciferase activity of each extract. The CAT values were expressed relatively to the pBLCAT2 plasmid, containing the CAT gene driven by the ubiquitous TK promoter, which was taken as the 100% value to compare CAT activity between cell lines, and were presented in bar graph. Each value is the average of a number of independent experiments indicated in parentheses. (A) Transfection experiments in murine cell lines: LIN-175 (megakaryocytic), MEL (erythrocytic), and NIH-3T3 (fibroblastic). (B) Transfection experiments in human cell lines: HEL (megakaryocytic), K562 (erythrocytic), and HeLa (epithelial).
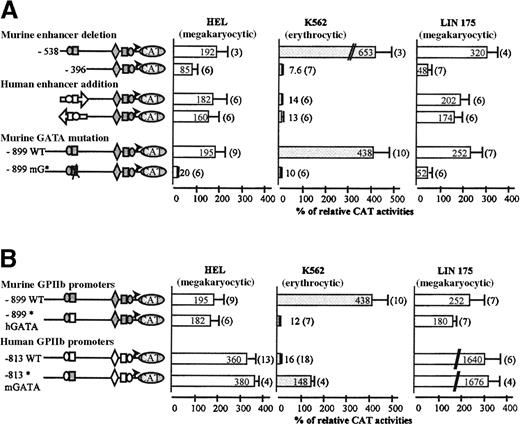
In murine megakaryocytic LIN-175 cells (Figure 2A), transfection of the 899 construct yielded 252% CAT activity, suggesting that this fragment contains all the elements essential for the transcriptional activity of the promoter. This fragment bears the −456 GATA and the −505 Ets binding sites (Figure 1), which is reminiscent of the −463 GATA and −515 Ets binding sites of the human GPIIb enhancer.19 Deletion of the −538/−396 region of the murine promoter resulted in a 5-fold decrease of the CAT activity, suggesting that this region is equivalent to the −598/−406 human enhancer region (Figure 3A). The transfected 899 construct yielded 5-fold lower activity in erythroid MEL (55%) compared with LIN-175 cells (252%), suggesting the existence of a repressor element controlling the megakaryocyte specificity, like in human GPIIb promoter (Figure 2A).23 Transfection of NIH-3T3 cells produced a low level of CAT activity, consistent with the megakaryocytic specificity of this promoter fragment in murine cell lines.
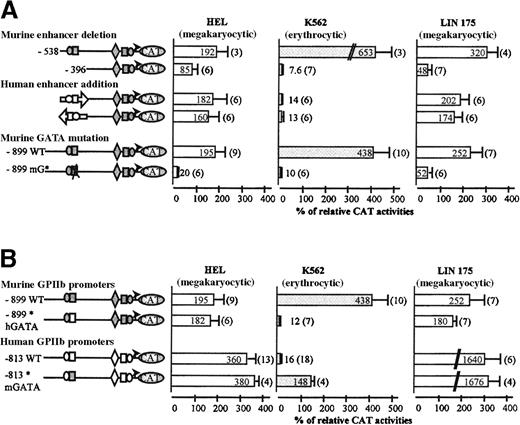
Role of the enhancer and GATA elements in the human and murine promoter activities.
Transcriptional activities were analyzed in HEL, K562, and LIN-175 cell lines as described in Figure 2. (A) The murine −538 and −396 GPIIb promoter constructs, extending from −538 and −396 to +32 respectively, were first analyzed. The human −598/−406 enhancer fragment generated by PCR and sequenced was inserted in direct or reverse orientation upstream from the murine −396 GPIIb promoter fragment. A disrupting mutation was introduced into the murine −456 GATA site of the murine −899 GPIIb promoter construct. Activity of this mutated promoter (−899 mG*) was compared with that of the wild-type (−899 WT) murine promoter. (B) The murine chimeric (−899* hGATA) construct with hGATA region (open box) and the human 813 chimeric (−813* mGATA) construct with mGATA region (gray-filled box) were transfected in HEL, K562, and LIN-175 cells. CAT activities are compared with that of the wild-type murine and human GPIIb promoter constructs as described in Figure 2.
Role of the enhancer and GATA elements in the human and murine promoter activities.
Transcriptional activities were analyzed in HEL, K562, and LIN-175 cell lines as described in Figure 2. (A) The murine −538 and −396 GPIIb promoter constructs, extending from −538 and −396 to +32 respectively, were first analyzed. The human −598/−406 enhancer fragment generated by PCR and sequenced was inserted in direct or reverse orientation upstream from the murine −396 GPIIb promoter fragment. A disrupting mutation was introduced into the murine −456 GATA site of the murine −899 GPIIb promoter construct. Activity of this mutated promoter (−899 mG*) was compared with that of the wild-type (−899 WT) murine promoter. (B) The murine chimeric (−899* hGATA) construct with hGATA region (open box) and the human 813 chimeric (−813* mGATA) construct with mGATA region (gray-filled box) were transfected in HEL, K562, and LIN-175 cells. CAT activities are compared with that of the wild-type murine and human GPIIb promoter constructs as described in Figure 2.
Because of strong conservation of cis-acting elements between the murine and the human GPIIb promoters and parallel tissue-specific regulation, we asked if the murine promoter would behave in a manner similar to the human promoter in human cell lines context. The murine 899 construct was transfected in the human HEL, K562, and HeLa cells and compared with the human 813 construct (Figure 2B). In HEL cells, the murine construct yielded an activity of 195%, standing within the same range than the human promoter (360%), confirming the activity of positive elements in the enhancer region. Interestingly, the murine 899 construct was highly active in K562 cells (438%), as opposed to the almost inactive human constructs (16%) (Figure 2B).32 23The same murine promoter fragment was much weaker in MEL cells (55%). The murine GPIIb promoter is thus deregulated in the human K562 cell context. This deregulation appears to be a specific feature of the erythro-megakaryocytic system, because, when transfected in HeLa cells, the murine 899 construct is inactive (1%). Conversely, we noted that the human 813 promoter was much more active in LIN-175 cells than in HEL cells (1640% versus 360%). However, this transcriptional activity remained megakaryocyte specific because its activity was low in mouse erythroid MEL cells and in nonhematopoietic NIH-3T3 cells, strongly suggesting that the megakaryocytic human promoter is adequately regulated in the murine context. This result contrasted with the deregulation of the murine promoter in human cell lines, which we decided to focus on.
