Abstract
The high-affinity human granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor (GMR) consists of an alpha (GMRα) and a common beta (βc) subunit. The intracellular domain of βc has been extensively characterized and has been shown to be critical for the activation of both the JAK/STAT and MAP kinase pathways. The function of the intracellular domain of GMRα, however, is not as well characterized. To determine the role of this domain in GMR signaling, an extensive structure-function analysis was performed. Truncation mutants α362, α371, and α375 were generated, as well as the site-directed mutants αVQVQ and αVVVV. Although α375β, αVQNQβ, and αVVVVβ stimulated proliferation in response to human GM-CSF, the truncation mutants α362β and α371β were incapable of transducing a proliferative signal. In addition, both α371 and αVVVV were expressed at markedly reduced levels, indicating the importance of residues 372 to 374 for proper protein expression. More importantly, we show that GMRα plays a direct role in the activation of the JAK/STAT pathway, and electrophoretic mobility shift assays (EMSA) indicate that both GMRα and βc play a role in determining the STAT5 DNA binding complex activated by the GMR. Thus, the intracellular domain of the human GMRα is important for activation of the JAK/STAT pathway and protein stabilization.
© 1998 by The American Society of Hematology.
GRANULOCYTE-MACROPHAGE colony-stimulating factor (GM-CSF) is a glycoprotein cytokine that induces the proliferation and maturation of immature myeloid progenitors and the functional activation of more mature myeloid cell types. GM-CSF elicits these diverse responses through the GM-CSF receptor (GMR). The high-affinity GMR is known to be composed of a specific ligand-binding alpha subunit (GMRα) and a common beta subunit (βc), which is also a component of the interleukins-3 (IL-3) and -5 (IL-5) receptors. Both subunits are members of the class 1 subgroup of the cytokine receptor superfamily and contain a number of conserved motifs, including two fibronectin type III domains, four spatially conserved cysteine residues, and a membrane-proximal WSXWS motif in the extracellular domain. They are also characterized by a single transmembrane domain and an intracellular domain of variable length. Although the intracellular domains of class 1 cytokine receptors possess no intrinsic enzymatic activity, they have been shown to stimulate rapid and reversible tyrosine phosphorylation of cellular signaling proteins upon ligand-mediated receptor activation. Some members, including βc, possess conserved domains known as box 1 and box 2. These domains have been shown to be crucial for the ability of the receptor to generate a proliferative response, and box 1 has been shown to be the site of JAK2 interaction.1 2 In addition, a proline-rich region is found within the intracellular domain of some members of the superfamily, including GMRα.
The GMRα subunit binds GM-CSF, both with a low affinity as a monomer, and at a high affinity when associated with βc. The human βc subunit is incapable of binding GM-CSF but is necessary for the formation of the high-affinity complex, and it has been shown to play an important role in GM-CSF–induced signal transduction. The βc subunit plays an integral part in the activation of the MAP kinase pathway. Upon receptor activation, the adapter protein, Shc, binds to βc at Tyr577 through its PTB domain.3 Shc is then able to interact with GRB2 and SOS, which leads to the activation of other more downstream molecules in the pathway.4Phosphorylation of βc and other signaling molecules in response to GM-CSF is dependent on JAK2,5 which interacts with βc through the box 1 region.1,2 Deletion of box 1 or use of a dominant-negative JAK2 leads to an inability to stimulate transcription of both c-fos and egr-1 and inhibits proliferation upon GM-CSF stimulation.5 6
Members of the JAK family of kinases are known to phosphorylate and thereby activate latent transcription factors, termed signal transducers and activators of transcription (STATs). These proteins are normally located in the cytosol, but upon receptor activation, they bind to the receptor, where they can then be phosphorylated by receptor-associated JAK kinases. Upon phosphorylation, STAT dissociates from the receptor and forms homodimers and heterodimers with other activated STATs. The dimer then translocates to the nucleus, where it is able to bind its cognate DNA binding site and activate transcription.7 The STAT family consists of STATs 1 through 6. Four forms of STAT5 are known to exist. STAT5a (p94) and STAT5b (p92) are encoded by two distinct genes but are 96% identical at the amino acid level and appear to have arisen from gene duplication.8 In addition, shorter isoforms of both STAT5a and STAT5b have recently been identified,9-12 which represent carboxy-terminal truncations of each gene that give rise to a 77-kD form of STAT5a and an 80-kD form of STAT5b. These truncated forms lack the transcriptional activation domain and have been shown to function as dominant-negative forms of the STAT5 genes.12-15 Dominant-negative forms of STAT5 reduced expression levels of cis, pim-1, osm,Id-4, and c-fos and inhibited the proliferative response of BaF3 cells to IL-3 stimulation, indicating a significant role for STAT5 in both IL-3 and GM-CSF signal transduction.8 12-15
Both IL-3 and GM-CSF are known to activate STAT5.8 In human peripheral blood monocytes, both forms of STAT5a and the 92-kD form of STAT5b are activated in response to GM-CSF stimulation. Upon maturation to the macrophage stage, GM-CSF has been reported to be unable to activate the p77 form of STAT5a. Immunoprecipitation experiments imply a preferential activation of STAT5a over STAT5b.10Activation and expression of STAT5 homologues has been shown to be cell type-specific.10,15 16 Here we present evidence that the cytokine receptors themselves are able to affect the formation of specific STAT DNA binding complexes.
As stated above, the intracellular domain of βc mediates both activation of the MAP kinase and JAK/STAT pathways. The role of GMRα in GM-CSF–activated signal transduction is not as well characterized. We and others have shown that the 54-amino acid intracellular domain of GMRα is absolutely required for GMR functionality.11,17-22 In addition, Weiss et al17 have shown that the 29 membrane proximal residues of this domain are sufficient for the GMR to stimulate a mitogenic response to GM-CSF. In this study, we sought to determine the exact residues in the GMRα intracellular domain that were important for GM-CSF signaling. By comparing parental 32Dcl3 cells with populations of cells expressing either βc or both GMRα and βc, we found that both GMRα and βc affect the homologue of STAT5 activated in response to GM-CSF. Using truncation and site-directed mutants of the GMRα intracellular domain, we determined that this domain is critical for activation of JAK2 and is able to determine which homologue of STAT5 is activated. In addition, we show that the intracellular domains of both GMRα and βc play a role in determining the type of DNA binding complexes formed by STAT5 homologues. We also found that the truncation mutants α362 and α371 were unable to transduce a mitogenic signal, unlike α375, αVQNQ, and αVVVV. Lastly, both α371 and αVVVV were expressed at considerably lower levels in 293T cells than the wild-type GMRα, indicating that amino acids 372-374 are important for proper expression of GMRα.
