Abstract
The human integrin β3 participates in a wide range of adhesive biologic functions and is expressed in a selected subset of tissues, but little is known about the cis-acting DNA elements or trans-acting factors responsible for this regulation. Using cell lines characterized for β3 expression, a number of upstream regulatory regions in the β3 gene were identified. (1) The three regions from −1159 to −584, −290 to −146, and −126 to −115 demonstrated positive, negative, and negative activity, respectively. (2) The region from −115 to +29 of the β3 gene was sufficient for cell-specific activity. Deletion of the sequence from −115 to −89 produced a 6- to 40-fold reduction in reporter gene activity in β3-expressing megakaryocytic cell lines (K562, Dami, and HEL), but only a 1.7- and 2.7-fold reduction, respectively, in β3-expressing endothelial and melanoma cell lines, and 1.3- and 2.8-fold reduction, respectively, in non–β3-expressing Chinese hamster ovary and 293 cell lines. This sequence also bound nuclear proteins in a cell-specific manner in electrophoretic mobility shift assays. Mutational analysis indicated that the sequence GAGGGG (positions −113 to −108) is a megakaryocytic cell line-specificcis-acting element. (3) The region from −89 to +29 promoted lower activity in all cell lines. We also provide evidence that a CCCACCC sequence at position −70 has transcriptional activity, most likely through the Sp1 transcription factor. These data supply the first detailed map of the transcriptional regulatory elements of the 5′ region of the β3 gene, define positive regulatory sequences with potent megakaryocyte preferential activity, and indicate that the ubiquitous transcription factor, Sp1, may augment β3 gene expression.
© 1998 by The American Society of Hematology.
THE BIOLOGIC IMPORTANCE of the integrin family of adhesive molecules is well-established, and these molecules participate in such diverse biologic processes as thrombus formation, angiogenesis, embryogenesis, inflammation, as well as tumor metastases.1 Many of these processes require the ability to upregulate or downregulate different integrin molecules. Integrins consist of αβ heterodimers and integrin β3 is able to pair with two α subunits: αIIb and αv, the classical fibrinogen and vitronectin receptors, respectively. When paired with αIIb, β3 expression is restricted to the megakaryocyte/platelet lineage, whereas the classical vitronectin receptor (VNR), αvβ3, is expressed in a number of tissues. The biologic importance of β3 protein is underscored by the inherited bleeding disorder, Glanzmann thrombasthenia, which results when the gene for β3 is mutated and not expressed.2 To date, nearly 20 β3 mutations have been described in Glanzmann thrombasthenia,3 but all have been in coding sequence or have resulted in defective RNA splicing, so that no clues have been provided as to transcriptional regulatory sequences.
β3 expression is controlled at the level of transcription, as demonstrated by the response to phorbol esters. Increased β3 mRNA levels have been demonstrated by Northern blot analysis and nuclear run-on experiments,4 and increased β3 was observed in cell-free translations of mRNA from K562 cells treated with phorbol 12-myristate 13-acetate (PMA).5 Although β3 stability and surface expression are in some measure dependent on a subunit expression (an example being the rapid β3 degradation in the nonexpressing αIIb thrombasthenia mutations), a variety of other factors and biologic processes are known to modulate β3 expression. For example, β3 is upregulated during the second half of the menstrual cycle and during pregnancy,6,7 and we have shown that this effect may be mediated by sex hormones.8 In a variety of different cells lines the expression of β3 is also regulated by basic fibroblast growth factor,9 vitamin D,10retinoic acid,11 the human homeobox gene HOX4A,12 and factors that promote angiogenesis.13,14 Although a vitamin D-responsive element has been identified in the avian gene,10 15 none of the necessary cis-acting elements for such regulatory effects have been identified in the human gene.
Because αIIb expression is restricted to the megakaryocyte/platelet lineage, the αIIbβ3complex is expressed exclusively in these cells. A substantial body of work exists on the transcriptional regulation of the αIIbgene,16-26 but comparisons to β3 regulatory sequences have been difficult due to limited information on the latter. On the other hand, αvβ3 is expressed in a number of tissues, including human osteoclasts,27endothelial cells,28,29 human placental syncytiotrophoblast brush border,30,31 monocyte-derived macrophages,32-34 cultured human embryonic fibroblasts,35,36 as well as a number of malignant cell lines. Upregulation of αvβ3 expression has been associated with malignant potential; for example, during the transition from dysplastic nevi to tumorigenic melanomas37and the neovascularization of tumors.13 However, although the tissue distribution of β3 has often been called widespread, this has not been assessed in a formal fashion. Some older studies used monoclonal antibodies (MoAbs) against αv to assess VNR expression and equated this with β3expression. Because αv can associate with β5,38,39 β6,40,41β8,42 and β3, specific markers must be used to assess its tissue distribution. A large report on platelet antigens used four different MoAbs to β3 and found that more than half of the tissues and cell lines examined were negative for β3 expression.43 This differential pattern of tissue expression for αIIb and β3 suggests that independent factors control gene transcription. On the other hand, unlike all other known integrin pairs, the genes for αIIb and β3 are physically linked on 17q21.32,44 45 raising the possibility of shared cis-acting elements coordinating gene expression in megakaryocytes. Regardless, the mechanisms controlling independent, shared, or even developmental expression of β3 are unknown.
We have previously cloned and partially characterized a portion of the β3 gene containing the 5′ region and signal peptide.46 47 We now report our characterization of the transcriptional regulatory elements of the 5′ region of the β3 gene and provide evidence for both significant tissue-specific and tissue- nonspecific cis-acting elements. These results provide detailed information on elements and a transcription factor affecting β3 gene expression and may supply insights into the regulation of expression of other β integrin genes.
MATERIALS AND METHODS
Reagents.
Purified Sp1 protein (Promega, Madison, WI), rabbit polyclonal IgG, 1C6 (anti-Sp1) MoAbs (Santa Cruz Biotechnology, Santa Cruz, CA), and the irrelevant antibody, mouse γ globulin (Pierce, Rockford, IL) were purchased. PMA (Sigma, St Louis, MO) was diluted to 1 mg/mL in dimethyl sulfoxide (Sigma) and frozen at −80°C. Radioisotopes were from Amersham (Arlington Heights, IL). MoAb T10 is specific for the human αIIbβ3 complex48 and was a generous gift of Dr Rodger McEver (Oklahoma Medical Research Foundation, Oklahoma City, OK); MoAb AP3 is specific for human β349 and was a generous gift of Dr Peter Newman (Blood Research Institute, Milwaukee, WI).
Cell lines and culture conditions.