Fusion of the human enhancer to the proximal murine promoter restores megakaryocyte specificity of the murine GPIIb promoter in K562 cells
Deletion analysis pointed to the −538/−396 region as an important regulatory element of the murine promoter (Figure 3A). Indeed the CAT activity decrease of the enhancer-deleted 396 construct, in HEL (2-fold) and in LIN-175 cells (7-fold), suggested that the murine −538/−396 sequence is active in human as well as in murine megakaryocytic contexts. Of more interest, when transfected in K562 cells, the murine 396 construct was almost inactive, suggesting that, in this cell line, the negative control of the enhancer strength is altered. To confirm this hypothesis, we then substituted the murine enhancer element by its human homologue (Figure 3A). The human enhancer was inserted either in direct or reverse orientations. When transfected in HEL or LIN-175 cells, both chimeric promoters displayed activities comparable to that of the wild-type murine GPIIb promoter. In LIN-175 cells, addition of the human enhancer to the enhancer-less construct induced a 4-fold increase of the CAT activity. These results confirm that the human enhancer is functional in the murine promoter context. Furthermore, when transfected in K562 cells, the chimeric constructs were almost inactive, indicating restoration of promoter tissue specificity by the human enhancer. Altogether these data suggest that deregulation of the murine GPIIb promoter in K562 cells involved the murine −538/−396 enhancer region.
GATA binding site located at position −456 is crucial for the enhancer activity of the murine GPIIb promoter
Sequence analysis of the murine −538/−396 fragment revealed a consensus sequence for a GATA binding site at position −456. In addition, the corresponding GATA sites of the human and rat GPIIb promoters were also shown to be potent cis-acting elements.19,28 Consequently, this site was examined for its potential transcriptional activity (Figure 3A). We introduced the mutation of two nucleotides (GA/TC) on murine −456 GATA site in the murine 899 construct (mG*). The same mutation on the hGATA site is known to abolish its enhancer activity.19 It also induced a decrease of the murine GPIIb promoter activity in LIN-175 cells (52% versus 252%) and in HEL cells (20% versus 195%). This mutation abolished the promoter activity in K562 cells (10% versus 438%), suggesting that transcriptional activity of the murine enhancer in the human K562 context involved the enhancer GATA sequence. The effect of this GATA mutation was comparable to that observed after deletion of the −538/−396 enhancer region.
Swapping of the murine −456 GATA region for its human counterpart is sufficient to restore megakaryocyte specificity of the murine GPIIb promoter
We designed a new chimeric mouse promoter in which 19 nucleotides in the vicinity of the mGATA sequence were substituted for their human equivalent (Figure 3B). This substitution generated 4 nucleotide differences 5′ to GATA, and 2 in 3′, without affecting the GATA site. The rest of the murine GPIIb promoter remained unchanged. This mutation induced an important drop in the CAT activity in K562 cells (12%), when compared with the wild-type murine promoter (438%) (Figure 3B). The activity of this GATA-mutated murine promoter was comparable to the wild-type human promoter (16%). Moreover, this mutant was almost as active as the murine wild-type promoter in HEL or in LIN-175 cells. Thus, substitution of the mGATA sequence for the hGATA rescues the megakaryocytic-specific activity of the murine GPIIb promoter in human cell lines. Conversely, the GATA-containing sequence of the human promoter was swapped for the murine corresponding sequence. This exchange induced a 9-fold increase in transcription of the resulting chimeric promoter in K562 cells (148%) when compared with the wild-type human promoter (16%). The activity of this chimeric promoter was not affected in HEL or in LIN-175 cells. These data are thus consistent with the involvement of a 19 bases GATA-centered sequence of the enhancer in erythro-megakaryocytic regulation.
Comparative EMSAs of the human −463 and the murine −456 GATA-containing sequences
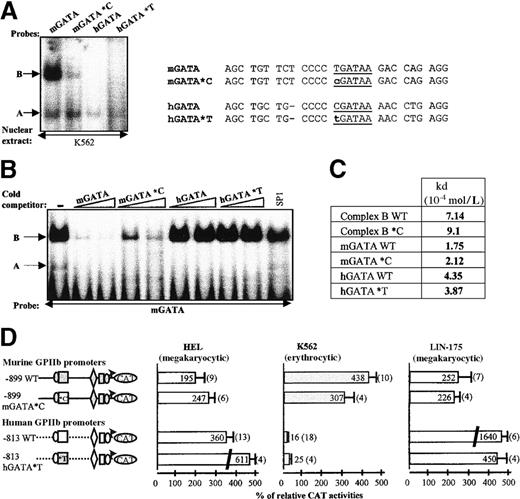
To check for the involvement of GATA DNA binding factors in the transcriptional activity of the murine GPIIb promoter, we carried out EMSAs by using probes encompassing the murine and the human enhancer GATA site. Human cord blood CD34+ hematopoietic cells were differentiated into almost pure erythroblasts or megakaryocytes (Figure4A). The RT-PCR experiment (Figure 4B) showed a band of 275 bp (GPIIb H), confirming that human GPIIb messenger RNA is expressed in megakaryocytes and HEL cells, whereas a faint band was detected in K562 cells, confirming low expression in these cells. No GPIIb signal was detected with RNA from erythroblasts. Similarly, a band of 170 bp (GPIIb M) corresponding to murine GPIIb was observed with LIN-175 RNA, and this band was not observed with MEL RNA. Nuclear extracts prepared from the megakaryocytic and erythroblastic primary cells were thus used in EMSA experiments to compare the DNA protein complexes obtained either with the human −463 or the murine −456 GATA sites (Figure 4C). The hGATA probe was previously shown to act as a specific binding sequence for GATA-1 protein.8 Indeed, one single band designated as A shifted with megakaryocyte extracts and in particular in erythroblasts known to produce high amounts of GATA-1. The same band was also detected with the mGATA probe. In addition, a slower migrating band designated as B and yielding a strong signal was observed with the murine probe but was not detected with the human probe.