MATERIALS AND METHODS
Generation of GMRα mutants.
Genes encoding the wild-type GMRα and βc subunits were cloned, as previously described.18 All mutants were created using PCR. Truncation mutants were amplified using the 5′ primer, α-14 (GAACCCTGTACAAGCTTCCTTCGG), in conjunction with α-12 (GGTTATCATTGCGGCCGCCTTAGATCTGTGGAACTG), α-20 (CTCCCAGATGATCGCGGCCGCCACCTAATGGTTATC), and α-31 (GGTAAGTTGCGGCCGCATCTACTCGTCTTCC) to create α362, α371, and α375, respectively. Site-directed mutagenesis of the VEDE region was performed using α-33 (GGTGAATTCCTCCCAGATGATCACGACTACCACCTCATGG) and α-34 (GGTGAATTCCTCCCAGATGATCTGGTTTTGCACCTCATGG), together with α-14 to amplify the αVVVV and αVQNQ fragments, respectively. The amplified αVVVV and αVQNQ fragments were then used to replace the 5′ region of the wild-type GMRα subunit between the HindIII andEcoRI sites with the mutant sequences. All mutants were confirmed by sequencing, using the Sequenase 2.0 kit (United States Biochemical, Cleveland, OH). Mutant and wild-type GMRα subunits were then ligated into the Hind III/Not I sites of the mammalian expression vector, pCDNA-3 (Invitrogen, Carlsbad, CA). A plasmid coding for wild-type βc was constructed as previously described.18
Cell lines.
The murine factor-dependent cell line, 32Dcl3 (generously provided by Joel Greenberger, University of Pittsburgh), was maintained in 1× Iscove's + 10% fetal bovine serum (FBS), L-glutamine, and antibiotics. Media was supplemented with 200 pmol/L murine GM-CSF (mGM-CSF; generously provided by Amgen, Thousand Oaks, CA). 32Dcl3 cells were electroporated with the wild-type β subunit and either wild-type or mutant α subunit constructs at 250 V, 960 microfarads (μF) using a BioRad Gene Pulser (BioRad Laboratories, Richmond, CA). Transfected cells were cultured for 24 hours in the presence of 200 pmol/L mGM-CSF. Cells stably expressing functional GM-CSF receptors were then selected for by growth on 200 pmol/L human GM-CSF (hGM-CSF; kindly provided by Amgen). Cells stably expressing βc (32Dβ8) were generated as previously described.18 Expression of GMRα and βc constructs was verified by flow cytometry. 32Dα362β cells were created by introduction of α362 into 32Dβ8 cells, and a clone stably expressing this mutant was isolated by limiting dilution subcloning. No stable cell line expressing α371β could be isolated by selection on hGM-CSF, subcloning by limiting dilution, or G418 selection.
Expression and molecular weight of the GMRα mutants were verified by expression in the human fibroblast cell line, 293T. Wild-type and mutant GMRα subunits were introduced into the cells using CaPO4 transfection. Expression was analyzed by fluorescence-activated cell sorter (FACS) and Western blot.
Flow cytometry.
Expression of the wild-type and mutant GM-CSF receptors was analyzed by flow cytometry. Flow cytometry was performed as previously described by Ronco et al18 using anti–GM-CSFRα and anti-βc (Santa Cruz Biotechnology, Inc, Santa Cruz, CA) for primary antibodies and phycoerythrin (PE)-conjugated goat anti-mouse F(ab′)2 (Caltag, Burlingame, CA) as the secondary antibody.
Growth curves and 3H-thymidine uptake assays.
Growth curves were performed using 32Dβ8 cells transiently expressing constructs encoding mutant and wild-type GMRα subunits. Transient transfectants were purified using Mini-MACS columns (Miltenyi Biotech, Auburn, CA) according to the manufacturer's instructions. Purified cells were washed 3× with 1× phosphate-buffered saline (PBS), resuspended at a final concentration of 4 × 104/mL, and 1-mL aliquots were plated in 24-well tissue culture plates. Individual aliquots were treated with either diluent (1× PBS + 0.5% bovine serum albumin [BSA]), 200 pmol/L mGM-CSF, or 400 pmol/L hGM-CSF. Cells were counted at 24-hour intervals by Trypan blue exclusion.
For 3H-thymidine uptake assays, stable cell lines were used, except cells expressing α371β, for which it was necessary to use transient transfectants purified using Miltenyi Mini MACS columns. Cells were resuspended to a final concentration of 5 × 106/mL in 1× Iscove's media + 10% FBS and plated in 96-well flat-bottomed microtiter plates for 24 hours. Each cell line was then treated with diluent, 200 pmol/L mGM-CSF or 400 pmol/L hGM-CSF for an additional 18 hours. The cells were then treated with 1 μCi [methyl-3H]-thymidine (NEN) for 6 hours, lysed with 5% Triton X-100, and harvested onto glass fiber filters. Samples were counted in liquid scintillation fluid by a Beckman LS 1800 (Beckman Instruments, Irvine, CA) scintillation counter.
Immunoprecipitation and immunoblotting.
32Dcl3 cell lines were deprived of factor for 12 hours, followed by treatment with diluent, 200 pmol/L mGM-CSF, or 400 pmol/L hGM-CSF. Cells (107 per treated cell line) were then obtained, washed in 1 mL 1× PBS + 1 mmol/L sodium vanadate, and lysed in 0.5 mL mild lysis buffer, 1% Triton X-100, 20 mmol/L Tris (pH 8.0), 137 mmol/L NaCl, 10% glycerol, and 20 mmol/L EDTA (1 mmol/L phenylmethylsulfonyl fluoride [PMSF], 1 μg/mL leupeptin, 1 μg/mL aprotinin, 0.7 μg/mL pepstatin, and 10 μg/ml soybean trypsin inhibitor) for 30 minutes. Lysates were spun at 14,000 rpm for 5 minutes at 4°C to remove nuclei and cell debris. Lysates were precleared by incubation with 2 μg of normal goat serum, normal rabbit serum, or mouse IgG (Sigma Biosciences, St Louis, MO) for 1 hour and shaking at 4°C. Protein G Sepharose (Pharmacia Biotech, Inc, Piscataway, NJ) was then added, and lysates were incubated for an additional hour with shaking at 4°C. Precleared lysates were incubated with agarose-conjugated anti-JAK2 antibody (Santa Cruz Biotechnology, Inc), anti-STAT5a antibody, or anti-STAT5b antibody (Santa Cruz Biotechnology, Inc) for 12 hours at 4°C. STAT5 immune complexes were precipitated using ImmunoPure Plus agarose-conjugated protein A from Pierce (Rockford, IL). The STAT5a antibody was raised by immunizing rabbits against amino acids 1-132 of STAT5a (generously provided by Dr K. Shuai, UCLA). This antibody specifically recognizes the native forms of both the full-length and truncated STAT5a and does not crossreact with STAT5b at the dilution used (Dr K. Shuai, unpublished results, March 1996). Immunoprecipitates were washed 6× with mild lysis buffer, followed by elution in 1× sodium dodecyl sulfate (SDS) loading buffer. Immunoprecipitated proteins were separated on 7.5% SDS-polyacrylamide gels and then transferred to Hybond ECL nitrocellulose membranes (Amersham, Arlington Heights, IL). Nitrocellulose blots were immunoblotted with anti-JAK2 (Upstate Biochemical, Inc, Lake Placid, NY) or 4G10 anti-phosphotyrosine (Upstate Biochemical, Inc) antibodies. The immune complexes were visualized using horseradish peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG antibodies (Amersham) and treatment with enhanced chemiluminescence (ECL) detection reagents (Amersham).