The β3-expressing megakaryocytic cell lines K562,50,51 Dami,52 and HEL53 and the non–β3-expressing HeLa cell line (American Type Culture Collection [ATCC], Rockville, MD) were cultured as described previously.54 The Dami cells were obtained from the ATCC in 1989 and demonstrate greater β3 expression than do our HEL cells, despite the likelihood of being a subclone of HEL. In some experiments, K562 cells were treated with PMA (100 nmol/L). The human microvascular endothelial cell (HMEC-1) line that expresses β355 was cultured in endothelial basal medium (MCDB 131; Clonetic Corp, San Diego, CA) supplemented with 10% fetal bovine serum (GIBCO BRL, Grand Island, NY), 10 μg/mL hydrocortisone (Sigma), and 10 ng/mL epidermal growth factor (EGF; Collaborative Biomedical Products-Becton Dickinson, Bedford, MA). WM793 is a β3-expressing melanoma cell line obtained from Dr Meenhard Herlyn (The Wistar Institute, Philadelphia, PA) that was grown in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO BRL) with 10% fetal bovine serum. Cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C. Two non–β3-expressing cell lines were used as controls: Chinese hamster ovary (CHO) cells were cultured in α-Minimal Essential Media (MEM; GIBCO BRL) containing 10% vol/vol fetal bovine serum (GIBCO BRL), and the transformed human embryonal kidney cell line 293 (ATCC) was cultured in Eagle’s minimal essential medium (GIBCO BRL) supplemented with 10% fetal bovine serum (GIBCO BRL).
Flow cytometry.
Cells were washed and suspended in phosphate-buffered saline (PBS) at a final concentration of 2 × 106/mL and incubated with T10 or AP3 at a final concentration of 1 μg/mL. Samples were washed twice in PBS and then incubated with fluorescein isothiocyanate (FITC)-labeled goat antimouse antibody for 60 minutes at 4°C. Samples were washed twice with PBS, centrifuged at 750g for 10 minutes, and then resuspended in 400 μL of PBS for analysis by flow cytometry.
Northern analysis.
Total cellular RNA was isolated from cell lines according to the method of Chomczynski and Sacchi.56 RNA samples (10 μg/lane) were electrophoresed in 1% agarose formaldehyde gels, transferred to nylon membranes, and hybridized with radiolabeled probes for human β328 and human fetal glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probes as previously described.54
Plasmid constructs.
Various regions of the β3 gene were cloned upstream of the luciferase reporter gene in the pGL2 Basic vector (Promega) or upstream of the CAT reporter gene in the PLCAT vector.46 In all β3 constructs, the 3′ end of the β3 gene sequence immediately flanked the ATG translation start codon (position +29 from the transcription start site). The plasmid pGL2 Promoter vector (Promega) that has an SV40 promoter was used as a positive luciferase control and for some normalization studies. The CMV-Sp1 expression plasmid and its parental vector without the Sp1 cDNA (CMV-empty) were the generous gifts of Robert Tjian (University of California, Berkeley, CA).
The −1159 internal deletion construct was generated by restriction digestion at the −146 Xba I and −31Not I sites, blunting the ends with T4 DNA polymerase, and religating. Mutations were introduced into the −146 wild-type template using the Site-Directed Mutagenesis kit (Clontech, Palo Alto, CA) according to the manufacturer’s recommendation. Primers for introducing mutations are listed in Table1. The mPw1 primer was used to generate the −146 mPw1mut construct; primer mMS7 was used to generate the −146 Sp1mutant construct. All constructs were sequenced to confirm that the mutation had been properly introduced.
CAT assays.
Several of the initial functional analyses were performed on the β3 gene by cloning various regions of the β3 genomic clone into plasmid PLCAT,46transiently transfecting constructs into K562 cells, and performing CAT assays as described previously.46 To account for variation in transfection efficiency, results were normalized to the luciferase activity obtained by cotransfecting 2 μg of pGL2-Promoter vector (Promega).
Luciferase assays.
K562, Dami, and HEL cells were harvested at a density of 1.5 to 2.0 × 105 cells/mL and electroporated (1 × 107 cells/sample) using a gene pulser (Bio-Rad, Richmond, CA) set at 500 μFD and 400 V with test (20 μg) and control pSV40-CAT (2 μg) plasmids. Cells were then incubated on ice for 10 minutes, resuspended in 25 mL of complete media, and incubated at 37°C and 5% CO2 atmosphere. CHO cells were transfected using a diethyl aminoethyl (DEAE) dextran method as described previously.57 Two hundred ninety-three cells were transfected using the CaCl2 method as described previously.58 HMEC-1 and WM793 cells were plated in 60-mm plates and transfected at 60% confluence with a total of 4 μg plasmid DNA, using 15 μL Lipofectin (GIBCO BRL) for 3 hours. Transfected cells were collected after 24 hours, washed twice with media, lysed, and analyzed using the Luciferase Assay System (Promega).
To account for variation in transfection efficiency, luciferase activity was normalized to CAT activity by cotransfection of the pSV40-CAT construct. CAT activity was measured as described previously.46 To assess the importance of various sequences within a given cell line, luciferase values that had been adjusted for transfection efficiency with CAT were divided by either the CAT-normalized luciferase activity of the pGL2-Basic (promoterless) or pGL2-Promoter (SV40 promoter) vectors (Promega). This fold increase over the pGL2-Basic and pGL2-Promoter vectors was not used for comparison between cell lines, because these arbitrary values were relatively high or low due to the different activities of the pGL2-Basic and pGL2-Promoter vectors in different cell lines. Rather, comparisons between cell lines were made by the relative changes in activities among different deletion constructs within a cell line.
Electrophoretic mobility shift assays (EMSAs).
Crude nuclear extracts were prepared from K562, Dami, HEL, HeLa, 293, HMEC-1, and CHO cells using the method of Andrews and Faller.59 Briefly, cells were lysed in buffer A (10 mmol/L HEPES-KOH, pH 7.9, at 4°C, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L dithiothreitol, 0.2 mmol/L phenylmethyl sulfonyl fluoride [PMSF]) and centrifuged, and the pellets were resuspended in buffer C (20 mmol/L HEPES-KOH, pH 7.9, 25% glycerol, 420 nmol/L NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 0.5 mmol/L dithiothreitol, 0.2 mmol/L PMSF). Cellular debris was removed by centrifugation, and supernatants were stored at −80°C.
DNA probes were prepared as follows. Single-stranded DNA oligonucleotides were slowly annealed to form double-stranded probes by incubating the sense and antisense strands at 95°C followed by a slow cool to room temperature. Double-stranded DNA probes (Table 1) were labeled with α32P-deoxycytidine triphosphate (dCTP; 3,000 Ci/nmol) or α32P-deoxyguanidine triphosphate (dGTP; 3,000 Ci/nmol) using DNA polymerase (Klenow fragment; Boehringer Mannheim, Indianapolis, IN). Crude nuclear extracts (5 μg protein/sample) were incubated with double-stranded32P-labeled oligonucleotide probes in binding buffer (10 mmol/L Tris-Cl, pH 7.4, 80 mmol/L KCl, 5% glycerol, 1 mmol/L dithiothreitol) for 15 minutes at room temperature. Poly dI:dC was used in gel shifts at 0.055 to 0.22 mg/mL to minimize background protein binding to probe. Competition experiments were performed by the simultaneous incubation with a 50-fold molar excess of unlabeled specific or irrelevant DNA probes (Table 1). Supershifts were performed using 2 μg of mouse Ig per reaction. Samples were electrophoresed on 6% polyacrylamide gels that were pre-run at 100 V for 1 hour and run for 2.5 hours at 250 V and 4°C. Gels were dried for 1 hour at 80°C and exposed to film (Eastman Kodak, Rochester, NY) for 12 to 96 hours at −80°C.