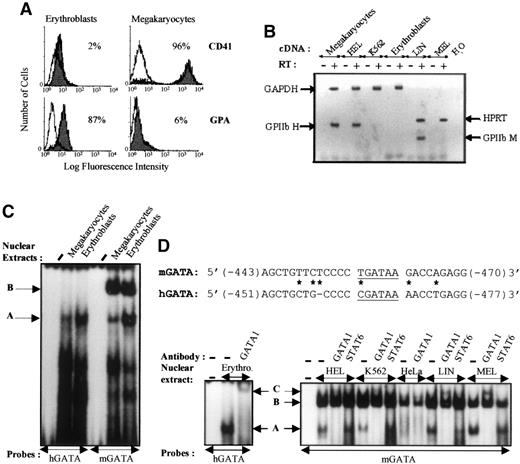
EMSA analysis of the hGATA and mGATA sites of GPIIb promoters.
(A) FACS analysis of erythrocytic (GPA) and megakaryocytic (CD41) cell surface antigen expression on CD34+ progenitor cells isolated from human umbilical cord blood and induced to differentiate into erythroid and megakaryocytic cells. After 12 days of culture, the cells were stained with FITC-antihuman GPA and PE-antihuman CD41 antibodies. Isotype-matched nonspecific antibodies were used as controls. Percentages of positive cells are indicated. (B) RT-PCR analysis of GPIIb expression. Total RNA was isolated from primary erythroid and megakaryocytic cells and from permanent cell lines (HEL, K562, LIN-175, and MEL). RT-PCR reactions were performed, including reverse transcriptase negative control (RT−) for each sample and H2O blank for each oligonucleotide primer. Amplification of GAPDH and HPRT was performed on each human and murine cDNA sample respectively, as an internal standard. (C) GATA-binding activity analysis. Cell specificity of the human −463 (hGATA) and murine −456 (mGATA) sites were analyzed with nuclear extracts from human megakaryocytes and erythroblasts. Sequence comparison between hGATA (previously described8) and mGATA probes is reported. The GATA-1 consensus binding sites are underlined. An asterisk represents nucleotide mismatch between the 2 sequences. Position of the DNA-protein complexes in the EMSA experiments are indicated by arrows (A and B). (D) Cell specificity and supershift assays. The mGATA sequence binding activity was analyzed with human nuclear extracts from HEL, K562, HeLa, and murine nuclear extract from LIN-175 and MEL, in the absence (−) or the presence of the indicated antibody (right panel). Positive control indicating positions of the DNA-protein complexes (arrows) was obtained with the human probe and nuclear extract from erythroid cells, in the absence (band A corresponding to GATA-1 binding) and in the presence (band C, supershift) of anti–GATA-1 antibodies (left panel).
EMSA analysis of the hGATA and mGATA sites of GPIIb promoters.
(A) FACS analysis of erythrocytic (GPA) and megakaryocytic (CD41) cell surface antigen expression on CD34+ progenitor cells isolated from human umbilical cord blood and induced to differentiate into erythroid and megakaryocytic cells. After 12 days of culture, the cells were stained with FITC-antihuman GPA and PE-antihuman CD41 antibodies. Isotype-matched nonspecific antibodies were used as controls. Percentages of positive cells are indicated. (B) RT-PCR analysis of GPIIb expression. Total RNA was isolated from primary erythroid and megakaryocytic cells and from permanent cell lines (HEL, K562, LIN-175, and MEL). RT-PCR reactions were performed, including reverse transcriptase negative control (RT−) for each sample and H2O blank for each oligonucleotide primer. Amplification of GAPDH and HPRT was performed on each human and murine cDNA sample respectively, as an internal standard. (C) GATA-binding activity analysis. Cell specificity of the human −463 (hGATA) and murine −456 (mGATA) sites were analyzed with nuclear extracts from human megakaryocytes and erythroblasts. Sequence comparison between hGATA (previously described8) and mGATA probes is reported. The GATA-1 consensus binding sites are underlined. An asterisk represents nucleotide mismatch between the 2 sequences. Position of the DNA-protein complexes in the EMSA experiments are indicated by arrows (A and B). (D) Cell specificity and supershift assays. The mGATA sequence binding activity was analyzed with human nuclear extracts from HEL, K562, HeLa, and murine nuclear extract from LIN-175 and MEL, in the absence (−) or the presence of the indicated antibody (right panel). Positive control indicating positions of the DNA-protein complexes (arrows) was obtained with the human probe and nuclear extract from erythroid cells, in the absence (band A corresponding to GATA-1 binding) and in the presence (band C, supershift) of anti–GATA-1 antibodies (left panel).
By using erythroblast extracts, we showed that the A band obtained with hGATA was supershifted by anti–GATA-1 antibodies, confirming that A complex involves GATA-1 protein (Figure 4D). When the mGATA probe was used, a band comigrating with A complex was detected with extracts from HEL, K562, LIN-175, and MEL cells, but not with HeLa cell extracts, suggesting that the murine −456 GATA sequence binds both the murine and the human GATA-1 proteins. This finding was further confirmed by interaction of this complex with anti–GATA-1 antibodies that recognize both proteins, whereas it was not affected by addition of anti-STAT6 antibodies used as negative control (Figure 4D). Moreover, B complex was observed in all nuclear extracts tested, including HeLa extracts, suggesting that it corresponds to a ubiquitous protein(s). B complex was not affected by the anti GATA-1 antibodies, showing that A and B are distinct proteins.
From our results, we can hypothesize that the abnormal activity of the murine promoter in K562 cells (1) is associated with the ability of the mGATA sequence to form B complex or (2) is linked to differences in affinity of GATA-1 binding to the murine and human GATA sequences. These hypotheses are not mutually exclusive. In the next experiments, we tested these 2 possibilities.