Antiphosphotyrosine and Western blotting.
Phosphotyrosine immunoblotting was performed as described by Ronco et al.18 Protein was quantitated using the BCA Protein Quantitation kit (Pierce), and results were obtained on a Molecular Devices (Menlo Park, CA) Emax microplate reader. For detection of STAT proteins, 15 mg of protein or 105 cells were separated on 7.5% SDS-polyacrylamide gels, transferred to Hybond ECL nitrocellulose membranes (Amersham), and probed with anti-STAT5a, anti-STAT5b, anti-STAT1 (generously provided by Dr K. Shuai), and anti-STAT3 (Zymed Laboratories, Inc [San Francisco, CA]; Santa Cruz Biotechnology, Inc; and generously provided by Dr K. Shuai) antibodies. Protein complexes were detected with horseradish peroxidase-conjugated anti-rabbit IgG or horseradish peroxidase-conjugated anti-mouse IgG and ECL detection reagents.
Electrophoretic mobility shift assays (EMSA).
Following treatment with diluent, 200 pmol/L mGM-CSF or 400 pmol/L hGM-CSF, 107 cells were lysed and prepared as previously described.23 The protein concentration of the lysates was determined using the BCA Protein Quantitation kit from Pierce. Twenty milligrams of each lysate was fractionated on 5.3% polyacrylamide gels, dried, and visualized by autoradiography. The DR probe corresponds to the γ-interferon activating sequence (GAS) element from the human Fcγ receptor I (FcγRI) gene,24 and the high-affinity Sis-inducible factor (HSF) probe corresponds to the human c-fos SIE.25 The γ-responsive element (GRE) probe was used a cold competitor. This probe corresponds to the GRE-1 sequence (CCTTACTATAAACTCCCCGTTTATGTGAAATGGA) of the migpromoter,26 and only the STAT1 homotetramer specifically binds to this element.
RESULTS
Construction and expression of the human GM-CSF receptor α subunit intracellular domain mutants.
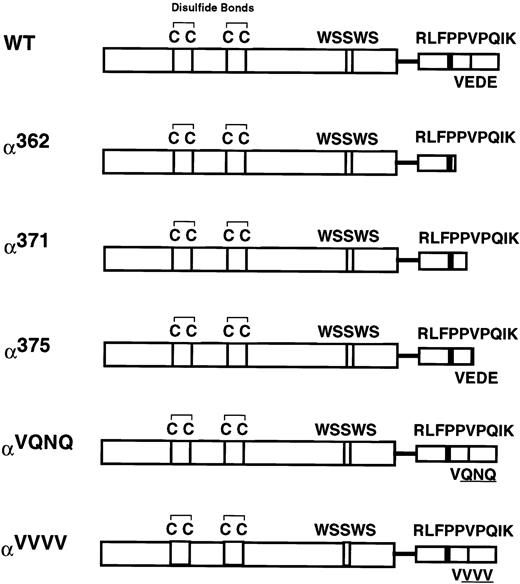
We previously showed that the 54-amino acid intracellular domain of the human GM-CSF receptor α subunit (GMRα) is required for GM-CSF–induced signal transduction.18,19 Weiss et al17 have shown that it is possible to truncate the GMRα intracellular domain down to 29 amino acids without having adverse effects on the ability of the GMR to transduce a mitogenic signal. In light of this, we sought to determine which residues in this short 29-amino acid region were critical to GMR function. We created three truncation mutants with intracellular domain lengths of 16 (α362), 25 (α371), or 29 (α375) amino acids by inserting a termination codon at the indicated position, using polymerase chain reaction (PCR; Fig 1). In addition, the experiments described below suggested that residues 372-374 (the 27th through 29th residues in the GMRα intracellular domain) were important for GMR functionality. We therefore sought to define the importance of this region by mutating it from the sequence Glu-Asp-Glu to Gln-Asn-Gln (αVQNQ) and Val-Val-Val (αVVVV) (Fig 1). αVQNQ was designed to conserve the biochemical nature of the residues as much as possible, whereas αVVVV was designed as a more disruptive mutation that replaces the wild-type charged, acidic residues with residues bearing small hydrophobic side chains.
Cartoon of the human GMRα subunit intracellular domain mutants. Depicted are motifs that are conserved within the cytokine receptor superfamily. The four spatially conserved cysteines and the WSXWS motif are located within the extracellular domain. A single transmembrane domain is depicted as a line, and the semiconserved proline-rich and VEDE regions are indicated in the intracellular domain. The wild-type hGMRα is 400 amino acids in length. Truncation mutants have a stop codon engineered at the amino acid number indicated. Mutagenesis of the VEDE region (residues 372 through 374) to the sequences VQNQ and VVVV is also depicted.
Cartoon of the human GMRα subunit intracellular domain mutants. Depicted are motifs that are conserved within the cytokine receptor superfamily. The four spatially conserved cysteines and the WSXWS motif are located within the extracellular domain. A single transmembrane domain is depicted as a line, and the semiconserved proline-rich and VEDE regions are indicated in the intracellular domain. The wild-type hGMRα is 400 amino acids in length. Truncation mutants have a stop codon engineered at the amino acid number indicated. Mutagenesis of the VEDE region (residues 372 through 374) to the sequences VQNQ and VVVV is also depicted.
