Abstract
CD164 is a novel 80- to 90-kD mucin-like molecule expressed by human CD34+ hematopoietic progenitor cells. Our previous results suggest that this receptor may play a key role in hematopoiesis by facilitating the adhesion of CD34+ cells to bone marrow stroma and by negatively regulating CD34+hematopoietic progenitor cell growth. These functional effects are mediated by at least two spatially distinct epitopes, defined by the monoclonal antibodies (MoAbs), 103B2/9E10 and 105A5. In this report, we show that these MoAbs, together with two other CD164 MoAbs, N6B6 and 67D2, show distinct patterns of reactivity when analyzed on hematopoietic cells from normal human bone marrow, umbilical cord blood, and peripheral blood. Flow cytometric analyses revealed that, on average, 63% to 82% of human bone marrow and 55% to 93% of cord blood CD34+ cells are CD164+, with expression of the 105A5 epitope being more variable than that of the other identified epitopes. Extensive multiparameter flow cytometric analyses were performed on cells expressing the 103B2/9E10 functional epitope. These analyses showed that the majority (>90%) of CD34+ human bone marrow and cord blood cells that were CD38lo/− or that coexpressed AC133, CD90(Thy-1), CD117(c-kit), or CD135(FLT-3) were CD164(103B2/9E10)+. This CD164 epitope was generally detected on a significant proportion of CD34+CD71lo/− or CD34+CD33lo/− cells. In accord with our previous in vitro progenitor assay data, these phenotypes suggest that the CD164(103B2/9E10) epitope is expressed by a very primitive hematopoietic progenitor cell subset. It is of particular interest to note that the CD34+CD164(103B2/9E10)lo/−cells in bone marrow are mainly CD19+ B-cell precursors, with the CD164(103B2/9E10) epitope subsequently appearing on CD34lo/−CD19+ and CD34lo/−CD20+ B cells in bone marrow, but being virtually absent from B cells in the peripheral blood. Further analyses of the CD34lo/−CD164(103B2/9E10)+ subsets indicated that one of the most prominent populations consists of maturing erythroid cells. The expression of the CD164(103B2/9E10) epitope precedes the appearance of the glycophorin C, glycophorin A, and band III erythroid lineage markers but is lost on terminal differentiation of the erythroid cells. Expression of this CD164(103B2/9E10) epitope is also found on developing myelomonocytic cells in bone marrow, being downregulated on mature neutrophils but maintained on monocytes in the peripheral blood. We have extended these studies further by identifying Pl artificial chromosome (PAC) clones containing the CD164 gene and have used these to localize the CD164 gene specifically to human chromosome 6q21.
© 1998 by The American Society of Hematology.
THE SIALOMUCINS are thought to play two key, but opposing, roles in vivo; the first as cytoprotective or anti-adhesive agents, and the second as adhesion receptors.1-4 Despite their common functions, these mucins encompass a heterogeneous group of secreted or membrane-associated proteins that vary in their apparent molecular weights from 50 to 3,000 kD and that share limited similarity/homology at the amino acid and nucleotide levels. They were identified initially on epithelial cells and, more recently, on endothelial cells and leukocytes. The epithelial-associated mucins include the MUC-1 to MUC-8 molecules, whereas the endothelial/leukocyte-associated mucins encompass CD34, CD43, CD45RA, CD164, GlyCAM-1, MAdCAM-1, TACTILE (CD96), and PSGL-1(CD162).1,3-5 Because of their structural heterogeneity, mucins have recently been defined by two criteria: (1) their high regional content of proline, threonine, and/or serine residues (20% to 55% of their amino acid compositions) and (2) their dense local concentrations of O-linked carbohydrates that are attached to these serine and threonine residues and that constitute one or more mucin-like domains.3,6 Such domains are a primary feature of molecules, such as PSGL-1, CD43 and CD164.5,7-9 However, for MAdCAM-1, CD34, and CD96, these mucin-like domains are interspersed with or linked to Ig-like structural motifs.3 10-13
For all the mucins, their high proline content and heavy O-linked glycosylation predict an extended filamentous conformation, with the membrane linked forms protruding above the glycocalyx. This provides an opportunity for their oligosaccharide sidechains and/or Ig-like domains to interact with ligands on opposing cells, bacteria, or viruses.3 It is now known that several of the leukocyte/endothelial-associated mucins mediate cell-cell adhesion, involving interactions between their mucin-like domains and the N-terminal C-type lectin domains of their appropriate L-, E-, or P-selectin ligands.4 These interactions may be enhanced by cooperativity between mucin-like domains and non-mucin motifs on the same molecule, thereby forming part of an adhesion cascade. In such cases, the mucin receptor/selectin ligand interactions would mediate the initial weak tethering of leukocytes to endothelium that precedes stronger integrin-mediated adhesion and subsequent transendothelial migration required for leukocyte trafficking.4 Although mucin receptors may be widely expressed, their function may differ on different cell types or on the same cell type under different states of activation. This functional diversity is dependent on the core peptide of the mucin and on the cell-specific expression of glycosyl transferases.1,4,14These, in turn, regulate the structure and presentation of the O-linked oligosaccharide sidechains, membrane anchorage, signal transduction abilities, and/or the trafficking of the mucin to the correct cellular domain.15-18 These alterations may then affect function.
Although a great deal of research has been directed toward the expression and function of the mucins on mature leukocytes and endothelial cells, there is still a paucity of information about their expression and function on human CD34+/hematopoietic “stem” and progenitor cells and on the associated stromal and endothelial cells that constitute the immediate stem-cell microenvironment. Previous studies have identified three of the mucin-like receptors, CD34, PSGL-1, and CD43, on primitive human bone marrow hematopoietic progenitor cells and/or associated microenvironmental stromal/endothelial cells.9,19-28 More recently, we have identified and cloned a fourth sialomucin, CD164, on human hematopoietic progenitor cells and bone marrow stromal reticular cells.5,7 On such cells, the four sialomucins have a variety of functions, with the specificity in receptor/ligand interactions depending on the structural characteristics of the mucin-like receptor. These functions include mediating,5,7,20,29,30 or regulating31,32hematopoietic progenitor cell adhesion and the negative regulation of their growth and/or differentiation.5,7,24,25 33-35
In this report, we have characterized four monoclonal antibodies (MoAbs) against the CD164 molecule. These MoAbs each recognize distinct epitopes on the CD164 molecule. We have examined the expression of these epitopes on CD34+ cells from cord blood and bone marrow. Because we have recently shown that antibody ligation of the 103B2/9E10 epitope can inhibit the proliferation of single human CD34+CD38lo/− hematopoietic progenitor cells in the presence of the cytokines, interleukin-3 (IL-3), IL-6, granulocyte colony-stimulating factor (G-CSF), and Steel factor in serum free medium and the adhesion of CD34+ cells to bone marrow stromal reticular cells,5 7 we also analyzed the distribution of this epitope on subsets of human bone marrow and cord blood cells and on their CD34+ cell precursors in some detail. Our results show that this epitope is present on phenotypically very primitive CD34+ progenitor cells, as defined by their coexpression of such surface antigens as AC133, Thy-1, FLT3 receptor, and c-kit receptor, but it exhibits differential expression on maturing B cells. Furthermore, using the CD164 cDNA as a probe, we have isolated PAC clones containing the CD164 gene and have used these to show, by fluorescence in situ hybridization (FISH) analysis, that the CD164 gene is localized to human chromosome 6q21.