Demonstration of differential affinity of the murine −456 and the human −463 GATA-containing sequences for human GATA-1 protein
The hGATA- and mGATA probes both bound GATA-1 protein. However, we observed that the murine (TGATAA) sequence is closer to the GATA consensus sequence ([T/G]GATA[G/A]) than the human (CGATAA) sequence. Thus the murine −456 GATA site may have a higher affinity than the hGATA site for human GATA-1, a possible explanation for the escape from megakaryocytic-specific restriction of the murine GPIIb promoter in the human K562 cell line. To test this hypothesis, we carried out EMSA experiments with cross-cold competition and Scatchard plot analysis to determine and compare the affinity of GATA-1 for the murine −456 and human −463 GATA sequences (Figure 5). Cold competition experiments were performed with the labeled human probe in the presence of an excess of either homologous or murine cold sequences (Figure 5A). Both cold sequences competed away the band A, confirming it corresponds to GATA-1, but competition with the murine sequence appeared to be slightly more effective than that observed with the human sequence. Competition was specific because it was not observed with a 200-fold excess of SP1 sequence. Similarly, the binding of GATA-1 to the labeled mGATA probe was competed away by an excess of both the human and murine cold sequences (Figure 5A). When comparing both competitors, we again observed a more effective decrease of the band intensity with the murine competitor. These results suggest that the murine sequence has a greater affinity for hGATA-1 than the human sequences. Interestingly, the B band, observed only with the murine probe, began to be competed away by a 20-fold excess of the murine competitor and was washed off with an 100-fold excess. Moreover, neither the hGATA-containing nor the SP1 sequences were able to compete this B band away, even at a 200-fold excess. This finding confirms that B interaction with the murine probe is specific, and not related to GATA-1.
Competition and Scatchard analysis of DNA binding activities of the hGATA and mGATA sequences.
(A) Competitive gel mobility shift assay. Nuclear extracts from K562 cells were incubated with labeled hGATA or mGATA probes, in the absence (−), or in the presence of 5-, 10-, 20-, 100-, and 200-fold molar excess of unlabeled hGATA or mGATA competitor, and 200-fold molar excess of SP1 cold competitor. A and B complexes are indicated by arrows. (B) Titration of GATA-1 and B complex with hGATA and mGATA probe in EMSA. Constant, nonsaturating amounts of K562 nuclear extract (10 μg) were incubated with increasing concentrations of hGATA and mGATA probes (0.02-6 ng) and were resolved in EMSA. The specific DNA-protein complexes indicated by the arrows (Bound A and Bound B) and the free probes at the bottom were quantified by phosphoImager. (C) Scatchard plot analysis. The ratio between bound and free DNA probe (B/F) versus specific DNA-protein binding concentration (bound [μmol/L]) was plotted, and the straight lines were drawn by fitting the data using a linear regression, with the Scatchard equation: B/F = −(1/Kd)[Bound] + Bmax/Kd, where Kd is the apparent equilibrium dissociation constant, and Bmax is the maximal number of binding sites. The Kd and Bmax values were calculated for each DNA/protein complex.
Competition and Scatchard analysis of DNA binding activities of the hGATA and mGATA sequences.
(A) Competitive gel mobility shift assay. Nuclear extracts from K562 cells were incubated with labeled hGATA or mGATA probes, in the absence (−), or in the presence of 5-, 10-, 20-, 100-, and 200-fold molar excess of unlabeled hGATA or mGATA competitor, and 200-fold molar excess of SP1 cold competitor. A and B complexes are indicated by arrows. (B) Titration of GATA-1 and B complex with hGATA and mGATA probe in EMSA. Constant, nonsaturating amounts of K562 nuclear extract (10 μg) were incubated with increasing concentrations of hGATA and mGATA probes (0.02-6 ng) and were resolved in EMSA. The specific DNA-protein complexes indicated by the arrows (Bound A and Bound B) and the free probes at the bottom were quantified by phosphoImager. (C) Scatchard plot analysis. The ratio between bound and free DNA probe (B/F) versus specific DNA-protein binding concentration (bound [μmol/L]) was plotted, and the straight lines were drawn by fitting the data using a linear regression, with the Scatchard equation: B/F = −(1/Kd)[Bound] + Bmax/Kd, where Kd is the apparent equilibrium dissociation constant, and Bmax is the maximal number of binding sites. The Kd and Bmax values were calculated for each DNA/protein complex.
Because of the differences observed in relative DNA-binding activity between the mGATA and the hGATA probes, we determined the dissociation rate constants (Kd) of each probe by Scatchard plot analysis. After separation of bound and free DNA molecules by EMSA (Figure 5B), the amount of DNA in each band was quantified, and the fraction of bound DNA (B/F) expressed as a function of the retarded DNA concentration (B μmol/L) was represented on a Scatchard plot (Figure5C). Experimental values obtained yielded 3 straight lines, corresponding to the 3 specific DNA-protein complexes, hGATA, mGATA, and B complex. Kd values were calculated for each complex, according to the Scatchard equation. These Kd values were 1.75 × 10−4 mol/L for mGATA, 4.35 × 10−4 mol/L for hGATA, and 7.14 × 10−4 mol/L for B complex. We thus confirmed that the mGATA probe binds GATA-1 with a slightly higher (2.5-fold) affinity than the human. This difference in affinity could be linked to differences in the core sequence of the 2 GATA sites or to other more distal nucleotides. This question was addressed in the next experiments. The Kd value of the B complex is within the same range of mGATA and hGATA, consistent with specificity of the murine probe/B complex interaction.
Comparative mutational analysis of the hGATAand mGATA-core sequences
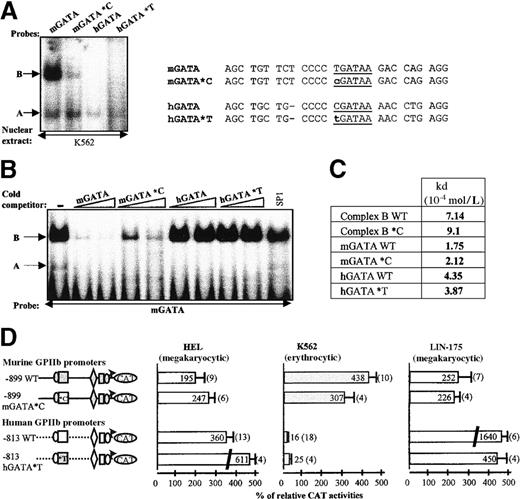
We first substituted the T located 5′ to the mGATA sequence by a C (mGATA*C probe, Figure 6A). We observed that GATA-1 binding was not affected by this substitution, whereas the intensity of the B complex binding decreased. Conversely, when the C located 5′ to the hGATA site was substituted by a T (hGATA*T probe), GATA-1 binding remained clearly detectable, and no B complex was detected. The T nucleotide 5′ to the mGATA site is thus involved in B complex interaction but is not sufficient per se.