To assay for expression, wild-type and mutant GMRα were transiently expressed in 293T fibroblasts. Cell surface expression was assayed by FACS, and all constructs were found to be present on the cell surface (data not shown). The cells expressed α362, α375, and αVQNQ at levels similar to those of the wild-type GMRα, whereas αVVVV and α371 were expressed at markedly lower levels, being only 60% and 7% of the level of the wild-type GMRα, respectively. Western blot analysis of lysates from the same 293T cell lines showed migration of wild-type and mutant GMRα according to their expected molecular weights. Wild-type GMRα, α362, α375, and αVQNQ were all expressed at high levels, but αVVVV was present at lower levels, and α371 was barely detectable (data not shown). These data indicate that α371 and αVVVV are expressed at significantly lower levels than the wild-type GMRα in 293T cells and that they are not accumulated within cellular compartments because of a defect in processing. This implies that they are instead being targeted for degradation, possibly due to improper folding of the proteins.
Stable expression of mutant GMRα subunits in CTLL2 cells.
We initially attempted to coexpress wild-type and mutant GMRα constructs with βc in the murine cytotoxic T-cell line, CTLL2. This cell line is an excellent system in which to study signaling activated by the human GMR due to the fact that it has been shown to express the requisite signaling molecules for GM-CSF mitogenic signal transduction yet is devoid of endogenous murine GMR subunits. We were, however, unable to efficiently introduce our constructs into this cell line and were therefore unable to create CTLL2 cell lines stably expressing the wild-type or mutant human GMR constructs. For this reason, we opted to use the factor-dependent murine myeloid cell line, 32Dcl3. These cells also contain the necessary signaling molecules to mediate mitogenesis in response to GM-CSF stimulation; they are also a well-established, outstanding system in which to study GM-CSF signal transduction. To create stable cell lines, 32Dcl3 cells were cotransfected with wild-type and mutant GMRα constructs along with the human βc subunit. Following transfection, cells were plated on 200 pmol/L hGM-CSF to select for a population of cells expressing functional human GM-CSF receptors (hGMR). Cells expressing wild-type GMRα, α375, αVQNQ, or αVVVV were readily selected by this method. α362 was transfected into 32Dβ8 cells, which stably express the human βc, and a clone of cells stably expressing α362β was isolated by limiting dilution subcloning. A population of cells stably expressing α371β could not be isolated using any method, and all subsequent experiments used purified populations of 32Dβ8 cells transiently expressing α371.
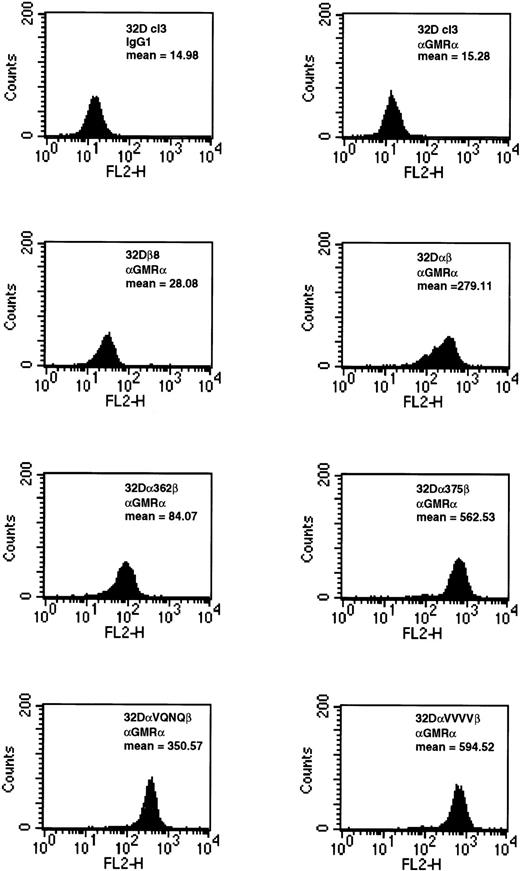
Relative expression levels of wild-type and mutant GMRα constructs in 32Dcl3 cells were similar to those seen in 293T cells. Cells expressing wild-type GMRα, α375, αVQNQ, or αVVVV were all expressed at high levels on the cell surface (Fig 2). The level of expression of α362 was marginally lower than those of other constructs. The fact that αVVVV is expressed at higher levels in 32Dcl3 cells indicates that we selected higher expressing populations or that the 32Dcl3 cells are able to express this construct more readily than 293T cells (Fig 2). Confirmation of βc expansion was determined by FACS (data not shown). Equilibrium binding analysis on 32Dα362β cells indicated that this mutant receptor bound GM-CSF with a high affinity (kd=17 pmol/L) and that high-affinity receptor numbers were at physiological levels (1,301 receptors per cell, data not shown). These results are consistent with previous data generated by us and others, showing that mutations in the intracellular domain do not affect high-affinity binding of GM-CSF to the GMR.11,17-21 27 It should be noted that cell lines made by stably expressing the various GMRα constructs in 32Dβ8 cells were indistinguishable from cells created by cotransfection in all assays performed.
Expression of wild-type and mutant GMRα subunits in 32Dcl3 cells. 32Dcl3 cells stably expressing wild-type and mutant GMRα were analyzed for cell surface expression by FACS. In the FACS analysis, indicated cells were stained with either IgG1isotype control or anti–GMRα subunit antibody (Santa Cruz Biotechnology, Inc) and analyzed as described in Materials and Methods.
Expression of wild-type and mutant GMRα subunits in 32Dcl3 cells. 32Dcl3 cells stably expressing wild-type and mutant GMRα were analyzed for cell surface expression by FACS. In the FACS analysis, indicated cells were stained with either IgG1isotype control or anti–GMRα subunit antibody (Santa Cruz Biotechnology, Inc) and analyzed as described in Materials and Methods.
Proliferation in response to human GM-CSF.
To elucidate the effects of the mutations on the ability of the GMR to stimulate a mitogenic response, growth curves and3H-thymidine uptake assays were performed. For3H-thymidine uptake, cell lines were deprived of factor for 24 hours and treated with diluent, 200 pmol/L mGM-CSF, or 400 pmol/L hGM-CSF for 18 hours, then incubated with 3H-thymidine for 6 hours. Parental 32Dcl3 and 32Dβ8 cells were both unable to respond to hGM-CSF (Fig 3). 32Dcl3 cells expressing the wild-type GMR, α375β, αVQNQβ, or αVVVVβ incorporated 3H-thymidine in response to hGM-CSF at levels comparable with mGM-CSF. In fact, the response for all of these cell lines was greater than that seen following treatment with mGM-CSF. This may be due to the fact that all of the constructs are overexpressed and, therefore, the number of hGMR subunits on the cell surface is greater than the number of endogenous mGMR subunits. 32Dcl3 cells expressing α362β and 32Dβ8 cells transiently expressing α371, on the other hand, were unable to respond to hGM-CSF, showing that neither receptor is functional.