MATERIALS AND METHODS
Cells and Cell Lines
Cell lines were cultured in RPMI 1640 or Iscove's modified Dulbecco's medium (IMDM) containing 10% to 15% (vol/vol) fetal calf serum (FCS; GIBCO-BRL, Paisley, Scotland). Heparinized human umbilical cord blood, normal peripheral blood, and normal bone marrow were drawn with informed consent of donors and with ethical permission from the Department of Transplantation Sciences, Southmead Hospital, Bristol, from local institutes or from the Medical Clinic, University of Tübingen, Tübingen, Germany.
Cell Isolation and Erythroid Cultures
Fresh human bone marrow, cord blood, and peripheral blood samples were fractionated on Ficoll-Hypaque (1.077 g/mL; Sigma Chemical Co, St Louis, MO). The light density cells were collected and any contaminating erythrocytes lysed in 0.147 mol/L NH4Cl for 30 to 60 minutes at room temperature. CD34+ cells were isolated using the Miltenyi Biotech (Bergisch Gladbach, Germany) Mini-MACS CD34 stem cell isolation kit as specified by the manufacturer. Cells were resuspended and cultured at 3,000 to 5,000 cells per mL in 24-well Falcon 3047 tissue culture plates (Becton Dickinson, Sunnyvale, CA) in defined StemBio-A.Erythro-CNRS serum-free media (StemBio-CNRS, Villejuif, France) containing optimal concentrations of the cytokines, IL-3, IL-6, Steel factor, and erythropoietin in a humidified incubator at 37°C in the presence of 4.5% (vol/vol) CO2 and 5% (vol/vol) O2 gas mixtures. Cells were obtained on days 5, 6, 9, and 13; the nucleated cell number determined; and cytospins phenotyped after centifugation onto slides as described below. In some instances, the cells were stained with antibodies and analyzed on the FACSCalibur (Becton Dickinson). Peripheral blood granulocytes (neutrophils) were isolated from 1 mL of Ficoll-hypaque–pelleted cells (density >1.077 g/mL) after a 20-second erythrocyte lysis with 20 mL of distilled water. After mixing, 20 mL of 0.308 mol/L ice-cold NaCl was added and the cells washed twice before staining.
Generation and Characterization of CD164 Antibodies
The CD164-specific MoAbs, 103B2/9E10 and 105A5, were generated5,7 after the immunization of mice with the erythromegakaryocytic cell line, MOLM-1; antibody 67D2 by immunization with the breast carcinoma cell line T47-D (obtained from the American Tissue Culture Collection); and antibody N6B6 by immunization with the pre–B-cell line Nalm-1 (obtained from the German Collection of Microorganisms and Cell Cultures), according to previously described methods.36 The 103B2/9E10 and 105A5 MoAbs were used for expression cloning. Comparative analysis revealed that both MoAbs recognized isolated FDCP-1 transfectant cell lines expressing human CD164. A detailed description of the isolation, sequencing, and generation of CD164 transfectant cell lines is described separately.7 In addition, the 67D2 and N6B6 MoAbs were also found to specifically recognize murine FDCP-1 cells expressing human CD164 but not the parental FDCP-1 cells (see Fig 1).
Antibodies and Antibody Conjugates
The isotypes of the MoAbs were determined by enzyme-linked immunosorbent assay (ELISA) (Boehringer-Mannheim, Mannheim, Germany). The murine 103B2/9E10 (IgG3 isotype), 105A5 (IgM isotype), 67D2 (IgG1 isotype,) and N6B6 (IgG2a isotype) MoAbs to human CD164 were used as culture supernatants or purified Ig preparations. The mouse anti-human CD34 MoAb (clone 43A1; IgG3) was generated by immunization with KG1A cells and then assigned to the CD34 cluster as described by Greaves et al.37 For immunohistochemistry, mouse MoAbs to human CD3 (clone 3D4; IgG1), glycophorin A (clone JC159; IgG1), glycophorin C (clone Ret40S; IgG1), and band III (clone Q1/156; IgG1) were purchased from Dakopatts (Copenhagen, Denmark) as culture supernatants or purified Ig fractions. For dual and multicolor fluorescence-activated cell sorter (FACS) analysis, the following mouse antibody conjugates, coupled with fluorescein isothiocyanate (FITC), phycoerythrin (PE), or PerCP were used: (1) CD3-PE or -FITC (clone SK7; IgG1); CD4-FITC (clone SK31,2; IgG1); CD8-FITC (clone SK1; IgG1); CD14-FITC (clone MOP9; IgG2b); CD20-PE or -FITC (clone L27; IgG1); CD19-PE or -FITC (clone SJ25C1; IgG1); HLA-DR-PE (clone L243; IgG2a); CD34-FITC, -PE, or -PerCP (clone 8G12; IgG1); CD38-PE (Hb-7; IgG1); and CD33-PE (clone p67.6; IgG1; all from Becton-Dickinson, San Jose, CA); (2) CD117-PE (clone 95C3; IgG1); CD65-FITC (clone 88H7; IgM); CD66b-FITC (clone 80H3; IgG1); and anti-glycophorin A-PE (clone 11E4B7; IgG1; all from Immunotech, Marseille, France); (3) CD90-PE (clone 5E10; IgG1; Pharmingen, Hamburg, Germany); (4) CD71 (clone T56/14; IgG1; Biotrend, Cologne, Germany); and (5) AC133-PE (clone AC133; IgG1; Amcell Corporation, Sunnyvale, CA). The CD135-PE (FLT3; clone SF1.340; mouse IgG1) was a kind gift from Drs O. Rosnet and A. van Agthoven, Marseille, France.38 As comparative negative controls, irrelevant antibodies (Dakopatts, Immunotech, or Becton Dickinson) of the same isotypes and with equivalent fluorescent tags or phosphate-buffered saline (PBS) were used in place of primary antibodies. Texas Red (TR)-, FITC-, or PE-conjugated isotype-specific secondary antibodies were purchased from Southern Biotechnology Associates Inc, Birmingham, AL, and FITC (Fab′)2 goat or rabbit anti-mouse Ig from Dianova, Hamburg, Germany or Dakopatts. All antibodies were used at 5 to 10 μg/mL per 107 cells/mL or at the concentrations recommended by the manufacturer.
Immunofluorescence Staining for Flow Cytometric Analysis and Cell Sorting
Single-color staining.
Cells were blocked with 10% (vol/vol) human AB serum (Behring, Marburg, Germany) or human gamma globulin (30% [vol/vol] Fc blocking reagent; Mitenyi Biotech) for 10 to 20 minutes at 4°C and then labeled with saturating concentrations of culture supernatants of the CD164 MoAbs, 103B2/9E10, 105A5, 67D2, or N6B6 MoAbs and counterstained with PE-conjugated goat anti-mouse isotype-specific Ig or with FITC-F(ab′)2 goat or rabbit anti-mouse Ig. To indicate background staining, isotype-matched control antibodies were used. For these and all subsequently described FACS analyses, positivity was determined by the use of arbitrary gates set against these negative isotype-matched controls.
Two-color staining of bone marrow and cord blood cells.
