Abstract
Interphase fluorescence in situ hybridization (FISH) studies of chromosomal region 13q14 were performed to investigate the incidence and clinical importance of deletions in multiple myeloma (MM). Monoallelic deletions of the retinoblastoma-1 (rb-1) gene and the D13S319 locus were observed in 48 of 104 patients (46.2%) and in 28 of 72 (38.9%) patients, respectively, with newly diagnosed MM. FISH studies found that 13q14 was deleted in all 17 patients with karyotypic evidence of monosomy 13 or deletion of 13q but also in 9 of 19 patients with apparently normal karyotypes. Patients with a 13q14 deletion were more likely to have stage III disease (P = .022), higher serum levels of β2-microglobulin (P = .059), and a higher percentage of bone marrow plasma cells (P = .085) than patients with a normal 13q14 status on FISH analysis. In patients with a deletion of 13q14, myeloma cell proliferation (Ki-67) was markedly increased (22.0% ± 6.9% compared with 15.6% ± 8.2% in patients without the deletion;P = .0008). Evaluation of bromodeoxyuridine incorporation in 5 patients revealed that both rb-1–deleted and rb-1–normal MM subpopulations were proliferative. The presence of a 13q14 deletion on FISH analysis was associated with a significantly lower rate of response to conventional-dose chemotherapy (40.8% compared with 78.6%; P = .009) and a shorter overall survival (24.2 months compared with > 60 months; P < .005) than in patients without the deletion. Multivariate analysis of prognostic factors confirmed the independent predictive value of 13q14 deletions for shortened survival. In conclusion, deletions of 13q14 are frequently detected by interphase FISH in patients with newly diagnosed MM, correlate with increased proliferative activity, and represent an independent adverse prognostic feature in MM.
Similar to observations in acute myelogenous leukemia, data have suggested that specific chromosomal abnormalities may determine the prognosis of patients with multiple myeloma (MM). Tricot et al1,2 found that partial or complete deletion of chromosome 13 and abnormalities involving 11q were associated with short event-free and overall survival in a large, uniformly treated patient population receiving tandem high-dose therapy followed by autologous stem-cell transplantation. The adverse prognostic role of monosomy 13 or deletion of 13q in MM was also reported by Seong et al.3 On the other hand, in 2 large cytogenetic studies of patients with MM, no significant association between specific chromosomal aberrations and prognosis was established.4,5These results, however, were all based on metaphase cytogenetic studies that revealed informative karyotypes in only 30% to 40% of patients with newly diagnosed MM. With molecular cytogenetic techniques, particularly interphase fluorescence in situ hybridization (FISH), chromosomal aneuploidy can be demonstrated in almost every patient with MM,6-9 thus making it necessary to define the true incidence of specific chromosomal abnormalities in MM and to reevaluate any correlations with clinical outcome and prognosis.
There is some evidence that deletions of 13q occur at a much higher frequency in MM than has been indicated by metaphase analysis (monosomy 13 or deletions of 13q in 15%-20% of patients).1-5 In 2 studies using a FISH probe specific for the retinoblastoma gene-1 (rb-1), the frequency of deletions was 52% (12/23 patients) and 33.3% (16/48 patients), respectively.10,11 Similarly, allelic loss of D13S319, a gene locus distal to rb-1 on 13q14.3, was detected by FISH in 10 of 24 patients (41.6%) with MM.12 Results reported by Perez-Simon et al11 also suggested short overall survival in patients with loss of rb-1 on FISH.
To establish the clinical importance of 13q14 deletions in MM, we performed interphase FISH studies with probes specific for rb-1 and D13S319 in a large group of patients with newly diagnosed MM. We found that despite the fact that FISH studies revealed 13q14 deletions in at least twice as many patients with MM as did metaphase analysis, the presence of a 13q14 deletion was the most important independent variable associated with inferior outcome after conventional-dose chemotherapy. Moreover, the presence of a 13q14 deletion was linked with an increased proliferative capacity of myeloma cells.