Effect of T/C substitution 5′ to the mGATA and hGATA sequences as assessed by EMSA and transfection analysis.
(A) EMSA. DNA binding activities of wild-type mGATA and hGATA probes are compared with that of mutated mGATA*C and hGATA*T probes in EMSA, using K562 nuclear extracts. A and B complexes are indicated by arrows. Probe sequences are described (right panel). (B) Competitive gel mobility shift assay. Nuclear extracts from K562 cells were incubated with labeled mGATA probe, in the absence (−), or in the presence of 100- and 200-fold molar excess of the unlabeled indicated competitor, and 200-fold molar excess of SP1 cold competitor. Bands A and B are indicated by arrows. (C) Titration of GATA-1 and B complex with mGATA*C and hGATA*T probes in EMSA. Binding affinity studies of mGATA*C and hGATA*T probes were performed by Scatchard plot analysis as described for wild-type mGATA and hGATA DNA probe in Figure 5. The Kd values were calculated for each DNA/protein complex. (D) Functional studies by transfection. In the murine −899 mGATA*C construct, the murineTGATAA sequence was replaced by humanCGATAA sequence. In the human −813 hGATA*T construct, the human CGATAA sequence was replaced by the murineTGATAA. The murine 899 mGATA*C with hGATA site (open box *C) and the human 813 hGATA*T construct with mGATA site (gray-filled box *T) were transfected in HEL, K562, and LIN-175 cells. CAT activities are compared with the wild-type murine and human GPIIb promoter constructs.
Effect of T/C substitution 5′ to the mGATA and hGATA sequences as assessed by EMSA and transfection analysis.
(A) EMSA. DNA binding activities of wild-type mGATA and hGATA probes are compared with that of mutated mGATA*C and hGATA*T probes in EMSA, using K562 nuclear extracts. A and B complexes are indicated by arrows. Probe sequences are described (right panel). (B) Competitive gel mobility shift assay. Nuclear extracts from K562 cells were incubated with labeled mGATA probe, in the absence (−), or in the presence of 100- and 200-fold molar excess of the unlabeled indicated competitor, and 200-fold molar excess of SP1 cold competitor. Bands A and B are indicated by arrows. (C) Titration of GATA-1 and B complex with mGATA*C and hGATA*T probes in EMSA. Binding affinity studies of mGATA*C and hGATA*T probes were performed by Scatchard plot analysis as described for wild-type mGATA and hGATA DNA probe in Figure 5. The Kd values were calculated for each DNA/protein complex. (D) Functional studies by transfection. In the murine −899 mGATA*C construct, the murineTGATAA sequence was replaced by humanCGATAA sequence. In the human −813 hGATA*T construct, the human CGATAA sequence was replaced by the murineTGATAA. The murine 899 mGATA*C with hGATA site (open box *C) and the human 813 hGATA*T construct with mGATA site (gray-filled box *T) were transfected in HEL, K562, and LIN-175 cells. CAT activities are compared with the wild-type murine and human GPIIb promoter constructs.
Next we showed that cold mGATA*C probe was less effective than the wild-type mGATA probe (mGATA WT) in competing B complex away (Figure6B). Neither the wild-type hGATA (hGATA WT) nor the hGATA*T probes were able to compete B complex away. All cold GATA probes washed the GATA-1 complex out. However, the Kd values of the complexes obtained with the mutated and wild-type probes are in the same range. This finding indicated that C/T substitution only slightly affects GATA-1 or B, protein affinity for these probes.
We then introduced these mutations into the human and the murine promoters, respectively (Figure 6D). In the murine promoter, the T/C substitution slightly lowered its high promoter activity in K562 cells (307% versus 438%) but did not affect it in HEL or LIN-175 cells. The C/T substitution also slightly increased the activity of the human promoter in K562 cells and induced a 2-fold increase (360% versus 611%) in HEL cells, maybe because of increased GATA-1 binding. Finally, the C/T mutation in the human promoter induced a drop of activity in the murine LIN-175 cells when compared with the wild-type human promoter (450% versus 1640%).
Taken together, our results may indicate that the slight differences in GATA-1 affinity for the GPIIb promoters as induced by the T/C substitutions may be responsible for altering promoter activity and cell specificity in K562 cells. However, because these mutations also affected B affinity, a role for B appears equally likely.