In some experiments (see Fig 3), bone marrow or cord blood cells were labeled with the CD164 MoAbs, 103B2/9E10 (IgG3), 105A5 (IgM), or N6B6 (IgG2a) and FITC-CD34 (8G12; IgG1) and then counterstained with PE-conjugated goat anti-mouse isotype-specific Ig. Alternatively, cells were stained with 67D2 (IgG1) plus CD34 (43A1; IgG3) before application of PE-conjugated anti-mouse IgG1 and FITC–anti-mouse IgG3. Peripheral blood mononuclear cells (see Fig 9) were labeled with 103B2/9E10 or an irrelevant IgG3− control antibody, together with CD4-FITC, CD8-FITC, CD3-FITC, CD20-FITC, or irrelevant FITC–isotype-matched controls before counterstaining with PE-conjugated anti-mouse IgG3.
Three-color staining of bone marrow and cord blood cells.
Flow cytometric analysis and cell sorting.
Cells were analyzed on a FACSCalibur using Cellquest software or analyzed and sorted on a FACS-Vantage flow cytometer (all from Becton Dickinson). The fluorescence of FITC, PE, and PerCP was excited with an argon ion laser at 488 nm and detected at emission wavelengths of 530 nm, 570 nm, and 670 nm, respectively. Cell sorting of CD34+CD164(103B2/9E10 epitope)+ and CD34−CD164(103B2/9E10 epitope)+ bone marrow cells was performed at 25 kHz using FITC-CD34 and the 103B2/9E10 MoAb followed by PE-conjugated anti-mouse IgG3. Sorted fractions were cytocentrifuged and stained with May-Grunwald-Giemsa solution for morphological analysis.
Dual-Color Immunofluorescence of Cytospins
All incubations were performed at room temperature for 30 minutes. After Fc receptor blockade as above, cytospins were incubated with 103B2/9E10, 105A5, or N6B6 MoAbs followed by FITC-conjugated isotype-specific secondary antibodies (1:25 dilutions in PBS). After washing in PBS, cells were incubated with CD3 (as a negative irrelevant IgG1 control), anti–glycophorin A, anti–glycophorin C, or anti–band III and developed with TR-conjugated goat anti-mouse IgG1 (1:50 dilution in PBS). The slides were mounted in fluorescent mounting medium (Dakopatts) containing 2% (wt/vol) 4,6-diamidine-2-phenylindole dihydrochloride (DAPI; Sigma) and viewed under an Olympus BX-60 fluorescence microscope (Olympus, London, UK).
Cross-Blocking Analysis of CD164-Specific MoAbs
For epitope mapping studies, MOLM-1 cells were incubated with either 103B2/9E10, 105A5, 67D2, N6B6 (all different isotypes), or with isotype-matched negative control antibodies all at concentrations of 5 μg/mL at 107 cells/mL for 30 minutes on ice. After washing, cells were incubated either with the same MoAbs to indicate positive or negative controls, respectively, or with test CD164 MoAbs that differed from the blocking MoAbs. In the final step, cells were stained with PE-conjugated anti-Ig that specifically identified the CD164 test MoAb and analyzed on a FACSCalibur flow cytometer. The percent blocking was calculated as follows: 100 − [(Median Fluorescence of Cells Stained With Test MoAb After Incubation with Blocking MoAb) − (Median Fluorescence of Cells Stained With Isotype-Matched Negative Control MoAb)] / [(Median Fluorescence of Cells Stained With Test MoAb After Incubation With Negative Control MoAb) − (Median Fluorescence of Cells Stained With Isotype-Matched Negative Control MoAb)] × 100.
Sensitivity of CD164 Epitopes to Vibrio choleraeNeuraminidase Treatment
Calu-1, KG1A, MOLM-,1 and BV-173 cells were incubated with 0.2 U/mL of neuraminidase from V. cholerae (Calbiochem, Heidelberg, Germany) for 60 minutes at 37°C in 250 mL PBS. After washing with ice-cold staining buffer, cells were blocked and labeled with each of the CD164 MoAbs or the appropriate negative isotype-matched control MoAb and counterstained with PE-conjugated isotype-matched anti-Ig. Cells were analyzed on a FACSCalibur, and percent binding was calculated as follows: [(Median Fluorescence of Neuraminidase-Treated Cells Stained With CD164 MoAb) − (Median Fluorescence of Neuraminidase-Treated Cells Stained With the Isotype-Matched Negative Control MoAb)] / [(Median Fluorescence of Untreated Cells Stained With CD164 MoAb) − (Median Fluorescence of Untreated Cells Stained With the Isotype-Matched Negative Control MoAb)] × 100.
CD164 cDNA and Probes
Two human CD164 cDNA clones (clone 105A5 and 103B2), isolated from a retroviral cDNA library of human bone marrow stromal cells39,40 by expression cloning with the 105A5 and 103B2/9E10 MoAbs,5,7 and subcloned into the pGEM-T vector (Promega, Southampton, UK) were transformed into XL2 Blue-MRF′ bacteria (Stratagene, Cambridge, UK). Large-scale DNA preparations of the two CD164 cDNA clones were purified on CsCl gradients,41 completely sequenced5,7 using oligonucleotide primers, and used to probe genomic libraries. The nucleotide and the predicted peptide sequences have been described previously.7 Initial screening of a human PAC library was performed with two CD164 cDNA probes, A and B, derived from the 105A5 cDNA clone in the pGEMT vector (Promega). The Sac 1/Spe1 probe A fragment contained 1 to 1907 bp of cDNA sequence, where bp1 indicates the translational start site. This encompasses the whole translated sequence plus part of the 3′UTR. The Spe1/Apa 1 probe B fragment comprised 1908 to 2867 bp of 3′UTR. Subsequent screening of the human PAC subclones and of Southern blots was performed with a 1.173 kbEcoRV/HindIII CD164 probe from the 105A5 cDNA (Probe C), which contained CD164 cDNA sequence, including a region spanning 1309 to 2487 bp of untranslated CD164 sequence in the 3′UTR. The restriction enzymes were purchased from Boehringer-Mannheim and endonuclease digestions performed in the buffers supplied with the enzymes and according to the manufacturer's instructions. Restriction fragments were isolated after electrophoresis on 1.5% to 2% (wt/vol) NuSieve agarose (FMC Bioproducts, Rockland, ME) in TAE buffer (40 mmol/L Tris-acetate buffer, pH 8.5, containing 2 mmol/L EDTA) and purification on Wizard PCR (polymerase chain reaction) preps columns (Promega) as detailed by the manufacturer. DNA concentrations were determined by agarose gel electrophoresis against known phage DNA quantitation standards (GIBCO-BRL). For labeling, 20 to 50 ng of each cDNA probe was labeled with 50 μCi α-32P–dCTP (Amersham Int, High Wycombe, Bucks, UK) using the T7 Quickprime kit (Pharmacia, Uppsala, Sweden) according to the manufacturer's protocol.
