Abstract
The IIb/β3 receptor is central to platelet aggregation. Biological studies of this receptor have been limited by the inability to reproduce IIb/β3 function in a cell system. Increasingly, efforts are being directed at studies of this receptor in mice models. The structure of murine (m) β3 has been reported. We now have sequenced the mIIb gene and found that it has the same size and organization as the human gene. The exon/intron borders are reported here, as are the distances between exons. mIIb protein is 1,033 amino acids (aa), 7 and 5 aa shorter than human (h) and rodent (r) IIb, respectively, with 79% and 90% homology, respectively. As part of the comparative analysis of the 3 known IIb chains included in this report, we found that a particular region of the IIb N-terminal β-propeller is highly conserved and speculate that it directly participates in ligand binding.
THE PLATELET-SPECIFIC integrin αIIb/β3 (GPIIb-IIIa, CD41, CD62) binds fibrinogen and other ligands after platelet activation.1,2 This receptor is densely packed on the surface of the platelet with 80,000 copies per platelet3 and also does not bind its ligand with high affinity until the platelet is activated and the receptors are “turned on.”4 Given the central role played by this receptor in thrombosis and the successful clinical application of anti-αIIb/β3–directed strategies to prevent thrombotic complications,5 a great deal of effort has been carried out to establish a functional assay system to study the αIIb/β3 receptor system ex vivo. These studies have focused on stable transfections in cell lines such as Chinese hamster ovary (CHO) or in lymphoid cells.6 7 Both lines suffer from the limited number of receptors on the surface and the artificial fashion in which these cells need to be activated. At best, these lines show binding in the nonactivated state and only modest increases after activation.
Attention has, therefore, focused on setting up more vigorous systems in in vivo models with an emphasis on murine studies. The mβ3 cDNA has been described8 and, subsequently, the mβ3 knockout has been produced with a phenotype similar to Glanzmann thrombasthenia.9 Furthermore, studies have been done with the first mβ3 knock-in, introducing a mutation into the mβ3 cytoplasmic tail (Y747F) and showing a mild decrease in platelet aggregation after agonist activation.10Additionally, studies have begun introducing human αIIb and β3 chains into primary megakaryocytic progenitors. For example, hβ3 constructs were introduced into mβ3 knockout marrow cells using a retroviral system, and the resulting megakaryocytes were rescued with regard to αIIb/β3 properties such as clot retraction, presumably secondary to mαIIb/hβ3 receptor complexes on the surface of the megakaryocytes.11 Such studies show that cross-species heterodimers readily form and are functional, although there are going to be important limitations in such studies, as it is known that αIIb/β3 receptors appear to have different species related properties such as in their sensitivity to RGD peptides.12 13
MATERIALS AND METHODS
Isolation of the mαIIb λ and bacterial artificial chromosome (BAC) clones.
Full-length rαIIb cDNA was random-primer labeled16 with32P-dCTP and used to screen a λFIX mouse 129SV genomic library (Stratagene, La Jolla, CA) using nitrocellulose filter lifts of the plated phage. Individual positive colonies were obtained using repeat rounds of dilutional plating, and these were grown in large scale and purified using a cesium gradient as previously described.16
DNA from the genomic clones was characterized by restriction digest and Southern blotting16 using again the rat αIIb cDNA as probe. A 6-kb BamHI fragment was subcloned from the original positive λFIX clone and subcloned into BamHI cut pBSK+ library (Stratagene).
The sequence in mαIIb Intron 12 was used to polymerase chain reaction (PCR)-screen a BAC mouse 129SV genomic library (Genome Systems, St Louis, MO), using the primers forward: 5′-ATGGACTTACCCCCATAGAT-3′ and reverse: 3′-ACTTCCCCGGGATTCTGCGC-3′ to give a 0.5-kb band. The BAC clone was then used to obtain the sequence through exon 15, and then a PCR-amplified region for exons 15 and 16 was randomly primer labeled and used to rescreen the original λFIX library to complete the characterization of the mαIIb gene.
mαIIb gene characterization.
The 5′-BamHI pBSK clone, the 5′- and 3′-λFIX clones, and the BAC clone described above were end sequenced with the appropriate primers (eg, T7, SP6, and T3 primers). Subsequent primers were generated based on the new data and used to prime the next round of sequencing reaction. All sequencing used fluorescenated dNTPs, and Sequenase (USB, Cleveland, OH) and an ABI 373A automated sequencer (PE Applied Biosystems, Foster City, CA). Sequences were stored and analyzed using MacDNasis (Hitachi Software, San Diego, CA) and the BLAST program at the National Center for Biotechnology, which hosts an internet site at URL: http://www.nlm.nih.gov/.
RESULTS AND DISCUSSION
Characterization of the mαIIb gene.