To optimize the detection of any potential DNA-protein interactions, several different mobility shift assay buffers were tested. In addition to the buffer described above, a second buffer (Incubation buffer; Hotfoot Buffers Kit; Stratagene, La Jolla, CA) was identified that was more sensitive to some of the DNA-protein interactions. The composition of this latter buffer is proprietary.
Computer search for DNA homologies.
We used the Basic Local Alignment Search Tool (BLAST) software through the National Center for Biotechnology Information (NCBI; Bethesda, MD) to search for nucleic acid homologies. We used the blastn program to search all nonredundant sequences in all the GenBank databases as well as those sequences in the eukaryotic promoter database (EPD).
RESULTS
Identification of megakaryocytic cell preferential positive activity in the 5′ region of the β3 gene.
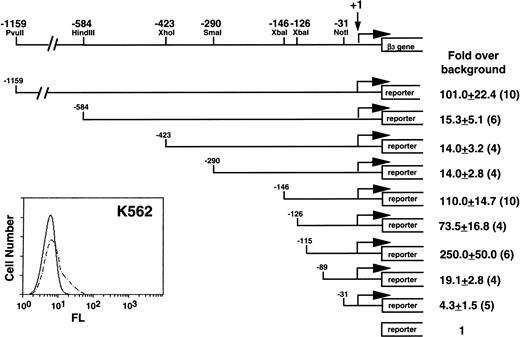
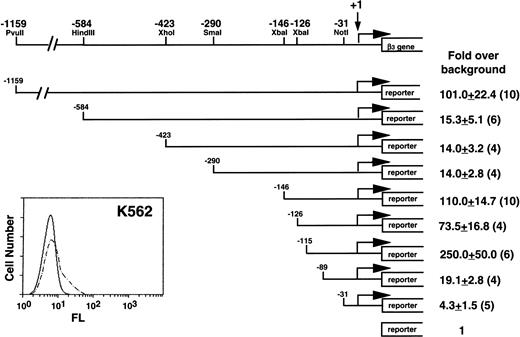
Our initial studies were performed in K562 cells, because they expressed β3 (Fig 1) and had the highest transfection efficiency of the megakaryocytic cell lines. A series of deletion constructs from positions −1159 to −31 relative to the transcription start site of β3 gene were used in transient transfection assays for reporter gene activity. Positive regulatory activity was shown in the region from −1159 to −584; negative activity was observed between −290 and −146 and between −126 and −115. The greatest positive activity was observed in the region from −146 to the first exon. There was substantial loss of activity when this region was deleted, although the −89 to −31 region retained a modest positive effect. The minimal promoter has arbitrarily been defined at the −31 position, because these sequences were still able to drive transcription.46 Subsequent studies focused on regions of positive activity downstream of −146.
Functional analysis of the 5′ region of the β3 gene in K562 cells. The insert at the lower left shows flow cytometry analysis of K562 cells. Fluorescence (FL) is shown on a logarithmic scale on the x-axis; cell number is shown on the y-axis. Dashed lines are the results using MoAb T10, which is specific for human IIbβ3. A restriction enzyme map with positions relative to the transcription start site of the β3 gene is shown above. Reporter gene constructs containing portions of the 1,159 bp upstream of the transcription start site were transiently transfected into K562 cells and assayed for reporter activity. The reporter gene used in these studies included both CAT and luciferase, and equivalent results were obtained with both reporters. Fold increase and the standard error of the mean over the promoterless construct (background) are indicated. The number of times each construct was tested is shown in parentheses. The Materials and Methods describes how values were normalized, emphasizing the importance of the relative changes, not the absolute fold increase.
Functional analysis of the 5′ region of the β3 gene in K562 cells. The insert at the lower left shows flow cytometry analysis of K562 cells. Fluorescence (FL) is shown on a logarithmic scale on the x-axis; cell number is shown on the y-axis. Dashed lines are the results using MoAb T10, which is specific for human IIbβ3. A restriction enzyme map with positions relative to the transcription start site of the β3 gene is shown above. Reporter gene constructs containing portions of the 1,159 bp upstream of the transcription start site were transiently transfected into K562 cells and assayed for reporter activity. The reporter gene used in these studies included both CAT and luciferase, and equivalent results were obtained with both reporters. Fold increase and the standard error of the mean over the promoterless construct (background) are indicated. The number of times each construct was tested is shown in parentheses. The Materials and Methods describes how values were normalized, emphasizing the importance of the relative changes, not the absolute fold increase.
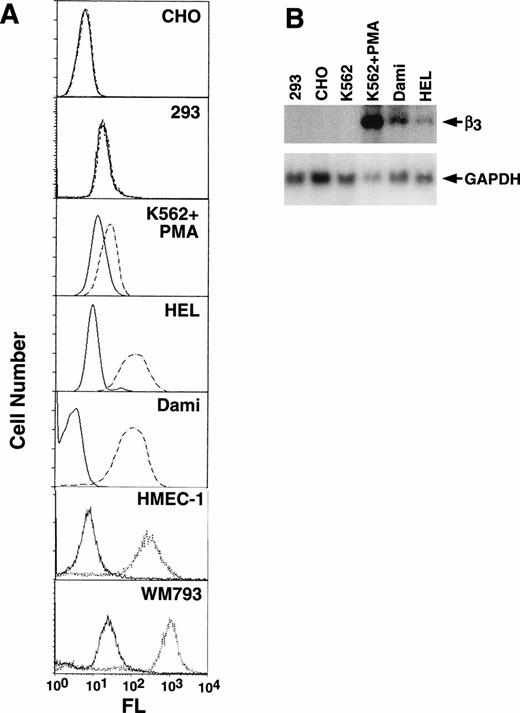
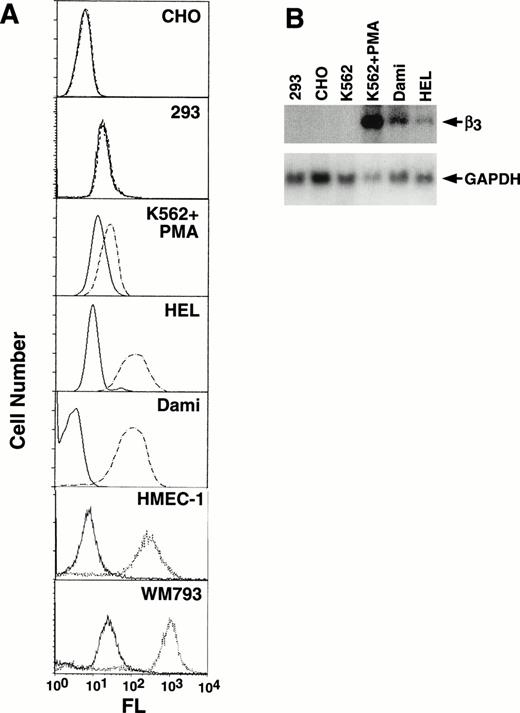
Because β3 is expressed in a variety of cell types, to assess tissue-specificity we characterized its expression in a number of cell lines (Fig 2A). The megakaryocytic cell lines, HEL and Dami, showed high levels of αIIbβ3surface expression, as did endothelial (HMEC-1) and melanoma (WM793) cell lines; no expression was observed in CHO and 293 cells. K562 cells showed increased expression with PMA induction (compare Figs 1 and 2A). Several of these cell lines were also studied by Northern analysis (Fig2B), which demonstrated undetectable levels of β3 mRNA in 293, CHO, and unstimulated K562 cells, but prominent expression in PMA-stimulated K562 cells and Dami cells and lower levels in HEL cells. As we and others have previously observed, uninduced K562 cells, which have megakaryocytic as well as erythroid properties, express only low levels of β3 protein, and mRNA is not easily observed by Northern blotting but can be detected by reverse transcription-polymerase chain reaction (data not shown).