Isolation and Subcloning of PAC Clones
The human PAC library derived from normal human male genomic DNA in the PCYPAC2N vector, containing 120,000 clones and grided on to 7 Hybond N filters, was kindly provided by the HGMP Resource Centre (Cambridge, UK) via Dr P. de Jong's group. Filters were hybridized with the α-32P–labeled human CD164 probes A and B as described below. Five identified positive clones were obtained from the HGMP Resource Centre and grown overnight in 2× TY broth containing 25 μg/mL kanamycin. These were digested with EcoR1, electrophoresed on a 0.7% (wt/vol) agarose gel, Southern blotted, and probed with α-32P–labeled CD164 probe C or analyzed by PCR using forward MGC-GP-F3 and reverse MGC-GP-B3 oligonucleotide primers (Oswell, Southampton, UK) and the PCR products sequenced as described below. The MGC-GP-F3 (455-479) and MGC-GP-B3 (1601-1577) primers were 5′-CCTCACAACCTGTGCGAAAGTCTAC-3′ and 5′-ACTCAAGACAGTCTGGTGG AAATCC-3′, respectively. Two positive clones (termed CD164 PAC1 and PAC5) were selected and digested with Pst 1 or BglII according to the manufacturer's instructions and used directly for ligation to pCRScript vector. The pCRScript (SK+) vector was digested with Pst 1 orBamH1 for 1 hour at 37°C in the appropriate buffers before the addition of 1 μL of shrimp alkaline phosphatase (Boehringer Mannheim) for 1 hour at 37°C. After heat inactivation at 68°C for 15 minutes, the enzyme-digested vector was precipitated on dry ice with 2 volumes of ethanol in the presence 0.3 mol/L sodium acetate buffer, pH 5.2. The resulting pellet was resuspended in double distilled water and used for ligations of the digested PAC fragments. In brief, all ligation reagents used were derived from the pMOS-Blue kit (Amersham). Fifty nanograms of enzyme-digested and phosphatased pCRScript vector was mixed with 170 to 350 ng enzyme-digested PAC clone, 1 μL 10× ligation buffer, 0.5 μL 100 mmol/L dithiothreitol (DTT), 0.5 μL 10 mmol/L adenosine triphosphate (ATP), and 0.5 μL T4 DNA ligase (4 U/mL) in a final volume of 10 μL and incubated at 15 to 20°C for 2 hours. The ligated samples were transformed into XL2-Blue MRF′ competent bacteria, plated onto L-broth agar plates containing 50 to 100 μg/mL ampicillin, and grown overnight at 37°C. Lifts were made on Hybond N filters, and the filters were probed with α-32P–labeled CD164 Probe C. Positive clones were picked off each master plate, grown in 10 to 15 mL of L-broth containing 50 to 100 μg/mL ampicillin, and characterized by restriction enzyme digestion, Southern blotting, and probing with α-32P–labeled CD164 Probe C or by PCR analysis using the MGC-GP-F3/MGC-GP-B3 primers and sequencing of the PCR products. Using oligonucleotide primers derived from the cDNA sequence, the CD164 gene was contained in a 23-kb region of the PAC clones. Sequencing of these fragments has identified 5 exons that encode the isolated CD164 cDNA, interspersed with introns of varying sizes and extending from the translational start site (bp1) into the 3′ UTR ( to position 2867 bp of the cDNA).42 Exons 1; 1 to 2; and 1, 2, and 3 corresponding to the CD164 cDNA were subcloned into the pEFBos-Ig mu vector,43 expressed as soluble recombinant proteins in 293T cells, and analyzed for reactivity with the 4 CD164 MoAbs or with isotype-matched negative controls by Elisa assays.44,45 A detailed description of the PAC clones, the genomic structure of CD164, and the epitope mapping using soluble recombinant CD164 protein domains are presented in separate papers.42 45
PCR Analysis of Somatic Cell Hybrids, PAC Clones, and Human Genomic DNA
DNA extracts from human x hamster or human x mouse somatic cell hybrids were kindly provided by the HGMP Resource Centre, Cambridge, UK. These (100 ng) were analyzed by PCR using the MGC-GP-F3 and MGC-GP-B3 primer pairs described above or the MGC-GP-F4 (1566-1591) and MGC-GP-B4 (2069-2045) primer pairs, 5′-GTACCTTGAAAGGA TTTCCACCAGAC-3′ and 5′-CAAGTGCGAAACTCAGCCACTATTG-3′, respectively (all from Oswell, Southampton, UK) to the CD164 cDNA sequence. PCR analysis was also performed on two human male and female genomic DNA preparations, human genomic DNA provided with the hybrid panel, the BglII subclone of CD164 PAC1, and the CD164 cDNA clones, 103B2/9E10 and 105A5. The PCR on the somatic cell hybrid panel was performed with 100 ng of each chromosome 1-22; chromosome X; chromosome Y DNA; plus mouse, human, and hamster DNA controls using the Expand Long Template System (GIBCO-BRL) and the following program: a hotstart of 95°C for 2 minutes, followed by 30 cycles of 95°C for 1 minute for denaturation, 65°C for 30 seconds for annealing, and 69°C for 4 minutes for extension; and a final extension cycle of 69°C for 10 minutes. Essentially the same protocol was used for PCR analysis of the PAC clones and subclones and the CD164 cDNAs. PCR analysis was, however, performed on the human genomic DNA samples using the Advantage Klentaq System (Clontech, Palo Alto, CA) and the following PCR program used: a hotstart of 95°C for 2 minutes, followed by 5 cycles of 95°C for 30 seconds and 72°C for 3 minutes, 5 cycles of 95°C for 30 seconds and 70°C for 3 minutes, 25 cycles of 95°C for 30 seconds and 68°C for 3 minutes, and a final extension cycle of 68°C for 7 minutes. The PCR products (10 μL) were analyzed by 2% (wt/vol) agarose gel electrophoresis in Tris borate EDTA (TBE) buffer using a 1-kb DNA ladder (GIBCO-BRL) as the molecular weight marker and blotted onto Nytran-N Nylon membranes (Schleicher and Schuell, Dassel, Germany). These membranes were prehybridized in 30 mL of 50% (wt/vol) formamide, 4× SSC, 0.001 mol/L EDTA, 0.05 mol/L sodium phosphate buffer (pH 7.2), 8% (wt/vol) dextran sulfate, 100 μg/mL of sonicated Herring sperm DNA (Sigma), and 25 μg/mL yeast tRNA (Sigma), 10× Denhardt's solution46 and 50 μg human placental DNA (Sigma) at 42°C for 1 to 18 hours before addition of the α-32P–labeled CD164 probes A, B, or C for 18 to 48 hours at 42°C. Filters were washed twice at 42°C in 2× SSC with 0.1% (wt/vol) sodium dodecyl sulfate (SDS) for 30 minutes each, 3 times at 65°C in 0.2× SSC containing 0.1% (wt/vol) SDS for 45 minutes each, and then exposed to Kodak X-OMat film (Eastman Kodak, Rochester, NY) with intensifying screens at −70°C as described.46
Automatic Sequencing
Miniprep or CsCl gradient prepared DNA (200 μL) was denatured at 37°C for 30 minutes with 20 μL 2 mol/L NaOH and 2 mmol/L EDTA and then ethanol precipitated. In some cases, the PCR products were directly sequenced after PEG precipitation. The PCR products (4 μL) or the DNA (0.5 μg) in 5 μL sterile double-distilled were added to 4 μL of ABI Prism Ready Dye de-oxy Terminator mix (Applied Biosystems, Perkin Elmer, Foster City, CA) with 0.025 μg M13 (Stratagene) or 3 pmol of CD164 forward or reverse primers (Oswell) in 2 to 3 μL sterile double-distilled water, and the sequencing reaction was performed according to the manufacturer's protocol before analysis on an ABI 373 Automatic Sequencer (Applied Biosystems). The sequences were analyzed using MacVector, Seqed, Assemblign, Analysis, and Sequencher software packages (Oxford Molecular, Oxford, UK), aligned to each other and to the CD164 cDNAs and a contig generated.