Patients and methods
Patients
For studies with FISH analysis, bone marrow cells from 104 patients with newly diagnosed MM (including 8 patients undergoing induction chemotherapy) and 15 patients with relapse of MM were used. Bone marrow aspirates were obtained from the posterior iliac crest or sternum and collected in a heparin-coated syringe. These procedures were performed for diagnostic purposes, and patients gave informed consent according to institutional guidelines to allowing an aliquot of the specimen to be used for research purposes. For the analysis of FISH results in the context of clinical and laboratory data, 97 consecutively seen patients were included (7 of the 104 patients were excluded because of severe medical conditions other than MM). These patients had a median age of 65.9 years (range, 33.0-86.4 years). Thirty-five patients (36.1%) were men and 62 (63.9%) were women. The paraprotein was IgG in 68 patients (70.1%), IgA in 23 patients (23.6%), and another subtype in 6 patients (6.3%). According to Durie and Salmon staging done at diagnosis, 21 patients (21.6%) had stage I MM, 16 patients (16.5%) had stage II, and 60 patients (61.9%) had stage III. In 10 patients, serum creatinine was > 177 μmol/L (10.3%).
Eighty-four patients received conventional-dose induction chemotherapy consisting of a melphalan-based regimen (either melphalan and prednisone or vincristine, melphalan, cyclophosphamide, and prednisone with or without carmustine) in 68 patients; vincristine, doxorubicin, and dexamethasone in 15 patients; and pulsed high-dose dexamethasone in 1 patient. Five patients eventually received high-dose melphalan (140 mg per square meter of body surface area) followed by autologous peripheral blood stem-cell support. Reasons patients did not receive chemotherapy were stage I MM without symptoms and no evidence of disease progression (10 patients) or early death (3 patients). Required criteria for an objective response were a reduction in the paraprotein level to at least 50% of the pretreatment value, resolution of preexisting hypercalcemia, and no increase in the number or size of lytic bone lesions.13
Interphase FISH
Mononuclear bone marrow cells fixed with Carnoy's solution (3:1 [vol/vol] methanol and glacial acetic acid) were used for FISH analysis. Experiments were performed as dual-color hybridizations combining the 13q14-specific probe (rb-1 or D13S319 as probes directly conjugated with Spectrum Orange; Vysis, Downers Grove, IL) with a centromere-specific reference probe (usually a chromosome 11 or 17α-satellite probe directly conjugated with Spectrum Green; Vysis). Prehybridization, hybridization, posthybridization washes, and DNA counterstaining were performed according to our previously reported standard protocol.14 15 The presence of ≥ 2 hybridization signals per cell with the reference probe in more than 90% of nuclei was required to fulfill the criteria of high hybridization efficacy. Only slides meeting these criteria were scored, and at least 200 cells with the morphologic appearance of plasma cells were counted. Cells were scored with use of an immunofluorescence microscope (Axioplan-2; Zeiss, Jena, Germany), and images were captured with a cooled charge-coupled device camera (Photometrics, Tucson, AZ), linked to a computer (Macintosh; Apple Computer, Cupertino, CA). Prints were obtained with a color printer (Phaser 440; Tektronix, Wilsonville, OR).
For FISH analysis of cells labeled with bromodeoxyuridine (BrdU),16 bone marrow cells were incubated with BrdU (Sigma, St Louis, MO; 20 μmol/L of BrdU in RPMI-1640) for exactly 60 minutes at 37°C and washed twice with phosphate-buffered saline (PBS). The cells were fixed in 70% ethanol for 30 minutes on ice and then fixed with methanol and acetic acid (3:1 [vol/vol]) for 20 minutes. Subsequently, cells were dropped onto clean slides and air dried. Prehybridization and hybridization were performed as described above. After the posthybridization washes (3 rinses in 50% formamide and 2 × SSC at 45°C and 2 rinses in 2 × SSC at 37°C), slides were incubated with fluorescein-isothiocyanate–labeled anti-BrdU (Becton Dickinson Immunocytochemistry Systems, San Jose, CA; final concentration, 10 μg/mL in PBS) for 60 minutes at room temperature. Slides were rinsed twice in PBS and counterstained with DAPI and Antifade (Oncor, Gaithersburg, MD). Cells incubated in medium without BrdU were treated identically and served as negative controls for the BrdU staining.
Additional experiments combined the FISH-BrdU technique with staining for cytoplasmic immunoglobulins. For this purpose, the technique described above was applied, except that after the posthybridization washes, slides were rinsed once in PBS and Tween 20 and stained with either goat antihuman κ or λ light-chain antibodies conjugated with 7-amino-4-methylcoumarin-3-acetic acid (Vector Laboratories, Burlingame, CA; dilution of 1:20 antibodies to PBS and Tween 20) for 30 minutes at room temperature. Staining with the anti-BrdU antibody was then done as described above. Cells with immunoglobulin-positive cytoplasm were selectively scored to determine the rate of BrdU-expressing myeloma cells in both the 13q14-deleted and the 13q14-normal cell populations. For each patient, at least 500 cells were evaluated for BrdU incorporation.