Characterization of cell line β3expression. (A) Flow cytometry analysis of CHO, 293, PMA-induced K562, HEL, Dami, HMEC-1, and WM793 cells. Fluorescence (FL) is shown on a logarithmic scale on the x-axis; cell number is shown on the y-axis. Dashed or dotted lines are the results using MoAb T10, which is specific for human IIbβ3 on CHO, PMA-induced K562, HEL, and Dami cells, or β3-specific MoAb AP-3 for 293, HMEC-1, and WM793 cells; solid lines are the negative control using mouse Ig as the primary antibody. FACS analyses were performed at different times such that baseline fluorescence differed among cell lines. The key finding was the shift in fluorescence using MoAbs specific for β3 or IIbβ3. (B) Northern blot analysis of 10 μg total RNA from selected cell lines probed with a full-length 2.6-kb β3 cDNA and exposed to film at −80°C for 14 hours (upper panel). The β3 mRNA signal in HEL cells is more easily seen on 24 hours of exposure. The filter was stripped and rehybridized with a GAPDH cDNA to assess loading equivalency (lower panel). Note that HEL and Dami cell β3 RNA levels correlate well with protein expression, whereas PMA-stimulated K562 cells do not, suggesting that PMA-stimulation has complex effects on β3 protein expression.
Characterization of cell line β3expression. (A) Flow cytometry analysis of CHO, 293, PMA-induced K562, HEL, Dami, HMEC-1, and WM793 cells. Fluorescence (FL) is shown on a logarithmic scale on the x-axis; cell number is shown on the y-axis. Dashed or dotted lines are the results using MoAb T10, which is specific for human IIbβ3 on CHO, PMA-induced K562, HEL, and Dami cells, or β3-specific MoAb AP-3 for 293, HMEC-1, and WM793 cells; solid lines are the negative control using mouse Ig as the primary antibody. FACS analyses were performed at different times such that baseline fluorescence differed among cell lines. The key finding was the shift in fluorescence using MoAbs specific for β3 or IIbβ3. (B) Northern blot analysis of 10 μg total RNA from selected cell lines probed with a full-length 2.6-kb β3 cDNA and exposed to film at −80°C for 14 hours (upper panel). The β3 mRNA signal in HEL cells is more easily seen on 24 hours of exposure. The filter was stripped and rehybridized with a GAPDH cDNA to assess loading equivalency (lower panel). Note that HEL and Dami cell β3 RNA levels correlate well with protein expression, whereas PMA-stimulated K562 cells do not, suggesting that PMA-stimulation has complex effects on β3 protein expression.
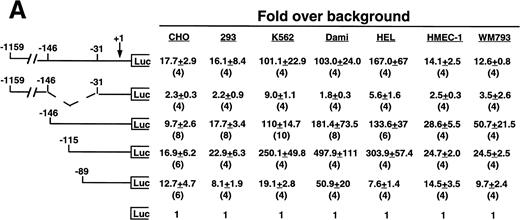
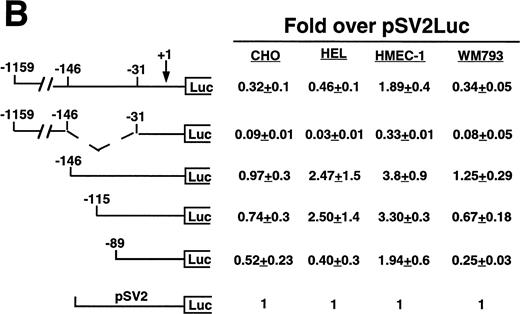
Cell line specificity of the −146 region was assessed using this spectrum of β3-expressing cell lines (Fig 3A). Because the background (promoterless) vector produced variable activity in the different cell lines, the absolute fold over background values cannot be compared across the different cell lines; rather, the relative changes in activities among different deletion constructs within a cell line were examined. The largest construct showed variable activity in both the megakaryocytic lines and in the nonmegakaryocytic lines. A mutant was generated in which the −146 to −31 sequence was removed from the −1159 construct. This mutant had greatly reduced activity in all cell lines, with the greatest decrease in the megakaryocytic cell lines, suggesting that the −146 to −31 sequence contained cell-specific activity not requiring the presence of the upstream negative and positive regulatory sequences. We observed modest and similar changes in activities in both megakaryocytic and nonmegakaryocytic cells upon deleting from −1159 to −146 and from −146 to −115. However, deletion from −115 to −89 resulted in a 13.0-, 9.8-, and 40-fold reduction in the megakaryocytic cell lines K562, Dami, and HEL, respectively. The β3-expressing endothelial and melanoma cell lines displayed a modest 1.3- and 2.8-fold reduction, respectively, which was similar to that seen the in non–β3-expressing CHO and 293 cell lines (1.7- and 2.7-fold reduction, respectively), suggesting megakaryocytic cell-preferential activity in the 26 bp between −115 to −89. To address whether the use of the promoterless construct as background might affect the results shown in Fig 3A, several of the key experiments were repeated, but this time normalized to a luciferase construct driven by a promoter of moderate activity (Fig 3B). In this case, the pSV2 construct showed variable and overall high activity in the different cell lines, leading to much lower fold changes. However, the key finding was unchanged: that deletion of the 26 bp between −115 and −89 caused a greater reduction in activity (6.2-fold) in the megakaryocytic HEL cell line as compared with a 1.4-, 1.7-, and 2.7-fold reduction in CHO, HMEC-1, and WM793 cells, respectively.
Cell line specificity of regulatory regions of the β3 gene. (A) The indicated constructs were transiently transfected into the β3-expressing megakaryocytic cells K562, Dami, and HEL, into the β3-expressing but nonmegakaryocytic cells HMEC-1 and WM793, and into non–β3expressing cell lines CHO and 293 and assayed for luciferase activity. Fold increase and the standard error of the mean over the promoterless construct (background) are indicated. The number of times each construct was tested is shown in parentheses. The Materials and Methods describes how values were normalized. (B) Additional reporter gene assays were performed four times and are presented as the fold increase over the pGL-2-Promoter with the standard error of the mean.