FISH
Metaphase spreads were prepared from phytohemagglutinin-stimulated normal human lymphocytes by use of standard techniques. Before hybridization, the slides were denatured in 70% (vol/vol) formamide and 2× SSC at 73°C for 3 minutes, then washed in 2× SSC and dehydrated through an ethanol series of cold 70% (wt/vol), 95% (wt/vol), and absolute ethanol. The BglII and Pst 1 subcloned PAC DNA in the pCRScript vector was biotinylated using the Bionick kit (GIBCO-BRL). Two hundred nanograms labeled probe was mixed with 5 μg Cot-1 DNA (GIBCO-BRL), precipitated, resuspended in 11 μL hybridization mix, denatured at 85°C for 5 minutes, and allowed to preanneal at 37°C for 30 minutes. The probe was then applied to a denatured slide and hybridized overnight. Slides were washed in 50% (vol/vol) formamide, 2× SSC pH 7 at 42°C, followed by 1× SSC at 60°C. Blocking solution (3% [wt/vol] BSA, 4× SSC and 0.1% [vol/vol] Tween 20) was applied and slides incubated at 37°C for 30 minutes. After incubation, avidin-FITC (diluted in 1% [wt/vol] BSA, 4× SSC, 0.1% [vol/vol] Tween 20) was applied and slides incubated at 37°C for 40 minutes. Slides were washed in 4× SSC, 0.1% (vol/vol) Tween 20 at 42°C and counterstained with 200 ng/mL DAPI, followed by 2 minutes in 2× SSC. Slides were mounted in Citifluor and images captured using a Photometrics KAF 1400-500 CCD camera (Photometrics, Tuscan, AZ) attached to a Zeiss Axioskop (Zeiss, New York, NY) epifluorescence microscope. Separate images of probe signals and DAPI banding patterns were pseudocoloured and merged using SmartCapture software (Vysis Inc, Chicago, IL).
RESULTS
The MoAbs 103B2/9E10, 105A5, 67D2, and N6B6 Specifically Detect CD164 and Recognize Distinct CD164 Epitopes
The predicted amino acid sequence of cDNA clones selected by expression cloning of a human bone marrow stromal cell cDNA library with the CD164 MoAbs, 103B2/9E10 and 105A5, were identical. The complete sequence is described separately.7 Parental FDCP-1 cells or FDCP-1 cells expressing these human CD164 cDNAs were stained with 67D2 and N6B6 MoAbs, as well as with 103B2/9E10 and 105A5, and analyzed by flow cytometry. All four MoAbs selectively recognized CD164 cDNA transfected, but not parental, FDCP-1 cells (Fig 1). Our more recent data42indicate that the identified CD164 cDNA is encoded by 5 separate exons, with the 103B2/9E10 and 105A5 MoAbs reacting with soluble recombinant protein derived from exon 1. In contrast, the N6B6 and 67D2 MoAbs do not react with soluble recombinant proteins derived from exon 1 or exons 1 and 2, but bind to a recombinant protein comprising the extracellular domains encoded by exons 1, 2 and 3, as summarized in Table 1. Detailed epitope analyses using these and other soluble CD164 recombinant constructs are described elsewhere.45 Reactivities of the four MoAbs with cell surface–expressed epitopes of CD164 were analyzed in more detail on a set of hematopoietic and nonhematopoietic cell lines (data not shown). Because all four MoAbs reacted strongly with the MOLM-1, Calu-1, KG1A, and BV-173 cell lines, these were used in further experiments. Cross-blocking experiments were performed to determine whether the 103B2/9E10 and 105A5 or the N6B6 and 67D2 MoAbs recognized identical or different epitopes on cell lines. As indicated in Table 1, prelabeling of MOLM-1 with the 103B2/9E10 and 105A5 MoAbs did not substantially block 67D2 or N6B6 binding, nor did 105A5 MoAb prelabeling prevent the 103B2/9E10 MoAb from binding. However, 67D2 almost completely blocked N6B6 staining, and vice versa. Partial blocking of 105A5 was observed after 103B2/9E10 prelabeling, but not vice versa. This partial inhibition might be caused by conformational changes of the CD164 molecule potentially induced by the 103B2/9E10 MoAb. These results support those obtained for exon mapping and show a close association between the 103B2/9E10 and 105A5 epitopes and between the 67D2 and N6B6 epitopes.
Binding of CD164 MoAbs to stable FDCP-1 cells expressing CD164. FDCP-1 cells stably transfected with CD164 cDNA were labeled with the N6B6, 67D2, 105A5, or 103B2/9E10 MoAbs or with irrelevant first MoAbs of the same isotype. The reaction was developed with FITC-conjugated anti-mouse Ig antibody as detailed in Materials and Methods, and the median fluorescence values were determined after FACSCalibur analysis. The results of one of three experiments are shown. The IgG negative control histogram contains the second FITC antibody only. The median fluorescence intensity values for the negative isotype matched controls were: no first MoAb = 3.4; IgG3 = 3.4; IgG2a = 3.4; IgM = 3.4; and IgG1 = 3.3. The median fluorescence for each CD164 MoAb was: 103B2/9E10 = 45.32; 105A5 = 26.42; 67D2 = 138.24; and N6B6 = 121.37.
Binding of CD164 MoAbs to stable FDCP-1 cells expressing CD164. FDCP-1 cells stably transfected with CD164 cDNA were labeled with the N6B6, 67D2, 105A5, or 103B2/9E10 MoAbs or with irrelevant first MoAbs of the same isotype. The reaction was developed with FITC-conjugated anti-mouse Ig antibody as detailed in Materials and Methods, and the median fluorescence values were determined after FACSCalibur analysis. The results of one of three experiments are shown. The IgG negative control histogram contains the second FITC antibody only. The median fluorescence intensity values for the negative isotype matched controls were: no first MoAb = 3.4; IgG3 = 3.4; IgG2a = 3.4; IgM = 3.4; and IgG1 = 3.3. The median fluorescence for each CD164 MoAb was: 103B2/9E10 = 45.32; 105A5 = 26.42; 67D2 = 138.24; and N6B6 = 121.37.
Epitope Mapping of CD164-Specific MoAbs
| . | % Inhibition of Test CD164 MoAb Binding . | CD164 MoAb Reactivity With Soluble Recombinant CD164 Domain Constructs-150 . | |||||
|---|---|---|---|---|---|---|---|
| Test CD164 MoAb | Blocking CD164 MoAb | Exon 1 | Exons 1 and 2 | Exons 1, 2, and 3 | |||
| 103B2/9E10 | 105A5 | N6B6 | 67D2 | ||||
| 103B2/9E10 | — | 0 | 3.5 | 3.5 | + | + | + |
| 105A5 | 60.4 | — | 0 | 0 | + | + | + |
| N6B6 | 21.4 | 0 | — | 90.1 | − | − | + |
| 67D2 | 13.5 | 3.5 | 98.3 | — | − | − | + |
| . | % Inhibition of Test CD164 MoAb Binding . | CD164 MoAb Reactivity With Soluble Recombinant CD164 Domain Constructs-150 . | |||||
|---|---|---|---|---|---|---|---|
| Test CD164 MoAb | Blocking CD164 MoAb | Exon 1 | Exons 1 and 2 | Exons 1, 2, and 3 | |||
| 103B2/9E10 | 105A5 | N6B6 | 67D2 | ||||
| 103B2/9E10 | — | 0 | 3.5 | 3.5 | + | + | + |
| 105A5 | 60.4 | — | 0 | 0 | + | + | + |
| N6B6 | 21.4 | 0 | — | 90.1 | − | − | + |
| 67D2 | 13.5 | 3.5 | 98.3 | — | − | − | + |
MOLM-1 cells were stained with one of the unconjugated CD164-blocking MoAbs or with the appropriate isotype-matched negative control MoAb before labeling with the second test CD164 MoAb, which differed from the blocking MoAb. After staining these cells with PE-conjugated anti-Ig that specifically reacted with the CD164 test MoAb, they were analyzed on the FACSCalibur and their median fluorescence intensities determined. Percent blocking was calculated from median fluorescence intensities as indicated in Materials and Methods.