Immunohistochemical staining for Ki-67
Proliferating cells were detected by staining with the Mib-1 monoclonal antibody (Immunotech, Marseille, France), which recognizes the Ki-67 antigen. Bone marrow cells fixed with Carnoy's solution were dropped onto sialin-coated slides, air dried, and fixed with heat according to the manufacturer's recommendations. Cells were then incubated with Mib-1 antibody (used at a dilution of 1:50 in PBS supplemented with 2% bovine serum albumin) for 45 minutes at room temperature and washed 3 times with PBS. For detection of bound antibody, we used a kit (Vectastain Elite ABC; Vector Laboratories), and the precipitate was visualized with use of diaminobenzidine (Fluka Chemie AG, Buchs, Switzerland). Finally, cells were counterstained with Mayer's hemalum. Cells incubated with an isotype-matched (IgG1) monoclonal antibody, but otherwise processed identically, were included as controls.
Determination of interleukin 6 (IL-6) messenger RNA (mRNA) expression
For this analysis, only samples containing at least 80% plasma cells were used. Otherwise, plasma cells were enriched by an immunomagnetic separation method after labeling with the B-B4 monoclonal antibody (CD 138; Immuno Quality Products, Groningen, the Netherlands) as previously described.6 RNA was isolated and complementary DNA (cDNA) was prepared as previously described.17 Five microliters of the large-scale cDNA preparation (equivalent to 200 ng of starting RNA template) was used for amplification of specific DNA sequences. Polymerase chain reaction (PCR) was done in a final volume of 50 μL containing 100 pmol/L of the specific primer set, 1.25 mmol/L of deoxytriphosphate nucleotides, 1 × PCR buffer, and 2 U of Taq DNA polymerase (all reagents from Promega, Southampton, UK). The IL-6–specific primer set was obtained from Clontech (Palo Alto, CA) and expected to generate a cDNA fragment of 628 base pairs. β-Actin was used as the internal control (primers from Clontech). Amplifications were done for 30 cycles. Each cycle contained a denaturation step at 94°C for 45 seconds, an annealing step at 60°C for 45 seconds, and an elongation step at 72°C for 2 minutes. After the last cycle, a final elongation step was done at 72°C for 7 minutes. Twenty microliters of the PCR products was separated electrophoretically on 2% agarose gels containing ethidium bromide. The products were then visualized with use of a UV transilluminator and photographed. The U-266 myeloma cell line (provided by K. Nilsson)18 was included as a positive control for IL-6; HL-60 and K-562 cells were used as negative controls.
Statistical analysis
Results were statistically evaluated by using the Fisher exact test, χ2 test, t test, and the nonparametric Kruskal-Wallis test, where appropriate. Kaplan-Meier estimates were obtained to evaluate survival times from the time of diagnosis, and differences between survival curves were analyzed by means of Breslow and Mantel-Cox tests. Multivariate analysis was done by using the Cox proportional hazards regression model. Stepwise evaluation of maximum partial likelihood ratio was used to identify prognostic variables.
Results
Interphase FISH analysis of 13q14 in MM
Using a probe specific for the rb-1 locus at 13q14, we observed a deletion in 48 of 104 patients (46.2%) with newly diagnosed MM (including 8 patients undergoing induction chemotherapy) and in 11 of 15 patients (73.3%) with relapse of MM. The frequency of chromosomally aberrant cells ranged from 12.5% to 90.1% (median, 32.8%; cut-off level for presence of an rb-1 deletion, 9.9%) in patients with newly diagnosed MM and from 39.2% to 73.2% (median, 53.1%) in patients with relapse of MM. Deletions were monoallelic, except in 1 patient with relapse, who had a monoallelic deletion of rb-1 in 19.3% of cells and a biallelic deletion in 26.2%.
Seventy-two patients, all of whom had newly diagnosed MM, were also studied with the D13S319 probe specific for 13q14.3. FISH analysis detected a deletion in 28 patients (38.9%), which was monoallelic in all cases (range of percentage of deleted cells, 10.8%-87.8%; median, 32.5%; cut-off level for deletion of D13S319, 9.3%). Only 1 patient with a deletion of D13S319 had a normal finding with the rb-1 probe; the remaining 27 patients had deletions of both rb-1 and D13S319. On the other hand, 3 patients with loss of rb-1 had a normal pattern for D13S319 on FISH analysis. On the basis of these findings, we conclude that deletions of 13q14 generally affect both loci investigated in this study; therefore, patients with a deletion at either locus were classified as having MM with a 13q14 deletion.