Cell line specificity of regulatory regions of the β3 gene. (A) The indicated constructs were transiently transfected into the β3-expressing megakaryocytic cells K562, Dami, and HEL, into the β3-expressing but nonmegakaryocytic cells HMEC-1 and WM793, and into non–β3expressing cell lines CHO and 293 and assayed for luciferase activity. Fold increase and the standard error of the mean over the promoterless construct (background) are indicated. The number of times each construct was tested is shown in parentheses. The Materials and Methods describes how values were normalized. (B) Additional reporter gene assays were performed four times and are presented as the fold increase over the pGL-2-Promoter with the standard error of the mean.
Identification of a cell-specific DNA-protein interaction associated with the −115 to −89 positive regulatory region of the β3 gene.
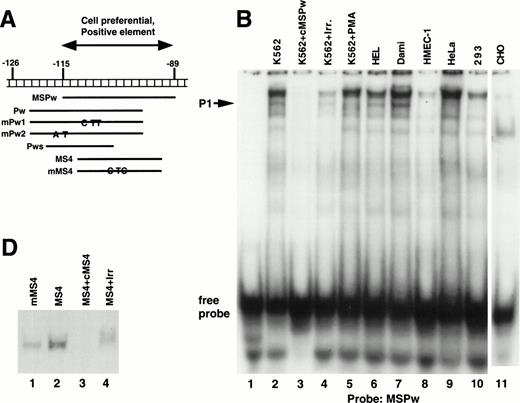
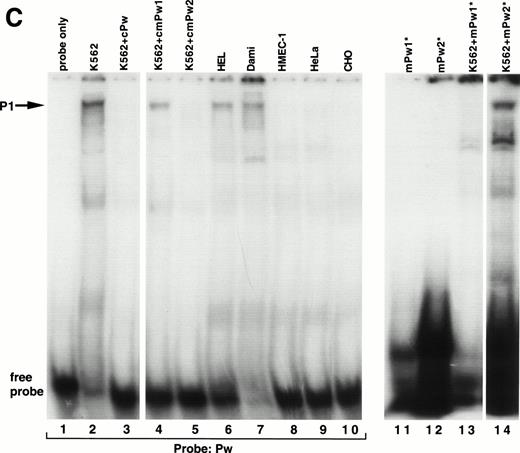
A series of EMSAs (probes listed in Table 1) identified DNA-protein interactions in the region of positive regulatory activity from −115 to −89. A 26-bp probe called MSPw (Fig 4A), which spanned the entire −115 to −89 region, demonstrated a complex pattern of shifted bands in EMSAs with crude nuclear extracts from K562 cells (Fig 4B, lane 2). Although these bands were competed by unlabeled MSPw (lane 3), the band labeled P1 was not competed by an irrelevant DNA probe (lane 4) and was only seen with nuclear extracts from K562, Dami, and HEL cells (lanes 2, 5, 6, and 7) but not in HMEC-1, HeLa, 293, or CHO cells (lanes 8 through 11). Much greater specificity was observed using probe Pw containing sequence shifted in a 5′ direction (Fig 4C). The Pw-generated megakaryocytic cell complex was presumed to be P1, because it comigrated with the complex seen using the MSPw probe when run on the same gel (data not shown). The lack of the P1 complex with HMEC-1 cells, which express β3, supported the functional studies and suggested that the −115 to −89 element functions in a megakaryocyte-specific fashion.
Cell-specific protein-DNA complex associated with a strong positive regulatory region (−115 to −89) of the β3 regulatory unit. (A) Location of the probes spanning a portion of the β3 regulatory unit that has cell-specific and potent positive regulatory function. In (B) and (C), cold specific and irrelevant oligonucleotide competitors (Table 1) were used in a 50-fold molar excess. Prefixes: c, unlabeled (cold) oligonucleotide; m, mutant oligonucleotide; cm, cold mutant oligonucleotide. (B) EMSA using32P-labeled MSPw oligonucleotide probe incubated with the indicated nuclear extracts. Nuclear extracts used in lane 5 were from K562 cells treated with PMA, which usually, but inconsistently, showed the P1 complex. The megakaryocytic cell-specific band is indicated as P1. The intensity of the P1 signal from PMA-treated K562 cells did not correlate with β3 RNA (see Fig 1B), suggesting that the −115 to −89 sequence is not the sole contributor to transcription. Irr., irrelevant DNA competitor (Table 1). (C) EMSA using32P-labeled Pw, mPu1, or mPu2 oligonucleotide probes incubated with the indicated nuclear extracts. The megakaryocytic cell-specific band is again shown as P1, because it comigrates with the MSPw complex (not shown). Radiolabeled probes are Pw in lanes 1 through 10, mPw1 in lanes 11 and 13, and mPw2 in lanes 12 and 14. (*)32P-labeled DNA probe. (D) EMSA using32P-labeled wild-type (MS4) and mutant (mMS4) oligonucleotide probes and K562 nuclear extracts. cMS4 is unlabeled MS4. Irr., irrelevant DNA competitor.
Cell-specific protein-DNA complex associated with a strong positive regulatory region (−115 to −89) of the β3 regulatory unit. (A) Location of the probes spanning a portion of the β3 regulatory unit that has cell-specific and potent positive regulatory function. In (B) and (C), cold specific and irrelevant oligonucleotide competitors (Table 1) were used in a 50-fold molar excess. Prefixes: c, unlabeled (cold) oligonucleotide; m, mutant oligonucleotide; cm, cold mutant oligonucleotide. (B) EMSA using32P-labeled MSPw oligonucleotide probe incubated with the indicated nuclear extracts. Nuclear extracts used in lane 5 were from K562 cells treated with PMA, which usually, but inconsistently, showed the P1 complex. The megakaryocytic cell-specific band is indicated as P1. The intensity of the P1 signal from PMA-treated K562 cells did not correlate with β3 RNA (see Fig 1B), suggesting that the −115 to −89 sequence is not the sole contributor to transcription. Irr., irrelevant DNA competitor (Table 1). (C) EMSA using32P-labeled Pw, mPu1, or mPu2 oligonucleotide probes incubated with the indicated nuclear extracts. The megakaryocytic cell-specific band is again shown as P1, because it comigrates with the MSPw complex (not shown). Radiolabeled probes are Pw in lanes 1 through 10, mPw1 in lanes 11 and 13, and mPw2 in lanes 12 and 14. (*)32P-labeled DNA probe. (D) EMSA using32P-labeled wild-type (MS4) and mutant (mMS4) oligonucleotide probes and K562 nuclear extracts. cMS4 is unlabeled MS4. Irr., irrelevant DNA competitor.