[methyl-3H] Thymidine uptake by 32Dcl3 cell lines expressing wild-type and mutant GM-CSF receptors. Indicated cell lines were plated as described in the Materials and Methods. Each cell line was stimulated with either diluent, 200 pmol/L mGM-CSF, or 400 pmol/L hGM-CSF. After exposure to 1 μCi [methyl-3H]-thymidine, cells were obtained and counted.
[methyl-3H] Thymidine uptake by 32Dcl3 cell lines expressing wild-type and mutant GM-CSF receptors. Indicated cell lines were plated as described in the Materials and Methods. Each cell line was stimulated with either diluent, 200 pmol/L mGM-CSF, or 400 pmol/L hGM-CSF. After exposure to 1 μCi [methyl-3H]-thymidine, cells were obtained and counted.
These results were supported by growth curve experiments. Miltenyi Mini MACS–purified populations of 32Dβ8 cells transiently expressing wild-type or mutant GMRα were tested in the growth curve experiments. All populations were more than 93% pure for expression of GMRα, and purification techniques selected only cells that expressed the GMRα constructs at high and roughly equivalent levels, as determined by FACS analysis. Because the various alpha subunits were all expressed in 32β8 cells, βc expression is equivalent and, thus, βc does not contribute to differences seen in proliferative potential. Transient populations were used to normalize the response of each receptor to α371β. These experiments again showed that 32Dβ8 cells were incapable of proliferating in response to hGM-CSF. This was also the case for both 32Dβ8 cells expressing α371 and α362β, indicating that sequences lying between residues 362 and 375 are required for induction of proliferation. In contrast, 32Dβ8 cells expressing the wild-type GMRα, α375, αVQNQ, or αVVVV were all able to stimulate a proliferative response when treated with hGM-CSF (data not shown). Thus, it appears that a minimal length of 29 amino acids is required for the hGMR to stimulate a proliferative response. The growth characteristics described above were similar, whether 200 or 400 pmol/L of hGM-CSF was used. Dose response experiments have previously shown that these concentrations give rise to a maximal proliferative response to GM-CSF.18 Similar growth curve results were obtained using stable cell lines for all mutants except α371β. Because of the extremely low expression of α371, it was possible only to assay for expression and proliferation, and no further experiments were performed on cells expressing this mutant.
Phosphorylation of STAT5 homologues by GMRα subunit mutants.
It has been previously shown that GM-CSF stimulates phosphorylation of a number of cellular proteins.18 We wanted to determine the effect the various intracellular domain mutations have on the ability of the hGMR to stimulate phosphorylation of these cellular proteins. Two major phosphoproteins having apparent molecular weights of 94 and 77 kD were induced in response to GM-CSF (data not shown). The phosphorylation of these proteins was induced by mGM-CSF in all cell lines. hGM-CSF was able to induce the phosphorylation of these proteins only in cells expressing the wild-type GMR, αVQNQβ, or, to a lesser extent, α375β and αVVVVβ. 32Dβ8 cells transiently expressing α362 were incapable of stimulating phosphorylation of these proteins in response to hGM-CSF, again indicating an inability to transduce a mitogenic signal. The size of the induced proteins corresponds to the known sizes of the full-length and the truncated forms of STAT5a, which are known to be activated in response to GM-CSF.8 10Reprobing of these filters with anti-STAT5 antibody verified that these bands represented STAT5 (data not shown). To assay for STAT protein expression levels, we probed Western blots of whole-cell lysates from 32D cell lines expressing hGMR subunits with antibodies to STAT1, STAT3, STAT5a, or STAT5b. We found that STAT1, STAT3, and both the full-length (p94) and the truncated (p77) forms of STAT5a and the 92-kD form of STAT5b are expressed in all cell lines (data not shown).
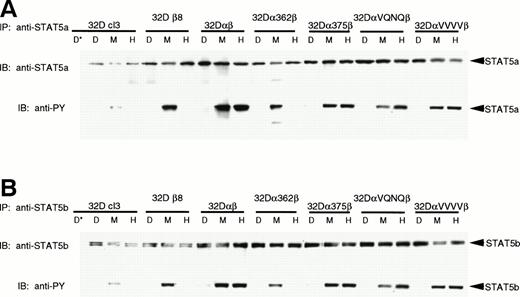
To further characterize the effects of the mutations on STAT5 phosphorylation, we immunoprecipitated STAT5a and STAT5b and assayed for tyrosine phosphorylation. Figure 4shows that both full-length STAT5a and STAT5b are present in all cell lines and are phosphorylated in response to mGM-CSF. In addition, cells lines expressing hGMR capable of stimulating proliferation (32Dαβ, 32Dα375β, 32DαVQNQβ, and 32DαVVVVβ) also phosphorylated the STAT5 proteins in response to hGM-CSF. Although our anti-STAT5a antibodies identify both full-length and truncated forms of STAT5a by Western blot and EMSA, we were unable to identify immunoprecipitation conditions that would allow us to immunoprecipitate the truncated form of STAT5a; we therefore were unable to use this method to determine whether this homologue is phosphorylated. Immunoprecipitation of STAT5b produced two very closely migrating bands representing different phosphorylation states of full-length STAT5b. Again, we were unable to immunoprecipitate the truncated form of STAT5b with our reagents.
Phosphorylation of STAT5a and STAT5b. Indicated 32Dcl3 cell lines were stimulated with diluent (D), 200 pmol/L mGM-CSF (M), or 400 pmol/L hGM-CSF (H), and lysed. Lysates were immunoprecipitated with anti-STAT5a (A) or anti-STAT5b (B) antibody or 2 mg mouse IgG or normal rabbit serum (*) as a negative control. Immunoprecipitates were subjected to SDS-PAGE (7.5% gel) and transferred to Hybond-ECL filters. Filters were probed with 4G10 antiphosphotyrosine antibody, stripped, and reprobed with anti-STAT5a or anti-STAT5b antibodies.
Phosphorylation of STAT5a and STAT5b. Indicated 32Dcl3 cell lines were stimulated with diluent (D), 200 pmol/L mGM-CSF (M), or 400 pmol/L hGM-CSF (H), and lysed. Lysates were immunoprecipitated with anti-STAT5a (A) or anti-STAT5b (B) antibody or 2 mg mouse IgG or normal rabbit serum (*) as a negative control. Immunoprecipitates were subjected to SDS-PAGE (7.5% gel) and transferred to Hybond-ECL filters. Filters were probed with 4G10 antiphosphotyrosine antibody, stripped, and reprobed with anti-STAT5a or anti-STAT5b antibodies.
Differential activation of STAT5 DNA binding complexes by GMRα subunit mutants.