Correlation between B complex interaction with GATA sequence and GPIIb promoter activity
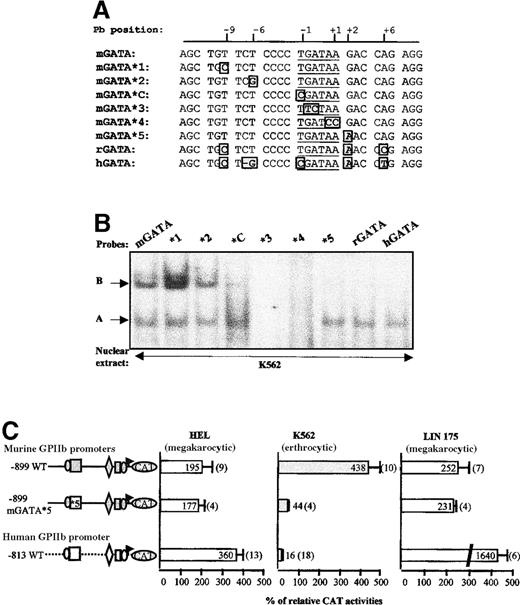
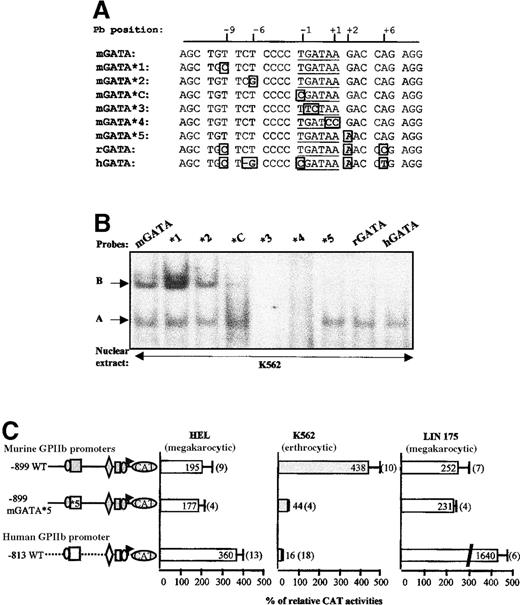
The major difference between the hGATA and the mGATA sequences is the specific interaction of B complex with the murine sequence. We thus tried to identify the nucleotides essential for this complex formation. Interestingly, the rat GPIIb promoter sequence exhibits only 3 nucleotide mismatches with the murine sequence in the GATA region (Figure 7A). Thus, we explored its DNA-binding activity by EMSA, using the rGATA probe, along with mouse and human probes (Figure 7B). Nine nucleotides upstream from the GATA sequence, the murine sequence contains a T instead of a C in the human or the rat sequences (Figure 7A). When this T was substituted by a C in the murine sequence (mGATA*1), B complex was enhanced in EMSA analysis (Figure 7B). In the next probe (mGATA*2), the T in position −6 was substituted by a G, as in the human sequence. This mutation had no effect on both GATA-1 and B complex formation. As already observed, the mutation of the upstream flanking nucleotide of the GATA sequence (T/C substitution, mGATA*C) affected B complex, that was, however, still visible. The next 2 mutations (mGATA*3 and *4) were located inside the GATA core sequence. As expected, GATA-1 did not bind to these sequences. These mutants were also unable to support B complex, suggesting that GATA is part of the B binding site. Finally, in mutant mGATA*5, the G downstream from the GATAA sequence was substituted by an A, as in the human and the rat sequences. Interestingly, this mutant was able to form a complex with GATA-1, whereas B complex was barely visible. Therefore, these mutation scanning experiments show that, although overlapping, GATA-1 and B binding sites are distinct.
Identification of the nucleotides involved in B complex formation.
(A) Scan of the murine enhancer GATA region. The sequence of mGATA*C (previously described) and of 5 mutated probes (mGATA *1 to *5), scanning the murine enhancer GATA region, were compared with that of the wild-type murine (mGATA), rat (rGATA), and human (hGATA) equivalent sequences. For each probe, nucleotides different from the mGATA sequence are boxed. (B) EMSA. The different probes described in panel A were then used in EMSA experiments with nuclear extracts from K562 cells. B and A complexes are indicated by arrows. (C) Functional studies by transfection. The nucleotide abrogating B complex formation as shown in panel B (mGATA*5) was introduced in the −899 murine GPIIb promoter construct. The CAT activity of this mutated promoter (−899 mGATA*5) was compared with that of the wild-type murine and human promoters by transfection of HEL, K562, and LIN-175 cells.
Identification of the nucleotides involved in B complex formation.
(A) Scan of the murine enhancer GATA region. The sequence of mGATA*C (previously described) and of 5 mutated probes (mGATA *1 to *5), scanning the murine enhancer GATA region, were compared with that of the wild-type murine (mGATA), rat (rGATA), and human (hGATA) equivalent sequences. For each probe, nucleotides different from the mGATA sequence are boxed. (B) EMSA. The different probes described in panel A were then used in EMSA experiments with nuclear extracts from K562 cells. B and A complexes are indicated by arrows. (C) Functional studies by transfection. The nucleotide abrogating B complex formation as shown in panel B (mGATA*5) was introduced in the −899 murine GPIIb promoter construct. The CAT activity of this mutated promoter (−899 mGATA*5) was compared with that of the wild-type murine and human promoters by transfection of HEL, K562, and LIN-175 cells.
The mGATA*5 mutation was then introduced in the murine GPIIb promoter (Figure 7C). This mutation, which prevents B complex formation while GATA-1 binding remains unaffected, induced an important decrease of the CAT activity in K562 cells (44% versus 438% for the murine wild-type promoter). This mutation did not affect the murine promoter activity in HEL and LIN-175 cells. Thus, the correlation between B-binding inhibition by mGATA*5 mutation and restoration of specificity of the murine GPIIb promoter in K562 strongly suggests involvement of B complex in the abnormal activity of the murine promoter in human K562 cells.
Discussion
Our goal is to characterize the general features of transcriptional control by megakaryocytes. An important body of evidence on the control mechanisms of tissue-specific transcription in megakaryocytes was brought about by studies of the GPIIb promoter in human cell lines23 or in rat bone marrow primary culture.49 In particular, it was shown that, although transcription of the GPIIb gene is under the control of erythro-megakaryocytic transcription factors, its expression is restricted to megakaryocytes.23 In this paper, we confirm that the murine GPIIb promoter is also megakaryocyte specific in murine cellular context. When compared, the human and murine GPIIb promoters display an overall 64% homology, suggesting a high conservation through evolution, presumably because of highly functionally relevant sequence domains. However, they exhibit some differences as well. We thus addressed the question of whether these differences could lead to transcriptional control alterations, which could provide us with clues for a better understanding of the underlying transcription control mechanism of a megakaryocytic specific promoter. We thus checked the activity of the human promoter in murine cell context and conversely of the murine promoter in human context.