The only previously characterized αIIb cDNAs were those of human and rat.14,15 Using the rat αIIb cDNA, we screened a 129SV murine genomic λFIX library. Sequencing of overlapping αIIb+ clones helped to provide the majority of sequence of the mαIIb gene and its surrounding locus. In addition, we PCR-screened an mBAC 129SV murine genomic library using primers derived from these λ sequences. A single αIIb+ BAC clone was obtained and sequenced to fill the remaining sequence. The complete sequence for the murine αIIb gene and cDNA are available through GenBank at the National Center for Biotechnology (accession nos.AF170316 and AF169829). Using restriction mapping of the mαIIb BAC clone, we have previously reported that the murine gene is flanked by the KIAA-O553 gene upstream and the Granulin gene downstream, and that this organization is conserved in the hαIIb locus.17 The genes themselves are also organized into very similar exons and introns of virtually identical size. A summation of the exon/intron borders of the mαIIb gene with the distance between the various exons is shown in Table 1. Since the promoter region, through the beginning of the coding region of the mαIIb gene had been previously published,18 the data begin with exon 1’s splice donor region. It should be pointed out that the hαIIb gene has a “GC” instead of the canonical “GT” at the splice donor sequence for exons 5 and 8. This variation is also present for the mαIIb gene (double underlined in Table 1). The 3′-untranslated region is poorly conserved except in the immediate region around the purported polyadenylation signal site.
Characterization of the mαIIb cDNA.
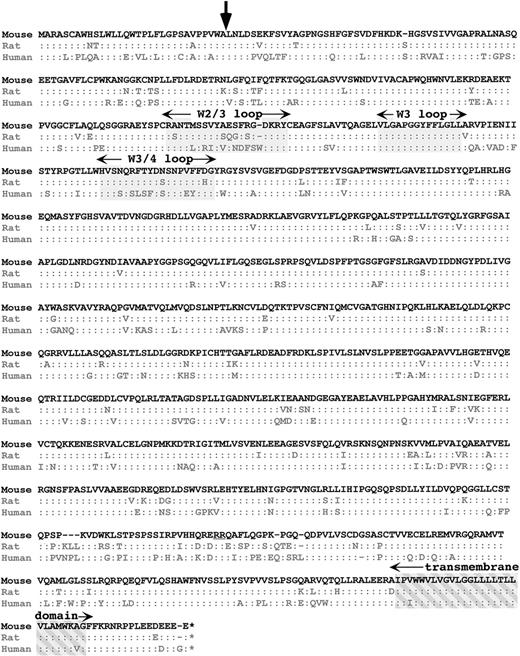
We extracted the mαIIb cDNA from the above sequence and have used these sequences for reverse transcriptase (RT)-PCR of murine platelet RNA with amplification of expected band size and have confirmed portions of the sequence. A comparison with rat and human protein sequence is shown in Fig 1. mαIIb is 1,033 amino acids (aa), 7 and 5 aa shorter than human and rodent αIIb, respectively. There is 79% and 90% homology between hαIIb and rαIIb with mαIIb, respectively, which is near average for these cross-species homology.15 As expected, the signal peptides have lower homology than the remaining mature proteins, but the region surrounding the start of the mature protein is well conserved (see arrow in Fig 1). The least-conserved region is near the cleavage site into the light and heavy chain. Although the cleavage site itself is conserved (underlined “RR” in Fig 1), homology with hαIIb and rαIIb is only 50% and 60%, respectively. In contrast, the transmembrane and cytoplasmic domains are highly conserved.
Cross-species comparison of mIIb with hIIb and rIIb. The single-letter amino acid sequence of mIIb is shown at the top and those of the other 2 species are shown below. A”:” refers to an identical homologous aa. A “–” refers to a missing aa. The start site for mature IIb is indicated by an arrow. The 3 regions of the N-terminal β-propeller regions discussed are grayed in and labeled. The transmembrane domain is crosshatched grayed in and is also labeled. The double-arginine cleavage site is underlined.
Cross-species comparison of mIIb with hIIb and rIIb. The single-letter amino acid sequence of mIIb is shown at the top and those of the other 2 species are shown below. A”:” refers to an identical homologous aa. A “–” refers to a missing aa. The start site for mature IIb is indicated by an arrow. The 3 regions of the N-terminal β-propeller regions discussed are grayed in and labeled. The transmembrane domain is crosshatched grayed in and is also labeled. The double-arginine cleavage site is underlined.
Studies of natural-occurring and directed mutations have suggested that the 3 upper surface loops of the N-terminal β-propeller of αIIb may be involved in ligand binding. Our cross-species comparison shows that there is 100% conservation at the middle of these 3 loops (grayed area for the “W3 loop” in Fig 1). However, there is poor conservation in the other 2 loops (“W2/3 loop” and “W3/4 loop” in Fig1). We would have expected a loop involved in ligand binding to be highly preserved. One possible explanation for this divergence could be that these differences in receptor loop structure help to accommodate for species differences in the primary structure of the fibrinogen ligand itself. Supporting this, it has been shown that the αIIb/β3 receptor of different species have different sensitivities to RGD peptide inhibition of fibrinogen binding so that rodent and murine platelets are much more resistant to RGD peptide inhibition than human platelets.12 Whether this difference in species RGD sensitivity is caused by the rapid evolution of sequences in the W2/3 and W3/4 loops remains to be investigated.
Supported in part by National Institutes of Health Grant No. HL40387.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Mortimer Poncz, MD, The Children’s Hospital of Philadelphia, 34th St and Civic Center Blvd, Philadelphia, PA 19104; e-mail: poncz@emailchop.edu.


 CAGCAGAGGGGGCGGGGTTCCTGGT
CAGCAGAGGGGGCGGGGTTCCTGGT GAGCTTGACAGTGAT
GAGCTTGACAGTGAT