Metaphase cytogenetic studies were performed in 49 patients. Among 30 patients with informative karyotypes, 17 had monosomy 13 or deletion of 13q, which was also detected by FISH analysis in all 17 cases. In addition, FISH analysis found a deletion of 13q14 in 9 of 19 patients with apparently normal or noninformative karyotypes. Thirteen patients with abnormal karyotypes but 2 normal chromosomes 13 were also disomic for 13q14 on FISH analysis.
Deletion of 13q14 and myeloma cell proliferation
In 74 patients newly diagnosed as having MM, staining with the Ki-67 monoclonal antibody was done to determine the growth fraction of myeloma cells. The percentage of Ki-67–positive plasma cells ranged from 2% to 33% (median, 19%). However, in 34 patients (45.9%) with a 13q14 deletion on FISH analysis, Ki-67 expression was significantly higher (mean ± SD, 22% ± 6.9%; 95% confidence interval [CI], 19.6%-24.4%) than in patients with normal chromosome 13 findings (15.6% ± 8.2%; 95% CI, 12.6%-17.7%;P = .0008).
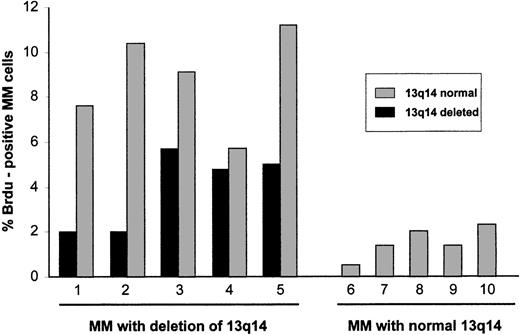
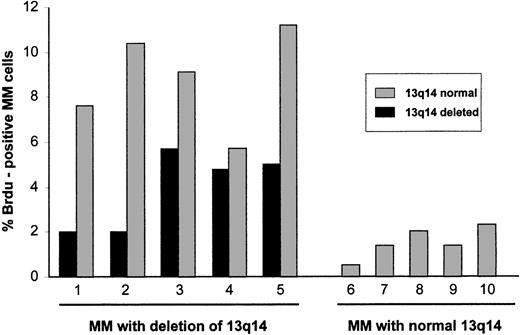
To obtain a direct assessment of the proliferative activity of myeloma cells with deletion of 13q14, interphase FISH results with the rb-1 probe and incorporation of BrdU were analyzed simultaneously (Figure1). In 5 patients with allelic loss of rb-1 (18.7%-72.0% of myeloma cells), positivity for BrdU was observed in 2.0% to 5.7% of rb-1–deleted cells, whereas in 5 patients with normal rb-1 status on FISH analysis (Figure2), positivity for BrdU was observed in 0.5% to 2.3% of myeloma cells. Of particular note, in patients with an rb-1 deletion, the fraction of myeloma cells (cytoplasmically positive for immunoglobulin) showing a normal hybridization pattern with the rb-1 probe was also proliferative, as determined by BrdU positivity in 5.7% to 11.2% of cells (patients 1-5 in Figure 2).