Soluble domain deletion constructs, derived from exons 1; 1 and 2; and 1, 2, and 3 were prepared after determination of the intron-exon structure of the CD164 gene,42 subcloned into the pEFBos-Ig mu vector,43 and expressed in 293T cells as soluble recombinant proteins as detailed elsewhere.45 The predicted peptide sequences for these soluble constructs are: Exon 1: DKNTTQHPNVTTLAPISNVTSAPVTSLPLVTTPAP; Exon 2: ETCEGRNSCVSCFNVSVVNTTCFWIECK; Exon 3: DESYCSHNSTVSDCQVGNTTDFCS with exon 1 predicted to contain nine O-linked glycosylation sites on the serine and threonine residues underlined. Potential N-linked glycosylation sites are indicated in bold lettering. The data represent a summary of the results of CD164 MoAb binding to these soluble recombinant proteins as determined by ELISA assays.45
Sensitivity of CD164 Binding to Sialidase Treatment
To further characterize CD164 epitopes, Calu-1 and KG1A cells were treated with V. cholerae sialidase, an enzyme that selectively hydolyzes N- or O-acyl-neuraminic acids, which are α2,3-, α2,6- or α2,8-linked to galactose, hex, NAc, or N- or O-acylated neuraminyl residues in oligosaccharides, before staining with the CD164 MoAbs. Table 2 shows that the epitopes detected by the 103B2/9E10, N6B6, and 67D2 MoAbs are relatively resistant to desialylation compared with the untreated positive controls, whereas binding of 105A5 is almost completely abrogated. Preliminary results also indicate that the 105A5 MoAb (in contrast to the other three CD164 MoAbs) does not bind to sialidase treated MOLM-1 and BV-173 cell lines, with labeling being reduced to 7.8% and 0%, respectively, of the untreated cells. These sialidase studies are in line with the exon localization and cross-blocking studies in showing that, although the epitopes recognized by the 103B2/9E10 and 105A5 MoAbs are closely associated, they are distinct from one another and are different from those recognized by N6B6 and 67D2.
Sensitivity of CD164 Epitopes to Sialidase
| Cell Line . | % CD164 MoAb Binding . | |||
|---|---|---|---|---|
| CD164 MoAb | 103B2/9E10 | 105A5 | N6B6 | 67D2 |
| Calu-1 | 93.8 ± 5.6 | 18.8 ± 1.1 | 88.9 ± 10.2 | 118.5 ± 1.5 |
| KG1A | 74.8 ± 8.7 | 0 ± 0 | 85.2 ± 6.4 | 94.6 ± 4.4 |
| Cell Line . | % CD164 MoAb Binding . | |||
|---|---|---|---|---|
| CD164 MoAb | 103B2/9E10 | 105A5 | N6B6 | 67D2 |
| Calu-1 | 93.8 ± 5.6 | 18.8 ± 1.1 | 88.9 ± 10.2 | 118.5 ± 1.5 |
| KG1A | 74.8 ± 8.7 | 0 ± 0 | 85.2 ± 6.4 | 94.6 ± 4.4 |
Calu-1 and KG1A cells were incubated with 0.2 U/mL of Vibrio cholerae sialidase for 60 minutes at 37°C. Cells were then washed and stained with each CD164 MoAb indicated or with an isotype-matched negative control MoAb and counterstained with PE- or FITC-conjugated isotype-matched anti-Ig. The median fluorescence for each cell group was determined by analysis on a FACSCalibur, and the percent binding determined as indicated in Materials and Methods. Values are means ± SD of three determinations.
Differential Expression of CD164 Epitopes on Normal Adult Peripheral Blood Cells and on Normal Bone Marrow and Cord Blood CD34+ Cells
Staining of Ficoll separated peripheral blood cells from normal donors with the CD164 MoAbs 103B2/9E10, 105A5, 67D2, and N6B6 and the separation of cell subsets based on forward and side scatter parameters revealed that peripheral blood lymphocytes were weakly cell-surface positive with 103B2/9E10, 67D2, and N6B6, whereas staining with the 105A5 MoAb resulted only in faint signals near background staining. Monocytes generally showed higher levels of CD164 MoAb binding than did lymphocytes. Median fluorescence intensity (MFI) values for CD164 staining of monocytes were 28.9 ± 6.8 for N6B6, 41.5 ± 15.6 for 67D2, 22.3 ± 27.1 for 103B2/9E10, and 7.5 ± 4.8 for 105A5, after subtraction of the MFI values for isotype-matched negative controls and where the values are means ± SD of three independent experiments. This compares with MFI values for lymphocyte staining of 7.9 ± 3.4 for N6B6, 8.3 ± 2.7 for 67D2, 2.7 ± 2.0 for 103B2/9E10, and 3.6 ± 2.0 for 105A5. Mature non-nucleated erythrocytes showed negligible staining with all four CD164 MoAbs (data not shown). Granulocytes were very weakly stained by the CD164 MoAbs, except for 105A5, which was completely negative. The relevant fluorescence profiles and MFI values of a representative experiment are shown in Fig 2.
Differential expression of CD164 epitopes on peripheral blood lymphocytes, monocytes, and granulocytes. Ficoll separated cells were stained with CD164-specific MoAbs, 103B2/9E10, 105A5, 67D2, and N6B6, selected for lymphocytes, monocytes, or granulocytes on the basis of forward and side scatter parameters and analyzed on a FACSCalibur flow cytometer. Representative histograms from one of three independent experiments are shown. Median fluorescence values after subtraction of the median fluorescence value of the negative isotype-matched control for each population in this experiment are as follows: for the lymphocytes: N6B6 = 8.01; 67D2 = 6.49; 103B2/9E10 = 5.01; 105A5 = 1.37; for the monocytes: N6B6 = 35.05; 67D2 = 58.51; 103B2/9E10 = 53.50; 105A5 = 2.09; for the granulocytes: N6B6 = 2.41; 67D2 = 4.41; 103B2/9E10 = 2.59; 105A5 = 0.31. Mature non-nucleated glycophorin-A+ erythrocytes did not stain with any of the CD164 MoAbs.
Differential expression of CD164 epitopes on peripheral blood lymphocytes, monocytes, and granulocytes. Ficoll separated cells were stained with CD164-specific MoAbs, 103B2/9E10, 105A5, 67D2, and N6B6, selected for lymphocytes, monocytes, or granulocytes on the basis of forward and side scatter parameters and analyzed on a FACSCalibur flow cytometer. Representative histograms from one of three independent experiments are shown. Median fluorescence values after subtraction of the median fluorescence value of the negative isotype-matched control for each population in this experiment are as follows: for the lymphocytes: N6B6 = 8.01; 67D2 = 6.49; 103B2/9E10 = 5.01; 105A5 = 1.37; for the monocytes: N6B6 = 35.05; 67D2 = 58.51; 103B2/9E10 = 53.50; 105A5 = 2.09; for the granulocytes: N6B6 = 2.41; 67D2 = 4.41; 103B2/9E10 = 2.59; 105A5 = 0.31. Mature non-nucleated glycophorin-A+ erythrocytes did not stain with any of the CD164 MoAbs.