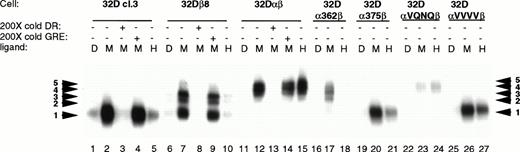
Since the mutations in the intracellular domain of GMRα affected the ability of both STAT5a and STAT5b to be phosphorylated in response to hGM-CSF, EMSA experiments were performed to see how this related to the ability of STAT5 to bind its cognate DNA binding site. EMSAs were performed on lysates from 32Dcl3 cells expressing wild-type or mutant GM-CSF receptors. Previous work has shown that in 32Dcl3 cells, a single gel shift complex is formed upon stimulation with IL-3.9 Likewise, mGM-CSF stimulates the formation of this same complex (complex 1; Fig 5). Expression of the human βc subunit in 32Dcl3 cells caused the formation of two more slowly migrating complexes (complexes 2 and 3), in addition to complex 1. Coexpression of the wild-type GMRα with βc resulted in the abrogation of these three complexes and formation of two new complexes (complexes 4 and 5) with even slower migration. Thus, it appears that not only βc but also GMRα plays a role in determining which STATs participate in the formation of DNA binding complexes in response to GM-CSF. Complete competition of all five induced complexes by 200-fold excess unlabeled DR oligonucleotide but not by excess GRE oligonucleotide showed the sequence specificity of these complexes.
STAT activation in 32D cell lines. 32D cell lines were stimulated with diluent (D), 200 pmol/L mGM-CSF (M), or 400 pmol/L hGM-CSF (H), and lysates were prepared and analyzed by EMSA using a32P-labeled DR probe. Specificity of binding was determined by competition with 200× cold DR probe and 200× cold GRE probe. The DR probe binds STAT5 and STAT1 with high affinity, whereas the GRE probe binds only the tetrameric form of STAT1 with high affinity.
STAT activation in 32D cell lines. 32D cell lines were stimulated with diluent (D), 200 pmol/L mGM-CSF (M), or 400 pmol/L hGM-CSF (H), and lysates were prepared and analyzed by EMSA using a32P-labeled DR probe. Specificity of binding was determined by competition with 200× cold DR probe and 200× cold GRE probe. The DR probe binds STAT5 and STAT1 with high affinity, whereas the GRE probe binds only the tetrameric form of STAT1 with high affinity.
Interestingly, the mutations in the intracellular domain of GMRα not only affected the amount of STAT5 complex formed, but also determined which specific complex was activated in response to GM-CSF. Cells expressing αVQNQβ activated the lowest mobility complexes (complexes 4 and 5) in response to both murine and human GM-CSF, similar to the wild-type GMR, but at greatly decreased levels. This decrease in the amount of complex formed is not due to lower receptor expression levels (Fig 2). Cells expressing α362β activated only the upper three complexes (3, 4, and 5), and only in response to mGM-CSF. Both α375β and αVVVVβ activated only the highest mobility complex (complex 1), but they were able to do so in response to both mGM-CSF and hGM-CSF. It is interesting to note that α375β causes the formation of reduced levels of STAT5 DNA binding complex in response to treatment with hGM-CSF. The differential complex formation seen in Fig 5 is not due to alternate activation of the STAT5 homologues, as evidenced in Fig 4. These results were obtained in multiple experiments using cell lines made on different occasions. Activation of STAT5 seems to correlate with proliferation, although the identity of the complex and relative amount of complex capable of binding to its cognate DNA binding element did not seem to matter.
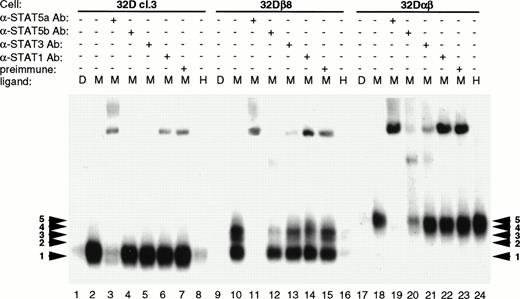
It has been shown that GM-CSF signals through both STAT5a and STAT5b, in addition to activating STAT1 and STAT3.28 The DR probe is known to be able to bind both STAT5 and STAT1 with high affinity, whereas the GRE probe binds only the tetrameric form of STAT1. To identify the components of the various complexes formed, EMSA was performed using antibody interference techniques. For this work, antibody to STAT5a, STAT5b, STAT3, or STAT1 was added to the lysates before incubation with radiolabeled probe. The antibodies bind to their respective antigen and prevent binding of the protein to the probe. As seen in Fig 6, antibody to STAT5a severely interferes with formation of all five complexes, indicating that the full-length or the truncated form of STAT5a is present in all of the complexes (Fig 6; lanes 3, 11, and 19). STAT5b antibody selectively interferes with binding of complexes 2, 3, and 4 to probe (Fig 6; lanes 4, 12, and 20). This indicates that STAT5b is associated with one or another form of STAT5a in these three complexes. No significant interference was seen using anti-STAT3 or anti-STAT1 antibodies (Fig 6; lanes 5, 6, 13, 14, 21, and 22). Both STAT1 and STAT3 have been shown to be activated in response to GM-CSF in human neutrophils.28 Because the DR probe binds STAT3 with only a low affinity, we performed EMSA with the same lysates using the HSF probe. This probe consists of the human c-fos SIE, which binds STAT1 and STAT3 with high affinity and STAT5 with low affinity. Using antibody interference EMSA, we found that STAT1 was activated to a minor extent, but no complex containing STAT3 was identified (data not shown). Thus, it appears that in 32Dcl3 cells, the hGMR preferentially activates the STAT5 homologues, with STAT1 only being activated to a small degree.
Identification of EMSA complex components by antibody interference. Parental 32Dcl3, 32Dβ8, and 32Dαβ cell lines were treated, and the lysates prepared as described in Materials and Methods. Lysates obtained from cells treated with 200 pmol/L mGM-CSF were treated with STAT5a, STAT5b, STAT3, STAT1, or preimmune serum before incubation with radiolabeled DR probe. EMSA was then performed on 5.3% polyacrylamide gels, which were visualized by autoradiography.
Identification of EMSA complex components by antibody interference. Parental 32Dcl3, 32Dβ8, and 32Dαβ cell lines were treated, and the lysates prepared as described in Materials and Methods. Lysates obtained from cells treated with 200 pmol/L mGM-CSF were treated with STAT5a, STAT5b, STAT3, STAT1, or preimmune serum before incubation with radiolabeled DR probe. EMSA was then performed on 5.3% polyacrylamide gels, which were visualized by autoradiography.