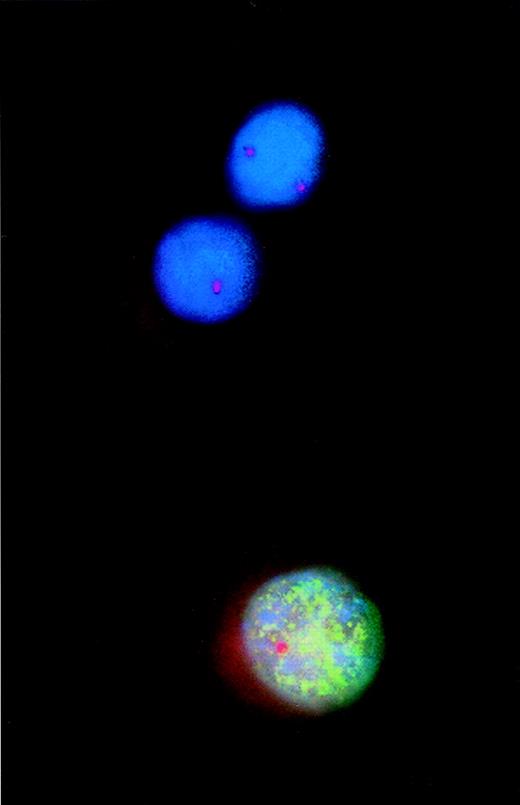
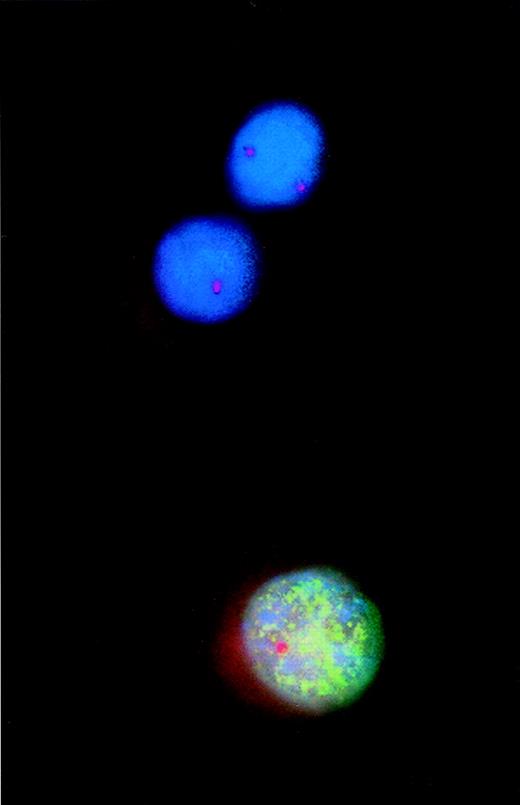
Combined analysis using interphase fluorescence in situ hybridization (FISH) and bromodeoxyuridine (BrdU) incorporation to assess proliferative activity of cytogenetically defined cell populations.
Incorporated BrdU was detected with a fluorescein isothiocyanate-labeled monoclonal antibody that produced the green fluorescent pattern of the nucleus. A Spectrum Orange-tagged probe specific for the retinoblastoma gene-1 (rb-1) was used for FISH analysis. Thus, of the 2 rb-1–deleted myeloma cells, 1 cell incorporated BrdU, indicating DNA synthesis and proliferation.
Combined analysis using interphase fluorescence in situ hybridization (FISH) and bromodeoxyuridine (BrdU) incorporation to assess proliferative activity of cytogenetically defined cell populations.
Incorporated BrdU was detected with a fluorescein isothiocyanate-labeled monoclonal antibody that produced the green fluorescent pattern of the nucleus. A Spectrum Orange-tagged probe specific for the retinoblastoma gene-1 (rb-1) was used for FISH analysis. Thus, of the 2 rb-1–deleted myeloma cells, 1 cell incorporated BrdU, indicating DNA synthesis and proliferation.
Proliferative activity of cytogenetically defined myeloma cells.
Five patients with a 13q14 deletion determined by FISH analysis with a rb-1 specific probe were studied for BrdU incorporation. Both rb-1–deleted and rb-1–nondeleted cells were found to be proliferative. Proliferation of these multiple myeloma (MM) cells was greater than that of MM cells from 5 patients with normal chromosome 13q14 on FISH analysis.
Proliferative activity of cytogenetically defined myeloma cells.
Five patients with a 13q14 deletion determined by FISH analysis with a rb-1 specific probe were studied for BrdU incorporation. Both rb-1–deleted and rb-1–nondeleted cells were found to be proliferative. Proliferation of these multiple myeloma (MM) cells was greater than that of MM cells from 5 patients with normal chromosome 13q14 on FISH analysis.
Deletion of rb-1 and IL-6 mRNA expression by myeloma cells
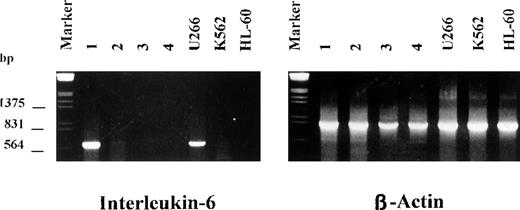
Because IL-6 is known to be a key molecule for myeloma cell proliferation and survival and normal rb-1 protein may function as a suppressor of the IL-6 gene promotor,19 it has been hypothesized that loss of rb-1 could contribute to autocrine growth of myeloma cells by means of up-regulation of IL-6. We therefore examined bone marrow plasma cells from 12 patients with MM and monoallelic deletion of 13q14 detected by FISH for IL-6 mRNA expression by using reverse transcriptase-PCR. As shown in Figure3, only 1 patient had a strong band for IL-6 expression, indicating that there was no correlation between IL-6 expression and rb-1 gene status in primary tumor cells from patients with MM.