The expression of the four CD164 epitopes was significantly higher on bone marrow and cord blood CD34+ cells than on peripheral blood mononuclear cells. When CD34+ cells were analyzed, staining was more consistent with the N6B6 and 67D2 MoAbs. In four independent experiments, 81.8 ± 9.9% and 81.5 ± 9.9% of CD34+ bone marrow cells expressed these respective epitopes. This is in contrast to more variable staining with the 103B2/9E10 and 105A5 MoAbs with 70.2 ± 26.3% and 63.3 ± 26.1% of CD34+ cells labeling, respectively. CD34+bone marrow cells expressing the 103B2/9E10 epitope of CD164 exhibited the highest MFI values. Examples of dot plots and MFI values for the CD34+CD164+ cell subsets defined with the different CD164 MoAbs are shown in Fig 3. Three-color immunofluorescence analysis indicated that the CD34+CD164(103B2/9E10)+ bone marrow cells coexpressed the N6B6 and 67D2 epitopes (data not shown). Some differential cell-surface staining with the CD164 MoAbs was also observed on cord blood. When cord blood mononuclear cells were dual-labeled with CD34 and the different CD164 MoAbs (Fig 3), approximately 90% of the CD34+ cells expressed the 103B2/9E10 (92.6 ± 2.8%), 67D2 (88.7 ±1 0.5%), and N6B6 (89.1 ± 8.9%) epitopes, whereas only 55.0 ± 11.6% of these cells were 105A5 epitope+, where values are means ±SD of three independent experiments. Again, the MFI values obtained for the CD34+CD164+ cord blood subset were higher when the 103B2/9E10 epitope was detected than for the other CD164 reactive epitopes (Fig 3).
Differential expression of CD164 epitopes on CD34+ bone marrow and cord blood cells from normal donors. Mononuclear cells were stained with CD164-specific MoAbs 103B2/9E10, 105A5, 67D2, and N6B6 together with CD34 as indicated in Materials and Methods and gated on forward and side scatter (top dot plots) before analysis on a FACSCalibur flow cytometer. In four independent bone marrow and three independent cord blood analyses, 4.6 ± 0.8% and 1.2 ± 0.1% of the scatter gated cells were CD34+, respectively. In the representative experiment shown, median fluorescence values for the CD34+CD164+ subsets of the human bone marrow (BM) gated cells were: N6B6 = 138.24; 103B2/9E10 = 212.88; 105A5 = 45.32; 67D2 = 198.10 and for the CD34−CD164+ gated subsets were: N6B6 = 294.97; 103B2/9E10 = 184.34; 105A5 = 220.67; 67D2 = 697.83. Median fluorescence values for the CD34+CD164+ subsets of human cord blood (CB) cells were: N6B6 = 148.55; 103B2/9E10 = 294.27; 105A5 = 37.86; 67D2 = 283.87 and for the CD34−CD164+ gated subsets were: N6B6 = 119.71; 103B2/9E10 = 69.78; 105A5 = 77.74; 67D2 = 273.84. Cells were also labeled with CD34-FITC or the CD34 MoAb, 43A1, plus an anti–IgG3-FITC secondary antibody, together with isotype-matched irrelevant control MoAbs for each CD164 MoAb used plus anti-isotype–specific PE-conjugated antibodies. Under these conditions, 0.08% to 0.12% of cells occured in the CD34+ Ig Isotype− gates, and 0.08% to 0.38% of cells occured in the CD34− Ig Isotype− gates.
Differential expression of CD164 epitopes on CD34+ bone marrow and cord blood cells from normal donors. Mononuclear cells were stained with CD164-specific MoAbs 103B2/9E10, 105A5, 67D2, and N6B6 together with CD34 as indicated in Materials and Methods and gated on forward and side scatter (top dot plots) before analysis on a FACSCalibur flow cytometer. In four independent bone marrow and three independent cord blood analyses, 4.6 ± 0.8% and 1.2 ± 0.1% of the scatter gated cells were CD34+, respectively. In the representative experiment shown, median fluorescence values for the CD34+CD164+ subsets of the human bone marrow (BM) gated cells were: N6B6 = 138.24; 103B2/9E10 = 212.88; 105A5 = 45.32; 67D2 = 198.10 and for the CD34−CD164+ gated subsets were: N6B6 = 294.97; 103B2/9E10 = 184.34; 105A5 = 220.67; 67D2 = 697.83. Median fluorescence values for the CD34+CD164+ subsets of human cord blood (CB) cells were: N6B6 = 148.55; 103B2/9E10 = 294.27; 105A5 = 37.86; 67D2 = 283.87 and for the CD34−CD164+ gated subsets were: N6B6 = 119.71; 103B2/9E10 = 69.78; 105A5 = 77.74; 67D2 = 273.84. Cells were also labeled with CD34-FITC or the CD34 MoAb, 43A1, plus an anti–IgG3-FITC secondary antibody, together with isotype-matched irrelevant control MoAbs for each CD164 MoAb used plus anti-isotype–specific PE-conjugated antibodies. Under these conditions, 0.08% to 0.12% of cells occured in the CD34+ Ig Isotype− gates, and 0.08% to 0.38% of cells occured in the CD34− Ig Isotype− gates.
The CD34+CD164+Subset Contains Phenotypically Primitive Cells
We have recently shown that the 103B2/9E10 epitope is expressed by clonogenic progenitors, pre–colony-forming unit–granulocyte-macrophage (pre–CFU-GM), colony-forming unit–granulocyte, erythroid, monocyte, megakaryocyte (CFU-GEMM), burst-forming unit–erythroid (BFU-E), and granulocyte-macrophage colony-forming cells (GM-CFC), and that exogenous addition of the 103B2/9E10 MoAb to single CD34+CD38lo/−cells cultured in serum-free media in the presence of IL-3, IL-6, G-CSF, and Steel factor can inhibit the proliferation of these cells by up to 65%. Within these cultures, 5 ± 3% of 103B2/9E10 treated cells compared with 14 ± 2% of cells treated with an irrelevant IgG3 MoAb proliferated by day 3, whereas 23 ± 1% compared with 50 ± 6%, respectively, of these same cells proliferated by day 10 when 100 to 200 single cells were analyzed.7 The 103B2/9E10 MoAb also partially inhibited the adhesion of CD34+ cells to bone marrow stroma.7To gain more insight into the phenotype of the most immature CD34+CD164(103B2/9E10 epitope)+ subset, CD34+ bone marrow and cord blood cells were stained with 103B2/9E10 and with selected MoAbs that define primitive CD34+ cell subgroups. Representative dot plots of human bone marrow–purified CD34+ cells stained with these markers are shown in Fig 4. The most primitive CD34+ cells are thought to occur in the CD90(Thy1)+, CD117(c-kit)+, CD135(FTL-3)+, AC-133+, CD38lo/−, CD33lo/−, and CD71lo/− subsets. It was therefore of interest that 49.9 ± 10.9%, 88.7 ± 6.4%, 94.8 ± 3.8%, and 54.3 ± 17% of the CD34+CD164(103B2/9E10 epitope)+ bone marrow cells expressed CD90, CD117, AC-133, and CD135, respectively, in four independent experiments. This represented greater than 90% of those CD34+ bone marrow cells that were CD90+ (96.9 ± 2.2%), CD117+ (98.3 ± 1.1%), AC133+(96.9 ± 1.8%), and CD135+ (90.2 ± 10.2%). The CD34+CD164(103B2/9E10)+ bone marrow cell subset also contained both HLA-DR+ and HLA-DRlo/− cells and the CD38lo/−subset (Fig 4). In these same studies, 10.3 ± 4.3% and 25.9 ± 16.7% of the CD34+ bone marrow cells were CD71lo/− and CD33lo/−, respectively. Of these, an average of 65% of the CD71lo/− and 89% of the CD33lo/−expressed the CD164(103B2/9E10) epitope. Similar results (data not shown) were obtained with cord blood, with essentialy all CD34+ cells that coexpressed CD90, CD117, AC133, and CD135 being CD164(103B2/9E10 epitope)+. These data indicate that CD164 is present on a very primitive hematopoietic CD34+progenitor cell subset in addition to early clonogenic cells.