As a first attempt at identifying the necessary sequences required for the P1 cell-specific DNA-protein interaction, two variations of the Pw probe (mPw1 and mPw2; Table 1 and Fig 4A) were synthesized that contained nucleotide substitutions around position −115. As seen in Fig 4C, mPw2 (lane 14), but not mPw1 (lane 13), resulted in the P1 shifted band seen with Pw (lane 2), indicating the mutations in mPw1 abrogated P1 binding. Competition experiments indicated that unlabeled mPw1 was unable to compete away the Pw-generated P1 band, whereas mPw2 did (Fig 4C, compare lanes 4 and 5). In data not shown, the short probe Pws (Table 1 and Fig 4A) bound no nuclear proteins, suggesting that non–tissue-specific factors bound to the 3′-most portion of probes MSPw and Pw. These data indicate that the tissue-specific shift required sequence around position −115 (present in the 5′ regions of MSPw and Pw). Because the short Pws probe contained the −115 region but did not bind either the specific or nonspecific proteins, it is possible that the tissue-specific shift either required the additional concomitant binding of the 3′ non–tissue-specific factor or a conformation of the DNA not present in the probe shortened at the 3′ end.
There is a consensus ets/PU.1 binding site from −108 to −103 in the β3 5′ region. Probe MS4 from this region specifically bound nuclear protein (Fig 4D), but when the consensus ets site was mutated (probe mMS4), the DNA-protein complex was still observed (lane 1), suggesting that ets/PU.1 protein does not bind this region. This conclusion is supported further by the fact that the mutant sequences in mPw1 abolished the DNA-protein binding despite retaining the intact ets site.
Functional analysis of the −113 region.
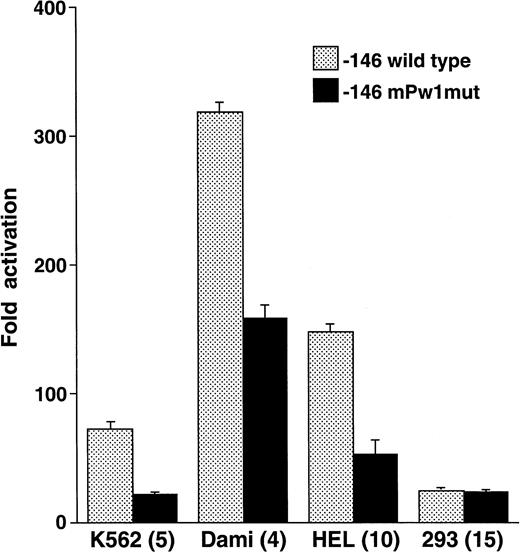
To test the functional significance of the DNA-protein interactions seen in Fig 4 and to define the extent of the megakaryocytic cell preferential activity, mutations were introduced between positions −115 to −107 and studied in reporter gene assays. Nucleotide substitutions corresponding to those in mPw1 resulted in an average 2.93-fold lower activity in the megakaryocytic cell lines K562, Dami, and HEL (Fig 5). No decrease was observed in 293 cells, consistent with the cell-preferential nature of the sequence mutated in mPw1. Point mutations at positions −115, −114, and −107 produced no significant change in reporter gene activity from the −146 wild-type construct (data not shown). These functional studies, together with the EMSAs discussed above, indicated that the minimal sequence conferring cell preferential activity was the 6-bp sequence from −113 to −108: GAGGGG.
The −112 to −109 sequence demonstrates cell preferential activity. The −146 construct was mutated at those positions that were altered in EMSA probe mPw1 and called −146 mPw1mut. Wild-type and mutant constructs were analyzed in the indicated cell lines for their luciferase activity and are displayed as fold activation over background with standard error bars. The number of times each experiment was performed is indicated in parentheses. The activity of the −146mPw1mut is only compared with the wild-type activity in those experiments when both constructs were used. Because there are some variations between experiments and because Fig 3summarizes numerous different experiments with the wild-type −146 construct, there are some differences in the activities of this construct between Figs 3 and 5.
The −112 to −109 sequence demonstrates cell preferential activity. The −146 construct was mutated at those positions that were altered in EMSA probe mPw1 and called −146 mPw1mut. Wild-type and mutant constructs were analyzed in the indicated cell lines for their luciferase activity and are displayed as fold activation over background with standard error bars. The number of times each experiment was performed is indicated in parentheses. The activity of the −146mPw1mut is only compared with the wild-type activity in those experiments when both constructs were used. Because there are some variations between experiments and because Fig 3summarizes numerous different experiments with the wild-type −146 construct, there are some differences in the activities of this construct between Figs 3 and 5.
The β3 promoter may interact with the Sp1 transcription factor.
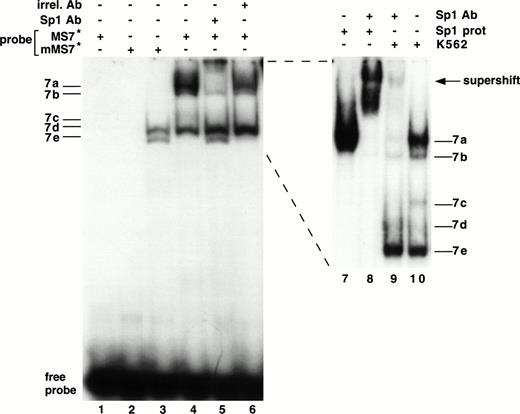
As shown in Fig 3, the sequence from −89 to +29 possesses substantial activity in seven different cell lines. Using EMSAs to study this region, no specific protein-DNA complexes were detected with the MS6 probe (Fig 6A) and nuclear extracts of K562 cells (data not shown). Five bands emerged in EMSAs using the MS7 probe and K562 nuclear extracts, and were named 7a through 7e (Fig6B). Bands 7a and 7b were most clearly seen as a doublet when the gel was run longer (see Fig 7, lanes 9 and 10). Bands 7d and 7e represent nonspecific protein-DNA complexes, because they were competed away with an irrelevant DNA probe. Bands 7a, 7b, and 7c were not competed with unlabeled irrelevant DNA probe but were competed away with unlabeled MS7, demonstrating that they were specific DNA-protein interactions. Because of their substantially greater intensity, we focused our attention on bands 7a and 7b (7c was not consistently observed). Using additional nuclear extracts (lanes 5 through 10), we also observed that bands 7a and 7b were most abundant in the megakaryocytic cell lines.
Demonstration of a DNA-protein complex in the MS7 region of β3. (A) Location of the MS6 and MS7 probes and the putative binding site for the Sp1 transcriptional factor. (B) EMSA using 32P-labeled MS7 DNA probe and the indicated nuclear extracts. Cold specific or irrelevant oligonucleotide competitors (Table 1) were used at a 50-fold molar excess. Irr., irrelevant DNA competitor; c, unlabeled oligonucleotide.
Demonstration of a DNA-protein complex in the MS7 region of β3. (A) Location of the MS6 and MS7 probes and the putative binding site for the Sp1 transcriptional factor. (B) EMSA using 32P-labeled MS7 DNA probe and the indicated nuclear extracts. Cold specific or irrelevant oligonucleotide competitors (Table 1) were used at a 50-fold molar excess. Irr., irrelevant DNA competitor; c, unlabeled oligonucleotide.