Expression of interleukin 6 (IL-6) messenger RNA by myeloma cells with deletion of 13q14.
A strong band for IL-6 was observed in only 1 of 14 patients. U266 cells were included as positive controls; HL-60 and K562 cells were the negative controls.
Expression of interleukin 6 (IL-6) messenger RNA by myeloma cells with deletion of 13q14.
A strong band for IL-6 was observed in only 1 of 14 patients. U266 cells were included as positive controls; HL-60 and K562 cells were the negative controls.
Correlation of 13q14 deletions with clinical and laboratory variables
Ninety-seven consecutively seen patients with MM (46 patients [47.4%] with a deletion of 13q14 on FISH analysis) were evaluable for analysis of clinical and laboratory results and follow-up data. Patients with a 13q14 deletion more frequently had stage III disease at diagnosis (P = .022), higher serum levels of β2-microglobulin (P = .059), and a higher percentage of bone marrow plasma cells (P = .085) than patients with a normal chromosome 13q14 status on FISH analysis (Table1). Otherwise, there was no correlation between deletion of 13q14 and standard laboratory findings. Of note, a deletion of 17p13 detected by FISH with a p53-specific probe15 was more common in patients with a deletion of 13q14.
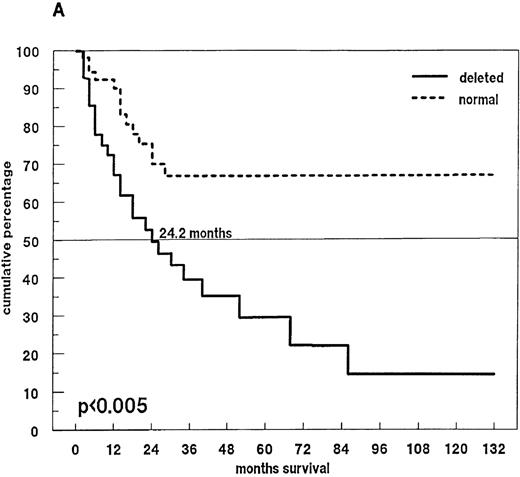
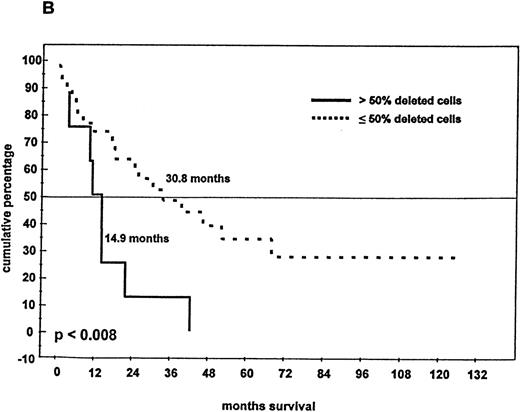
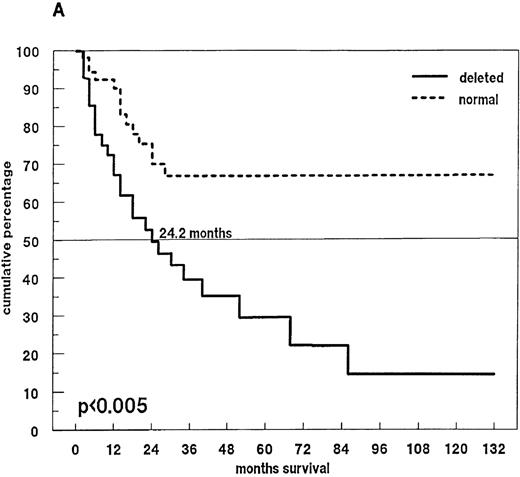
Eighty-one patients received conventional dose-chemotherapy as induction treatment, and an objective response was documented in 54 (66.7%). In the patients with a 13q14 deletion on FISH analysis, the response rate was only 40.8%, whereas patients with disomic findings for 13q14 had a response rate of 78.6% (P = .009). When overall length of survival from diagnosis was analyzed, a significant difference was again observed between patients with and without a deletion of 13q14 (24.2 months compared with > 60 months;P < .005; Figure 4A). In the 46 patients with a deletion of 13q14, we also observed that survival was influenced by the percentage of deleted myeloma cells: the presence of a 13q14 deletion in more than 50% of myeloma cells was associated with a median overall survival time of 14.9 months, whereas patients with the deletion in no more than 50% of myeloma cells had a median overall survival time of 30.8 months (P < .008; Figure 4B).