When transfected in murine context, the human promoter preserved its megakaryocytic cell specificity because it is much more active in LIN-175 cells than in MEL or NIH-3T3 cell lines. We observed a high activity of the human enhancer in the murine megakaryocytic context that we have found to be linked to the human −463 GATA site (Albanese et al, unpublished data). However, this GATA hyperactivity does not affect regulation of cell specificity, because the human promoter was only weakly active in murine erythroid MEL cells. Next we studied the murine GPIIb promoter and showed that it is active and megakaryocyte specific in murine cell lines, like the human promoter in human cell lines. However, the murine GPIIb promoter lost its tissue specificity when transfected in the human K562 cell line in which it exhibited even a higher expression level than in HEL cell line, whereas the human GPIIb promoter was almost inactive in K562 cells. The murine promoter was inactive in KG1 cells (not shown) and in HeLa cells, indicating that the deregulation observed with this promoter only affects the erythro-megakaryocytic system.
By progressive deletion analysis of the murine GPIIb promoter, we localized an enhancer region (not shown), homologous to the erythro-megakaryocytic human enhancer. Deletion of this enhancer from the murine promoter led to a drop in activity in K562 to a level comparable to the human. The same drop in activity in K562 cells was also obtained when this murine enhancer region was replaced by its human counterpart, whereas, in LIN-175 or HEL megakaryocytic context, this chimeric promoter essentially behaved like the wild-type murine promoter. These data thus confirmed the involvement of the murine enhancer in cross-species erythroid deregulation. The activity of the human GPIIb promoter is dependent on 2 essential GATA and Ets elements.19,50 In the murine promoter, point mutation disruption analysis showed that the erythro-megakaryocytic enhancer activity was dependent on the −456 GATA site but not on the −505 Ets (Albanese et al, unpublished data). This result is different from the concerted engagement of both elements required for full activity of the human enhancer19 (see discussion below). Nevertheless these results clearly suggest that deregulation of the murine GPIIb promoter in the human K562 context is essentially supported by the GATA site within the enhancer region.
To confirm the role of the GATA site per se in the deregulation, we substituted the 19 nucleotide-long region centered around the murine −456 GATA site for the corresponding human sequence. The resulting chimeric hGATA/murine promoter exhibited features identical to those of the murine promoter substituted with the entire human enhancer region, because it was (1) almost inactive in K562 cells and (2) active in LIN-175 and HEL cells. Thus, our results clearly demonstrate that the human −463 GATA region can correct the abnormal activity of the murine GPIIb promoter in K562 cells. Conversely, the murine −456 GATA sequence in the human promoter, although not affecting megakaryocytic regulation in HEL, induced a strong deregulation of the human promoter in K562 (9.2-fold increase over wild type), mimicking the murine deregulation. This result pointed to the 19 base-long GATA-centered sequence as a critical element in transcriptional deregulation of the murine GPIIb promoter in the human K562 context.
To compare the DNA binding activities of the murine −456 and the human −463 GATA sites, we carried out EMSA experiments. We first confirmed that both sequences bound GATA-1 whether in HEL, K562, LIN-175, or MEL cells. In addition to GATA-1, the murine −456 GATA formed an additional complex (termed B) of higher molecular weight. The B complex was observed in all nuclear extracts tested, including HeLa cells, suggesting B complex is ubiquitously expressed. B complex did not interact with anti–GATA-1 antibodies, indicating it is different from GATA-1. The B complex was specific to the murine −456 GATA-containing sequence, because it was washed off by cold murine −456 GATA oligonucleotide but not by the human −463 GATA oligonucleotide. Moreover, B complex was also detected in erythroid and megakaryocytes from primary cultures of human cord blood hematopoietic cells, strongly suggesting it was not an artifact linked to permanent leukemic cell lines.
This high activity of the mGATA sequence can either be due to a greater affinity for GATA-1 or linked to the B complex formation. EMSA experiments followed by Scatchard plot analysis showed that, although standing within the same range, the human probe exhibited a 2.5-fold higher Kd and, therefore, lower affinity for GATA-1 than the murine probe using K562 nuclear extracts. Interestingly, the murine −456 GATA sequence (TGATAA) matches the GATA canonical sequence ([T/A]GATA[G/A]) more closely than the human −463 GATA sequence (CGATAA). To test whether this nucleotide difference might affect GATA-1 affinity, we performed EMSA experiments and functional studies using swapped GATA sequences in which the T was replaced by a C in the murine probe (mGATA*C) and the C by a T in the human (hGATA*T). These mutations affected only slightly the GATA-1 affinity for the hGATA and mGATA sequences without affecting significantly the murine and human GPIIb promoter activities, strongly suggesting that deregulation in K562 may not be accounted for solely by GATA-1 affinity differences.
Another effect of the T to C substitution in the mGATA sequence was a reduction in binding affinity of the B complex. To test its potential involvement in deregulation, we identified the nucleotides necessary for B complex formation. Among several positions, the G nucleotide located two bases downstream from GATA appeared unique in that its mutation into an A (the conserved nucleotide in the non-B binding human and rat sequences) disrupted B but not GATA-1 binding. Moreover, when introduced in the murine promoter, deregulation of this promoter in K562 cells was fully corrected, without affecting activity in HEL and LIN-175 cells. This finding provides strong evidence for involvement of B complex formation in the loss of specificity of the murine promoter in K562 cells.
Interestingly, K562 and HEL cells exhibit a very similar phenotype, because they both display erythrocytic and myeloid markers.53,54 Undifferentiated K562 cells also express megakaryocytic markers but at a low level. This level is, however, significantly increased when these cells are treated with phorbol esters, while simultaneously the expression of the erythroid and other myeloid markers decrease.55 In these conditions of induction, the expression of endogenous GPIIb gene and of a reporter gene driven by GPIIb promoter increases, confirming that K562 cells undergo megakaryocytic differentiation under phorbol ester treatment. Moreover, without any differentiating treatment, HEL cells express a high level of GPIIb, suggesting that these cells display more megakaryocytic features than undifferentiated K562 cells. Whether K562 cells have a more immature phenotype than HEL cells or whether they are more engaged into the erythroid lineage is not clear. In this context, the fact that the homologous murine and human GPIIb promoters behave differently in 2 very similar cell lines confirms that the transcriptional control of the megakaryocyte specific expression is tightly regulated.