Activation of JAK2 by GMRα subunit intracellular domain mutants.
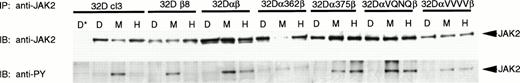
We next looked at the ability of the various mutants to activate JAK2 in response to GM-CSF stimulation. It is possible that the mutations allowed JAK2 to be activated but somehow directly prevented activation of STAT5 in response to hGM-CSF. Alternatively, lack of STAT5 activation could be due to an inability of the mutant GMR to stimulate phosphorylation of JAK2, which would abrogate all downstream signaling. As seen in Fig 7, 32Dcl3 and 32Dβ8 cells were able to stimulate phosphorylation of JAK2 in response to only mGM-CSF. Cell lines expressing functional human GMR (32Dαβ, 32Dα375β, 32DαVQNQβ, and 32DαVVVVβ), on the other hand, were able to activate JAK2 in response to both murine and human GM-CSF. However, 32Dcl3 cells expressing α362β were able to stimulate phosphorylation of JAK2 only in response to mGM-CSF, albeit at lower levels. Truncation of the intracellular domain of GMRα therefore leads to an inability of the GMR to activate JAK2. This abrogates all downstream signaling, leading to an inability to mount a proliferative response upon treatment with GM-CSF.
Activation of JAK2 by wild-type and mutant GM-CSF receptors. Indicated 32Dcl3 cell lines were stimulated with diluent (D), 200 pmol/L mGM-CSF (M), or 400 pmol/L hGM-CSF (H), and lysed. Lysates were immunoprecipitated with anti-JAK2 antibody from Santa Cruz Biotechnology, Inc and normal goat serum (*) as a negative control. Immunoprecipitates were subjected to SDS-PAGE and transferred to Hybond-ECL filters. Filters were probed with 4G10 antiphosphotyrosine antibody, stripped, and reprobed with anti-JAK2 antibody.
Activation of JAK2 by wild-type and mutant GM-CSF receptors. Indicated 32Dcl3 cell lines were stimulated with diluent (D), 200 pmol/L mGM-CSF (M), or 400 pmol/L hGM-CSF (H), and lysed. Lysates were immunoprecipitated with anti-JAK2 antibody from Santa Cruz Biotechnology, Inc and normal goat serum (*) as a negative control. Immunoprecipitates were subjected to SDS-PAGE and transferred to Hybond-ECL filters. Filters were probed with 4G10 antiphosphotyrosine antibody, stripped, and reprobed with anti-JAK2 antibody.
DISCUSSION
Binding of GM-CSF to its high-affinity receptor induces rapid tyrosine phosphorylation of cellular signaling proteins, causing subsequent transcriptional activation of a number of primary response genes such as c-myc, fos, and jun. Previous studies have delineated a role for βc in GM-CSF signaling. The role of GMRα in GM-CSF signaling, however, is not nearly as clear. Although the necessity in the intracellular domain of this subunit for GM-CSF signaling has been shown,11 17-22 the exact mechanism by which it functions has yet to be elucidated.
In this study, we sought to further define the residues within the GMRα intracellular domain that are required for GM-CSF–induced signaling. Our results indicate that residues 372-374 (EDE) of GMRα are important for proper expression of the protein. Truncation of GMRα at residue 375 (α375) had no effect on receptor expression, but deletion of four more residues (α371) resulted in a dramatic decrease in expression. In addition, mutation of residues 372-374 from the sequence VEDE to all valines (αVVVV) lowered expression levels to 60% of the wild-type level in 293T cells.
In agreement with Weiss et al,17 our observation that neither α362β nor α371β was able to transduce a proliferative signal, whereas α375β could, implies the absolute necessity of the membrane-proximal 29 amino acids, not only for protein stabilization, but also for signaling. Within this region lies not only the VEDE region, but also the proline-rich region. Such proline-rich regions are known to function as docking sites for SH3-containing proteins. It is possible that deletion or mutation of the VEDE region prevents binding of the SH3-containing protein to the GMRα proline-rich region and thus disrupts signaling in response to GM-CSF. JAK2 is known to be activated within 3 minutes of GM-CSF stimulation, and it is thought to be the primary event in activation of secondary messengers.1 Cells expressing α362β were not able to activate JAK2 or any of the STAT5 homologues in response to hGM-CSF, indicating that the signal is disrupted at a point upstream of JAK2 activation. These results show the importance of the GMRα intracellular domain in activating signal transduction pathways. Chimeric proteins fusing the intracellular domain of βc to the extracellular and transmembrane domain of both the GMRα and IL-5Rα subunits to the βc intracellular domain have indicated that dimerization of the βc intracellular domain is all that is required for a proliferative signal to be transduced.29-31 The goal of our experiments has been to understand the mechanism of signaling by high-affinity wild-type receptors.
Previous evidence has been presented by Lia et al11 that GMRα intracellular domain truncation mutants are dominant negative over the wild-type GMRα and inhibit the proliferative response to GM-CSF. Some of the results that we obtained for α362β indicated that α362 acted as dominant negative over the murine GMRα. Both JAK2 activation and STAT5 DNA binding complex formation in response to mGM-CSF were greatly decreased when compared with functional human receptors. Also, growth response to mGM-CSF was only approximately 50% of that seen for the other receptors. In addition, 32Dα362β cells were found to be of irregular shape and relatively unhealthy when compared with other cell lines by visual inspection. The dominant negative theory would also explain why we were unable to obtain a stable cell line expressing α371β. These observations again attest to the importance of GMRα in GM-CSF signaling.
We also show that the intracellular domains of both GMRα and βc play a role in determining which STAT5-containing DNA binding complexes are formed in response to GM-CSF. Previous work by Azam et al9 showed that 32Dcl3 cells activate only a single high-mobility complex in response to murine IL-3, and addition of the erythropoietin (Epo) receptor into these cells shifted this complex to a much lower mobility. We obtained similar results with parental 32Dcl3 cells, which also produced a single high-mobility complex in response to mGM-CSF stimulation. Addition of βc produces the formation of two intermediate mobility complexes, and expression of both βc and GMRα gives rise to the formation of two lower mobility complexes and abrogates the formation of all smaller complexes. Antibody interference techniques identified all complexes as containing either the full-length or truncated version of STAT5a, which has been previously described.10-12 The second, third, and fourth highest mobility complexes contain STAT5b as well as STAT5a. Thus, each subunit appears to alter the ability of different STAT5 DNA binding complexes to be formed in response to GMR activation. Although previous work has shown that GM-CSF is also capable of activating STAT1 and STAT3,28 we obtained only minor activation of STAT1, and no complex was identified that contained STAT3.