Survival and 13q14 deletion.
(A) Overall survival of 97 patients with MM with (n = 46) and without (n = 51) a 13q14 deletion on interphase FISH analysis. Patients with a normal 13q14 status had a significantly longer median survival than those with a deletion of 13q14 (> 60 months compared with 24.2 months; P < .005). (B) Survival analysis of the 46 patients with a 13q14 deletion, grouped according to the percentage of deleted MM cells detected by FISH analysis. Median survival time of MM patients with > 50% deleted cells (n = 9) was significantly shorter than that of patients with ≤ 50% deleted cells (n = 37) (14.9 compared with 30.8 months; P < .008).
Survival and 13q14 deletion.
(A) Overall survival of 97 patients with MM with (n = 46) and without (n = 51) a 13q14 deletion on interphase FISH analysis. Patients with a normal 13q14 status had a significantly longer median survival than those with a deletion of 13q14 (> 60 months compared with 24.2 months; P < .005). (B) Survival analysis of the 46 patients with a 13q14 deletion, grouped according to the percentage of deleted MM cells detected by FISH analysis. Median survival time of MM patients with > 50% deleted cells (n = 9) was significantly shorter than that of patients with ≤ 50% deleted cells (n = 37) (14.9 compared with 30.8 months; P < .008).
We performed a multivariate analysis of prognostic factors that included deletion of 13q14, Durie and Salmon stage, patient age, type of paraprotein (IgA compared with non-IgA), extent of bone marrow infiltration by plasma cells, plasma cell morphologic features (graded according to the method of Bartl et al20), hemoglobin, platelet count, albumin, creatinine, calcium, lactate dehydrogenase, C-reactive protein, and β2-microglobulin as categorical and continuous variables (where appropriate). In this analysis, disease stage (P = .002), deletion of 13q14 (P = .004), β2-microglobulin level (P = .007), and age (P = .04) were independent predictive variables for shortened survival. In the subgroup of 74 patients in whom data on cellular proliferation (Ki-67) were available in addition to the variables listed above, Ki-67 (P = .002) and deletion of 13q14 (P = .031) were independent unfavorable variables for overall survival.
Discussion
Our results obtained by interphase cytogenetic studies confirm and extend previous observations suggesting that deletions of chromosome 13q are strong prognostic indicators in MM.1-3,11 In agreement with studies using interphase FISH10-12 and comparative genomic hybridization,21-23 in which chromosome 13 abnormalities were found in 30% to 79% of patients, we observed deletions of 13q14 in 46% of patients with newly diagnosed MM. A comparison of results from FISH analysis and metaphase cytogenetics studies in a subset of our patients indicated that failure to obtain metaphases from myeloma cells is responsible for underestimation of the true incidence of this abnormality in MM by conventional cytogenetic analysis.
Patients with a 13q14 deletion frequently had stage III disease, and compared with patients with a normal chromosome 13 status on FISH analysis, they tended to have a higher percentage of bone marrow plasma cells and elevated serum levels of β2-microglobulin at diagnosis. It is important to note, however, that a deletion of 13q14 was commonly observed in patients with stage I MM (9 of 26 patients [34.6]), and data from other studies24 (and unpublished data) also indicated that deletions of 13q14 have already occurred in plasma cells from patients with monoclonal gammopathy of undetermined importance. It is therefore reasonable to conclude that development of this chromosomal abnormality is a rather early event in the pathogenesis of plasma cell malignant diseases. An additional observation demonstrating the importance of 13q14 deletions for the biologic features of MM is its association with the increased proliferative capacity known to be correlated with the clinical course of MM.25 26 Patients with MM and a deletion of 13q14 had a significantly higher growth fraction (determined by Ki-67 staining) than those with normal chromosome 13 status.