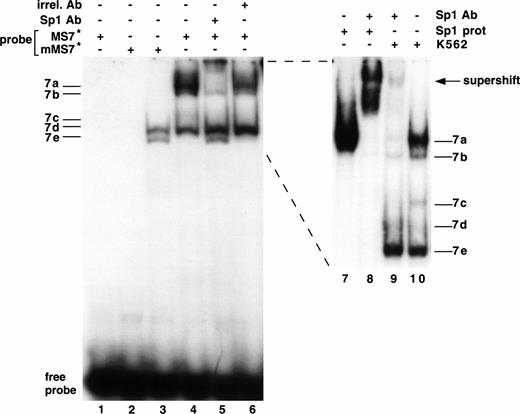
A binding site for the Sp1 transcription factor was present in the MS7 DNA probe, and we performed a series of studies to determine if this site (between −73 and −66 bp) was functional (Fig 7). Several experiments were highly suggestive that bands 7a and 7b contained Sp1. (1) Bands 7a and 7b disappeared when the probe was mutated (mMS7, Table 1) at sites known to destroy Sp1 binding60 (Fig 7, lane 3). (2) Bands 7a and 7b were supershifted with an MoAb specific for Sp1 (lanes 5 and 9) and not with an irrelevant mouse IgG (lane 6). (3) A comigrating band was seen when purified Sp1 protein was used in an EMSA with MS7 (compare lanes 7 and 10, Fig 7); the experiment in lane 8 confirms the position of the supershift as well as authentic Sp1 reactivity with probe MS7.
Interaction of Sp1 with the β3 promoter. EMSA using 32P-labeled mMS7 or MS7 oligonucleotide probes incubated with K562 nuclear protein or purified Sp1 protein as indicated above by the “+.” Sp1-purified protein was used at 1.5 μg/sample. Lanes 7 through 10 are from a different gel that was run longer. The appearance of the 7b band was inconsistently observed with purified Sp1 and this probe. Anti-Sp1 antibody or the irrelevant mouse Ig (irrel. Ab) was added to samples as indicated by the “+” above. Prefix “m” indicates mutant oligonucleotide, as described in Table 1.
Interaction of Sp1 with the β3 promoter. EMSA using 32P-labeled mMS7 or MS7 oligonucleotide probes incubated with K562 nuclear protein or purified Sp1 protein as indicated above by the “+.” Sp1-purified protein was used at 1.5 μg/sample. Lanes 7 through 10 are from a different gel that was run longer. The appearance of the 7b band was inconsistently observed with purified Sp1 and this probe. Anti-Sp1 antibody or the irrelevant mouse Ig (irrel. Ab) was added to samples as indicated by the “+” above. Prefix “m” indicates mutant oligonucleotide, as described in Table 1.
Functional evidence for Sp1 activity.
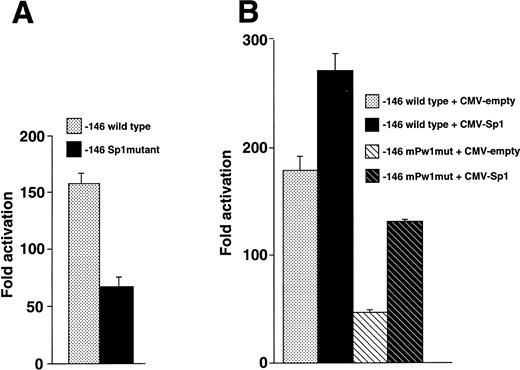
We introduced nucleotide substitutions into the −146 luciferase construct that destroyed the Sp1 binding site and observed 2.2-fold less activity compared with wild-type (Fig8A), suggesting that this region binds a positive regulatory factor in vivo. A series of cotransfection experiments were also performed to assess whether the Sp1 transcription factor interacted with the −113 region (Fig 8B). Overexpression of Sp1 resulted in increased expression, further implicating an Sp1 function in megakaryocytic cells. The Sp1 stimulated activity did not require the wild-type sequence at positions −113 to −109 (note the increase when Sp1 is overexpressed with the −146mPw1 construct) and merely had an additive effect.
Sp1 functions at the −70 position of the β3 promoter. (A) Sp1 mutational analysis. HEL cells were transfected with equivalent amounts of −146 wild-type and Sp1 mutant constructs, assayed for luciferase activity, and expressed as fold activation over background. The mutant construct contained the 2-bp substitution in probe mMS7 that destroyed the Sp1 site at position −70. Each experiment was performed five times. (B) Cotransfection of Sp1 enhances expression. HEL cells were transfected with the indicated plasmids and assayed for luciferase activity. Each experiment was performed four times. Twenty micrograms of −146 wild-type or −146 mPw1mut and 4 μg of CMV-Sp1 or CMV-empty (total plasmid DNA, 24 μg) were transfected in each experiment.
Sp1 functions at the −70 position of the β3 promoter. (A) Sp1 mutational analysis. HEL cells were transfected with equivalent amounts of −146 wild-type and Sp1 mutant constructs, assayed for luciferase activity, and expressed as fold activation over background. The mutant construct contained the 2-bp substitution in probe mMS7 that destroyed the Sp1 site at position −70. Each experiment was performed five times. (B) Cotransfection of Sp1 enhances expression. HEL cells were transfected with the indicated plasmids and assayed for luciferase activity. Each experiment was performed four times. Twenty micrograms of −146 wild-type or −146 mPw1mut and 4 μg of CMV-Sp1 or CMV-empty (total plasmid DNA, 24 μg) were transfected in each experiment.
DISCUSSION
Although integrin β3 is expressed in numerous tissues, our studies have identified a sequence in the 5′ regulatory portion of the β3 gene modulating expression in a megakaryocytic-specific manner. We found that the GAGGGG sequence at positions −113 to −108 augmented reporter gene expression and bound nuclear proteins to a greater degree in megakaryocytic cells than in nonmegakaryocytic cells, regardless of whether they expressed β3. We also obtained evidence suggesting that the Sp1 transcription factor binds to and upregulates expression through the −70 position of the β3 transcriptional regulatory unit and that this interaction appeared especially prominent in megakaryocytic cells. Thus, β3 expression is regulated by specific and nonspecific factors. The latter, when bound in particular combinations and specific locations, may perhaps contribute to tissue-specific gene expression. Our data also provide rationale for a significant degree of low-to-modest levels of β3 expression in a spectrum of different tissues and at the same time support a hypothesis that megakaryocytes express higher concentrations than other tissues.
Functional studies identified distal sequences upstream of the β3 gene with positive and negative regulatory activity, and the region from −115 to −89 displayed megakaryocytic cell- specific activity. K562, Dami, and HEL cells showed 6- to 40-fold reduced activity when this sequence was deleted (Fig 3), whereas there was only a 1.7- and 2.7-fold reduction, respectively, in β3-expressing endothelial and melanoma cell lines; CHO and 293 cells showed a 1.3- and 2.8-fold reduction, respectively. The minimal sequence required for a megakaryocytic cell-specific DNA-protein interaction was positions −113 to −99 (GAGGGGAGGAAGCGC), and the GAGGGG sequence behaved as a megakaryocyte-specific element (MSE), because mutations in this sequence affected activity in only the megakaryocytic lines. In addition, there was no detectable binding of HMEC-1 nuclear proteins to the GAGGGG sequence (Fig 4C). There are at least several possible explanations for the minimal activity of the MSE in β3-expressing endothelial and melanoma cell lines, such as (1) these cells contain cell specific factors that bind to enhancers not contained within our constructs, and (2) β3expression could be regulated in these cells by nontranscriptional mechanisms. Although our data indicate that a nuclear protein expressed at high levels in megakaryocytic cell lines and not in nonmegakaryocytic cells binds to the MSE of the β3 gene, non–tissue-specific factors also bound to several other adjacent sequences, and additional studies are required to determine whether these are involved in binding to the MSE.