Our results suggest that the human and the murine promoters share canonical functional features, namely GATA-1 as a major transactivating factor. However, they also differ significantly in that they probably use different GATA-1 partners. Supporting this hypothesis is our observation that the enhancer Ets binding site shown to be crucial for the human enhancer activity47,50 was inactive in the murine enhancer (Albanese et al, unpublished data). From our experiments, it is tempting to speculate that in the case of the mouse GPIIb promoter GATA-1/B complex interactions may be an alternative to GATA-1/ETS cooperation for the human GPIIb promoter in human cell lines. FOG is another important GATA partner that has been shown to be required for both erythroid and megakaryocytic gene activation programs.17 18 Whether FOG is part of B complex remains to be assessed, as well as the molecular interplay between B complex, GATA-1, and FOG in murine and human megakaryocytes.
Interestingly, although apparently not required for human GPIIb transcriptional regulation in human cell lines, B complex is present and active in these cells, suggesting that it could regulate other sets of human genes. It would thus be of interest to identify the protein(s) that is (are) included in B complex, to gain access to its (their) potential target(s) and its (their) exact role(s) in transcription machinery. B complex may be a new transcription factor because GATA-1 was the only DNA binding factor that scored significantly when data banks were searched using the B-specific TGATAAGAC sequence. Moreover, because this factor is abundantly expressed in different cell lines, it should be amenable to purification and further characterization.
Acknowledgment
We are indebted to Dr M. Poncz (Philadelphia, PA) for providing us with the rat promoter genomic sequence.
Supported by an ARC fellowship to P.A.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Georges Uzan, INSERM U. 506, Hôpital Paul BROUSSE, 14 Avenue Paul Vaillant Couturier, F-94800 Villejuif Cedex 08, France; e-mail: guzan@infobiogen.fr.


![Fig. 5. Competition and Scatchard analysis of DNA binding activities of the hGATA and mGATA sequences. / (A) Competitive gel mobility shift assay. Nuclear extracts from K562 cells were incubated with labeled hGATA or mGATA probes, in the absence (−), or in the presence of 5-, 10-, 20-, 100-, and 200-fold molar excess of unlabeled hGATA or mGATA competitor, and 200-fold molar excess of SP1 cold competitor. A and B complexes are indicated by arrows. (B) Titration of GATA-1 and B complex with hGATA and mGATA probe in EMSA. Constant, nonsaturating amounts of K562 nuclear extract (10 μg) were incubated with increasing concentrations of hGATA and mGATA probes (0.02-6 ng) and were resolved in EMSA. The specific DNA-protein complexes indicated by the arrows (Bound A and Bound B) and the free probes at the bottom were quantified by phosphoImager. (C) Scatchard plot analysis. The ratio between bound and free DNA probe (B/F) versus specific DNA-protein binding concentration (bound [μmol/L]) was plotted, and the straight lines were drawn by fitting the data using a linear regression, with the Scatchard equation: B/F = −(1/Kd)[Bound] + Bmax/Kd, where Kd is the apparent equilibrium dissociation constant, and Bmax is the maximal number of binding sites. The Kd and Bmax values were calculated for each DNA/protein complex.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/4/10.1182_blood.v96.4.1348/4/m_h81600019005.jpeg?Expires=1769290621&Signature=Kjx7KqvZ-UJin9in5z3TqjakPR7oWkOVOORb2M-qn56YcfjAmK5wIDM8bsyrH6OKVfqzH9Rywcjz3tk9DiwbUCaOYIuzyeX2Ya~LTsHdi5z23sjeyiClz~uCPDvLKfL29~L6ZLrCMw-Aqzp3JrJS2~A8B0TtYPQ9ug508K-nj5FG2forP12WXGenl2rDcuDj4gj7UH7QqrssKffJj1MKhXpSnmqkwftvB0cSej3JKERO5ykprFJdFHfIiY-wHCOy-ZFz~~VArLh8K~TDx5ov38bkvufA1G~md9gSEzHEQuVfztcSSqxJU4nNmEWTf453LLqO~wY9PDkeXxHSwVahGw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Fig. 5. Competition and Scatchard analysis of DNA binding activities of the hGATA and mGATA sequences. / (A) Competitive gel mobility shift assay. Nuclear extracts from K562 cells were incubated with labeled hGATA or mGATA probes, in the absence (−), or in the presence of 5-, 10-, 20-, 100-, and 200-fold molar excess of unlabeled hGATA or mGATA competitor, and 200-fold molar excess of SP1 cold competitor. A and B complexes are indicated by arrows. (B) Titration of GATA-1 and B complex with hGATA and mGATA probe in EMSA. Constant, nonsaturating amounts of K562 nuclear extract (10 μg) were incubated with increasing concentrations of hGATA and mGATA probes (0.02-6 ng) and were resolved in EMSA. The specific DNA-protein complexes indicated by the arrows (Bound A and Bound B) and the free probes at the bottom were quantified by phosphoImager. (C) Scatchard plot analysis. The ratio between bound and free DNA probe (B/F) versus specific DNA-protein binding concentration (bound [μmol/L]) was plotted, and the straight lines were drawn by fitting the data using a linear regression, with the Scatchard equation: B/F = −(1/Kd)[Bound] + Bmax/Kd, where Kd is the apparent equilibrium dissociation constant, and Bmax is the maximal number of binding sites. The Kd and Bmax values were calculated for each DNA/protein complex.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/4/10.1182_blood.v96.4.1348/4/m_h81600019005.jpeg?Expires=1771244749&Signature=AaFHa3-Z2jUf8qHSj2qfzutjtVKMync1JtSCcd2AjzGCR-TcMvPTfYdUt6x02pN1oTP3UC7tryMWyde0Ppa0vz-DDrSK1I6VA--pHrU2onxM20bV2WHYiH5Hl7luWv5oeK1oNUFCfeVEBzmprs3otnm4LpGi3VdOhjjzPYghEOAEiuTRu-LuMyZdZaWDKncin0RGYKRkSoOyJJP4rVzszUdRU31Gx32WutMauGCcaXSgTY209ppNdPv9Qt0AnxDnnXFBrCwpmLbrjYlkCNKGVA8TsgWBRSTC8-4vezgFo9BcFx3ICqnSFeHwLEEgeIIxURwLlIZNc~2ISKAyYRMB-g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)