The proliferative state of MM cells was further investigated by an analysis combining incorporation of BrdU and FISH. When 5 patients with a deletion of 13q14 were studied in detail, BrdU incorporation was found not only in the rb-1–deleted cells but also in the population of MM cells showing 2 signals with the rb-1 probe. This suggests that the presence of a 13q deletion in even a fraction of myeloma cells has consequences for the entire myeloma cell population. Although we did not find evidence of enhanced IL-6 expression by 13q-deleted myeloma cells, it may still be possible that 13q-deleted cells produce a cytokine supporting the growth of the nondeleted myeloma cell population. It appears that, over time, the 13q-deleted MM cells are biologically more relevant than the remaining cell populations, since the proportion of myeloma cells carrying the deletion is higher at relapse than at diagnosis and patients with a deletion in the majority (≥ 50%) of MM cells have the worst survival. This heterogeneity of clonal cells already present at diagnosis may be one reason why currently available treatment strategies are less effective in patients with MM who have deletion of 13q14 than in other patients with MM.
Results obtained in 97 consecutively seen patients indicated that deletions of 13q14 were significantly correlated with a poor response to conventional-dose chemotherapy and short survival (24.2 months compared with > 60 months). Inferior outcome of patients with a rb-1 deletion on FISH analysis was also reported by Perez-Simon et al,11 who found a median survival time of only 14 months in such patients, compared with 60 months in patients with a normal chromosome 13. In their study, however, no significant correlation between 13q deletion and response to conventional-dose chemotherapy was observed, probably because of the small number of patients studied. In agreement with their study,11 our study provides evidence for the independent prognostic importance of 13q14 deletions in MM, despite their increased frequency of detection by FISH analysis. Thus, the reported association between monosomy 13 or deletion of 13q (detected by metaphase cytogenetics) and prognosis1-3 is unlikely to be just a reflection of cellular proliferation. This idea is further supported by the observation by Perez-Simon et al11and us (this study) that myeloma cell proliferation and deletions of 13q14 are independent prognostic factors for survival.
Allelic loss may indicate the location of a tumor suppressor gene involved in the pathogenesis of a malignant disease. In the current study, both loci at 13q14 were found to be deleted in a monoallelic fashion, in agreement with previous reports.10-12Additional mechanisms could lead to inactivation of the second allele that results in a complete loss of normal function of the affected gene. As far as the rb-1 gene is concerned, mutational inactivation appears to be a rare event in MM. Among nongenetic mechanisms leading to inactivation of the rb-1 gene product, IL-6 was shown to promote myeloma cell growth by means of phosphorylation of the rb-1 protein.27 It is worth noting that monoallelic deletions of rb-1 were found to have no effect on expression of rb-protein in MM.28 It is therefore reasonable to assume that 13q harbors another tumor suppressor gene besides rb-1 with biologic importance in MM. For example, in B-cell chronic lymphocytic leukemia, which is also characterized by frequent allelic loss of 13q14, a candidate tumor suppressor gene was suspected distal to rb-1, in the vicinity of D13S319.29-31 There is some evidence that the same may hold true for MM: in a study using comparative genomic hybridization, a commonly deleted region corresponding to 13q14q21 was observed in 30% of 25 patients with MM.21 Similarly, interphase FISH studies with a set of 13q-specific probes suggested biallelic loss of regions adjacent to rb-1 in some patients with MM,32 again indicating the presence of a yet-unidentified gene with tumor suppressor function on or close to 13q14. It is also conceivable that concomitant deletions of rb-1 and a second gene locus on 13q14 may contribute to the pathogenesis of this MM subtype with a poor outcome.
In summary, we found a deletion of 13q14 in 46% of patients with untreated MM and that this deletion represents a strong independent prognostic variable for poor response to standard-dose chemotherapy and shortened survival. In addition, the observation of 13q14 deletions in a sizable fraction of patients with stage I MM (and monoclonal gammopathy of undetermined significance) and the demonstration of a marked proliferative capacity of the 13q14-deleted cells imply that deletion of 13q14 may be one critical event in the pathogenesis of MM. Because patients with a 13q14 deletion also have a poor outcome after high-dose treatment,33 there is a need for innovative therapeutic concepts in this particular risk group of patients with MM.
Supported by the Austrian Fonds zur Förderung der Wissenschaftlichen Forschung (Grant P12432-MED) and the ICP Program (Molecular Medicine) of the Austrian Ministry of Research and Transport.
Reprints:Johannes Drach, University of Vienna, Department of Internal Medicine I, Division of Clinical Oncology, Währinger Gürtel 18-20, A-1090 Vienna, Austria; e-mail:johannes.drach@akh-wien.ac.at.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.