We performed extensive searches of DNA databases for homologies to the GAGGGG sequence and the best matches were with genes matching via their CpG islands (none of which had any other relation to transcriptional activity or to megakaryocyte genes) and with three genes in the promoter database: wheat histone H3, the human c-sis/platelet derived growth factor 2 (PDGF2), and a human skeletal actin. Of these, only the region of the c-sis gene had been shown to affect gene expression, although the specific sequence identical to the β3 gene had not been studied.61 62 To assess whether this homologous sequence may be important in the context of other genes, we prepared a 28-bp DNA probe from the c-sis/PDGF2 gene corresponding to probe Pw in the β3 gene that contained the homologous region in its center. However, this probe did not gel shift any specific bands with K562 nuclear extracts (data not shown), suggesting coincidental homology or that other cell extracts or binding conditions will be required to detect a protein interaction.
Functional studies showed that the −146 construct displayed overall positive and megakaryocytic-preferential expression, and those experiments shown in Figs 1 and 3A suggested a negative element in the −126 to −115 region. EMSAs using this region showed very complex patterns, perhaps because of a 20-bp direct repeat at position −146, and mutations demonstrate that it has negative activity in both HEL and 293 cells (data not shown). A tissue-specific, negative regulatory sequence has been described with the β-globin gene,63 but further studies will be required to determine if this is present in the β3 gene. Another example of the complexity of DNA-protein interactions at the MSE was the consistently observed reduction in the P1 protein complex with probe Pw and nuclear extracts from PMA-stimulated K562 cells (Fig 4C). Thus, although PMA treatment of K562 cells induces β3 RNA and protein expression (Fig 2), such overall positive effects on expression must operate through another site, while possibly suppressing the DNA-protein interaction at −113 to −108. Taken together, this transcriptional regulatory unit of the β3 gene contains both positive and negative elements and multiple nuclear factors are likely acting on them to regulate the complex pattern of β3 expression.
Sp1 is a zinc finger transcription factor that has been shown to regulate expression in other integrin genes, including αIIb,25 α2,64 and four of the leukocyte integrins, CD11a,65CD11b,66 CD11c,65 and CD18.67 The β3 gene has at least four potential Sp1 binding sites and the promoter is GC-rich, TATA-less and lacks a consensus Inr element, which are often features of Sp1-responsive genes. Typically, the most important Sp1 element is 40 to 70 bp upstream of the transcription start site,68 so we focused on the only site in this region of the β3 gene, ie, that at position −70. This binding site does not conform to the classical Sp1 consensus binding site (GGGCGG), but rather has a sequence observed in a variety of other genes (CNCACCC [N = A, T, or C]). In addition to Sp1, this sequence will also bind CACD or EKLF, which is felt to be an erythroid-specific transcription factor related to the Kruppel family of nuclear proteins.60,69 There are several lines of evidence against EKLF binding to this site. (1) The MoAb used in the gel shift assay reacts specifically with the carboxyl terminus of Sp1 and does not cross-react with Sp2, Sp3, Sp4, or EKLF. (2) Sp1 is expressed in K562 cells,70 whereas EKLF is not.71 (3) An EKLF knock-out mouse had no megakaryocyte phenotype.72 In addition, the data in Figs 7 and 8 provide strong functional, biochemical, and immunologic evidence that Sp1 is most likely the major factor binding to this sequence at position −70 of the β3 gene. This site is in a region of modest transcriptional activity (Fig 3) and does not require or cooperate with the MSE, although the two sites have additive effects (Fig 8).
Sp1 is ubiquitously expressed and as such might not, by itself, be expected to demonstrate the increased abundance of complexes that we observed with DNA in megakaryocytic cell lines compared with nonmegakaryocytic lines (Fig 6B). In the case of CD11c, Sp1 has been shown to participate in tissue-restricted expression,65,73but there are other possible explanations for the more abundant Sp1-DNA complexes observed with megakaryocytic cell lines, such as partial degradation of nuclear proteins in the nonmegakaryocytic cells or a protein complexed to Sp1 in these cells that enhances (in megakaryocytic cells) or prevents (in non–β3-expressing cells) Sp1 binding to the CCCACCC sequence. Sp1 can also mediate transcription through the CCACCC sequence via an indirect stimulation with the retinoblastoma (RB) gene product.74 Although we cannot exclude the possibility that there are proteins positively or negatively modulating Sp1 binding at position −70, we presently have no evidence that either occurs.
Few general principles regarding integrin gene expression have been elucidated,75 although the promoters of most integrin genes studied to date tend to be GC-rich and lack TATA and CAAT boxes, and only a few have Inrs.46,76-80 Repressor elements are features common to integrins,16,81,82 and the β3 gene contains at least two: one between −290 and −146 and another between −126 and −115. The β2 promoter has been studied in some detail: two members of the Ets family (PU.1 and GABP) are required for cell-specific expression and both Sp1 and retinoic acid receptor binding sites have been identified.79,83-85 Sequence analysis of the 5′ regions of α integrin genes has shown binding sites for Sp1, GATA-1, AP1, AP2, and Ets, although few functional studies have been performed.77,78,80,86,87 The αIIb gene is expressed solely in megakaryocytic cells, and this expression is controlled at least in part by Sp1 and factors binding to GATA-1 and Ets sites.25,88 The β3 gene has an ets/PU.1 site at −105, but our EMSA studies suggest there is most likely no ets/PU.1 binding (Fig 4C and D). There are several GATA-1 sites in the 5′ region of the β3 gene (in studies not shown, a GATA-1 expression construct was not able to transactivate through these), but none in sequences that showed the greatest cell-specificity in these studies. Thus, as would be expected, some aspects of β3 expression are mediated via different mechanisms than αIIb. On the other hand, a 22-bp sequence of the αIIb gene containing the GAGG portion of this sequence has been shown to bind to megakaryocytic cell nuclear proteins by DNaseI footprinting and EMSAs.23 Prandini et al20 mutated the last 2 bp of this sequence (GAGG to GATT) and demonstrated a marked decrease in promoter activity in HEL and K562 cells, although nonmegakaryocytic cells were not tested. Thus, it will be interesting in the future to determine whether the same megakaryocyte-specific transcription factor acts on these sites in both the αIIb and β3 genes. Such a mechanism could contribute to some degree of coordinated gene regulation.
Supported by Grant No. HL51457 from the National Institutes of Health (Bethesda, MD) and by the Rogers-Wilbur Foundation.
Address reprint requests to Paul F. Bray, MD, Ross 1015, Johns Hopkins University School of Medicine, 720 Rutland Ave, Baltimore, MD 21205.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.