Each GMRα mutant had a distinct effect on which STAT5 complex was formed in response to receptor activation. The effect seen by the various GMRα mutants on STAT5 activation implies that in addition to βc, the intracellular domain of GMRα affects not only the stoichiometry of the STAT5 homologues present in the DNA binding complexes but also which homologues that are contained within these complexes. Analysis of STAT5 phosphorylation states indicates that the differential complex formation is not due to altered tyrosine phosphorylation of the full-length STAT5 homologues. The ratio of truncated to full-length STAT5a and STAT5b does not seem to vary either. Therefore, the differential DNA binding complexes formed do not seem to be due to the amount of activation or the quantity of the STAT5 homologues. Instead, the GMR itself or some other unknown factor must be responsible. Currently, the mechanism by which receptors might alter STAT complexes is unknown. Previous work has shown that STAT proteins bind to phosphorylated tyrosine residues via their SH2 domains. The binding sites for STAT5 have been identified for the Epo receptor and have the consensus sequence, XXYXXLD, with an acidic residue at −1 or −2 and a hydrophobic residue and +2.32The GMRα intracellular domain contains only one tyrosine residue, which does not conform to the sequences identified for the Epo receptor; therefore, STAT interaction at this site is unlikely. Although no evidence has been presented showing that STAT proteins interact with other proteins through SH3 domains, it is possible that STAT5 may interact with the proline-rich region of GMRα. Alternatively, GMRα indirectly affects the STAT5 interaction with tyrosine residues present within the intracellular domain of βc. This would directly affect the number and possibly the type of STAT present in the GMR signal transduction complex, and thereby alter the DNA binding complexes formed. Lastly, we propose that alteration of the GMR may affect the binding and/or activation of a STAT-interacting protein that is either a complex component or regulates complex formation. A number of proteins, including PIAS3 and p48, have been shown to interact with STAT proteins and mediate diverse functions, ranging from negative regulation to sequence-specific DNA binding.33-35 Alteration of the primary sequence or the subunit content of the GMR may affect the ability of the STAT-interacting protein to perform its function and leads to the differential complex formation seen in Fig 5. Further studies will be necessary to deduce the exact mechanism by which differential STAT complex formation occurs.
Interaction between murine and human GMR subunits has been previously observed.22 Our results not only corroborate these observations but also infer the existence of a GMR heteromultimer. EMSA analysis showed that the STAT5 DNA binding complexes formed by the GMR were determined by the identity of the human GMR subunits present in the cell. Stimulation of a given cell line with murine or human GM-CSF resulted in an identical pattern of STAT5 DNA binding activity, indicating a direct interaction between the murine and human GMR subunits. If this interaction was not occurring, the complex formed in response to mGM-CSF would be expected to remain constant in all the cell lines and not vary according to the presence of the human subunits. Our results indicate that both GMRα and βc are interacting with the murine GMR. Both the hGMRα and βc subunits must be present to transduce a signal in response to hGM-CSF. Likewise, both the murine subunits must interact to transduce a signal in response to mGM-CSF. Thus, at least a heterotetramer consisting of all four subunits must form to obtain the results seen in Fig 5. The presence of human βc or wild-type or mutant GMRα would delineate which homologue of STAT5 would preferentially interact with the receptor complex, and thus determine the homologue(s) activated in response to murine or human GM-CSF. It has been previously hypothesized that the GMR might in fact be an oligomeric complex consisting of α2β2 or a complex consisting of an as yet unidentified subunit, along with the GMRα and βc.36,37In addition, cross-linking studies have indicated the presence of not only GMRα and βc dimers, but also GMRα2/βc2 heterotetramers.11Molecular modeling studies also support the hypothesis that the GMR exists as a heterotetramer.38 Unusual flexibility by the ligand-binding region of the GMRα would be required for the GMR to bind to GM-CSF as only a heterodimer. Instead, a more likely scenario exists with the GMR present as a heterotetramer in which much more normal ranges of flexibility would be required.
We have previously presented evidence supporting the existence of a preformed complex consisting of GMRα and βc.18 The results presented here indicate that this preformed complex does not consist of a heterodimer but is instead composed of multiple subunits. In this case, murine and human subunits would randomly associate and dissociate with each other on the cell surface. Binding of either murine or human GM-CSF to the preformed heteromultimers would activate these chimeric heteromultimers and activate signaling.
ACKNOWLEDGEMENT
We thank Ke Shuai and Johanna ten Hoeve for expert technical assistance, critical reading of the manuscript, and thoughtful discussions. We are indebted to Emily Chou and Felix Shin for endless hours of technical assistance. We are also grateful to Wendy Aft for careful preparation of the manuscript. Flow cytometry was performed with the help of Negoita Neagos in the UCLA Jonnson Comprehensive Cancer Center Flow Cytometry Core Laboratory.
Supported by National Institutes of Health Grant No. RO1 CA40163. S.E.D was supported by the Tumor Immunology Training Grant, CA9120-19. The UCLA Jonnson Comprehensive Cancer Center Flow Cytometry Core Laboratory is supported by the JCCC Core Grant No. CA-16042.
Address reprint requests to Judith C. Gasson, PhD, Director, UCLA Jonsson Comprehensive Cancer Center, 8-684 Factor Building, Box 951781, Los Angeles, CA 90095-1781.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 3. [methyl-3H] Thymidine uptake by 32Dcl3 cell lines expressing wild-type and mutant GM-CSF receptors. Indicated cell lines were plated as described in the Materials and Methods. Each cell line was stimulated with either diluent, 200 pmol/L mGM-CSF, or 400 pmol/L hGM-CSF. After exposure to 1 μCi [methyl-3H]-thymidine, cells were obtained and counted.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/3/10.1182_blood.v92.3.867/5/m_blod41514003x.jpeg?Expires=1769207960&Signature=40b4blAPG1hRoN3Z5jg2LhT4BB2~LtVqQezCWRUh3E4KzqZmsr8OjRswNqW~8efCwUV4rn6SCVCRrV5yYytfFo-QW1Eh~~IH-~Djcan0oZDrovH3XZ821nUsumZQfh0fDz0iqZQ6wQfx7lE1v22fd1K0d9Eg1Jj4gazNb660mC860ZdvJOmgSzSk~1AcqQtJBSG182rsMupt6JvPTyy0jLzTE~HIOeS9~hliINCFDYZaXVpkvFn7FnTsSb6~-Jx99rHjfT5FhK15zTkjxXoBW9POOdRGLIaxnip~wlv6z-lnk57z5W~Ryx-GDVNF2drQ4k8A4SoMbUZZwngSxM7lhQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)