Triple-color analysis of primitive markers on CD34+CD164(103B2/9E10 epitope)+ bone marrow cells. CD34+ bone marrow cells were labeled with 103B2/9E10 plus CD34-PerCP and with each of the antibody PE conjugates indicated before staining with FITC-F(ab)2 goat anti-mouse IgG3. Plots are two-color dot plot displays from a representative experiment of CD34 purified cells gated on forward and side scatter parameters (left top dot plot) and on PerCP-CD34+ fluorescence and side scatter (middle dot plot). The arbitrary gates indicated on the dot plots were determined with appropriate isotype-matched negative controls. Examples of the IgG1/IgG3 isotype negative controls (top right dot plot) and of CD164(103B2/9E10 epitope)/anti–IgG3-FITC staining plus labeling with a nonbinding PE-conjugated anti–glycophorin A conjugate (bottom right dot plot) are indicated. Similar results were obtained for the IgG1/IgG2a isotype negative controls.
Triple-color analysis of primitive markers on CD34+CD164(103B2/9E10 epitope)+ bone marrow cells. CD34+ bone marrow cells were labeled with 103B2/9E10 plus CD34-PerCP and with each of the antibody PE conjugates indicated before staining with FITC-F(ab)2 goat anti-mouse IgG3. Plots are two-color dot plot displays from a representative experiment of CD34 purified cells gated on forward and side scatter parameters (left top dot plot) and on PerCP-CD34+ fluorescence and side scatter (middle dot plot). The arbitrary gates indicated on the dot plots were determined with appropriate isotype-matched negative controls. Examples of the IgG1/IgG3 isotype negative controls (top right dot plot) and of CD164(103B2/9E10 epitope)/anti–IgG3-FITC staining plus labeling with a nonbinding PE-conjugated anti–glycophorin A conjugate (bottom right dot plot) are indicated. Similar results were obtained for the IgG1/IgG2a isotype negative controls.
CD164 Expression Is Maintained on Nucleated Erythroid Cell Subsets, but Is Lost on Terminal Erythroid Differentiation
The localization of day-14 BFU-E within the CD34+CD164(103B2/9E10 epitope)+ fraction of human bone marrow5 is consistent with the finding that a proportion of CD34+CD164(103B2/9E10 epitope)+ cells coexpress the CD33, CD117, HLA-DR, and CD71 markers (Fig 4). Because the CD164/103B2/9E10 epitope was present on the BFU-E but absent from the mature erythrocytes in peripheral blood, and as the whole CD34− erythroid lineage from the proerythroblast stage to the erythroblast, normoblast, and mature non-nucleated erythrocyte stages can be defined by the regulated expression of three markers, CD71, CD117, and the erythroid specific marker glycophorin A,47 we used these markers to determine at which stage in the erythroid lineage this CD164 epitope was lost. Within the CD34− erythroid lineage, CD117 and CD71 are expressed on both the proerythroblasts and erythroblasts, with glycophorin A appearing on some erythroblasts, being strongly expressed on the more differentiated normoblasts, and then maintained to the mature erythrocyte stage. Although CD71 is maintained on normoblasts and then lost from reticulocytes and mature erythrocytes, CD117 expression is lost at the normoblast stage.47 When human bone marrow cells were defined by the erythroid/lymphoid/blast cell light scatter parameters shown in Fig 5, 52.9 ± 9.2% of these cells were CD34−CD164(103B2/9E10 epitope)+, where values are means ±SD of four independent evaluations. When these cells were examined for their coexpression of erythroid associated markers, averages of 93%, 82%, and 8% of these cells were CD71+, glycophorin-A+, and CD117+, respectively, in two independent experiments (Fig 5). In addition, a mean of 88% of the glycophorin A+ nucleated erythroid cells within these bone marrow samples were 103B2/9E10+. Similar results with cord blood (Fig 5) confirmed that this prominent CD34−CD164+ cell population consists predominantly of nucleated erythroid cells, although most but not all glycophorin A+ nucleated cells express the 103B2/9E10 epitope. This was confirmed in a separate experiment by morphological analysis of sorted CD34lo/−CD164(103B2/9E10 epitope)+ bone marrow cells, which revealed that a large proportion of these cells belonged to the erythroid lineage (9% lymphocytes, 13% monocytes, 1% promyelocytes, 49% polychromatic normoblasts, 10% basophilic normoblasts, 3% erythroblasts, and 13% proerythroblasts).
CD34−CD164(103B2/9E10 epitope)+ bone marrow and cord blood cells contain nucleated erythroid cells. Ficoll separated bone marrow or cord blood mononuclear cells were labeled with 103B2/9E10 together with the PE-antibody conjugates indicated on the dot plots before staining with FITC-F(ab)2 goat anti-mouse IgG3 and flow cytometric analyses. Plots are two-color displays of cells gated on the lymphoid/erythroid/blast cell gate in Fig 3. The arbitrary gates shown on the dot plots were determined with negative control isotype-matched irrelevant antibodies as described in Materials and Methods.
CD34−CD164(103B2/9E10 epitope)+ bone marrow and cord blood cells contain nucleated erythroid cells. Ficoll separated bone marrow or cord blood mononuclear cells were labeled with 103B2/9E10 together with the PE-antibody conjugates indicated on the dot plots before staining with FITC-F(ab)2 goat anti-mouse IgG3 and flow cytometric analyses. Plots are two-color displays of cells gated on the lymphoid/erythroid/blast cell gate in Fig 3. The arbitrary gates shown on the dot plots were determined with negative control isotype-matched irrelevant antibodies as described in Materials and Methods.
To analyze at which stage of erythroid development the CD164(103B2/9E10) epitope was expressed, CD34-enriched human umbilical cord blood cells were expanded in liquid culture under conditions known to generate erythroid cells. Cells were collected at days 5, 6, 9, and 13 of culture and their total nucleated cell number determined. In two separate experiments, 100- to 200-fold and 400- to 600-fold expansions in cell numbers were observed in these cultures at days 9 and 13, respectively. Cells were then centrifuged onto slides for fixation and staining with the CD164 MoAbs and with the erythroid specific markers, glycophorin C, glycophorin A, and band III, with glycophorin C being expressed on BFU-E and proerythroblasts before the appearance of the other markers on erythroblasts and normoblasts. At day 5, the cultured cells were mostly glycophorin A, glycophorin C, or band III negative (>98.5%). However, these cells stained strongly in the Golgi region for all CD164 MoAbs (95% to 99%). By days 6 to 9 of culture, the majority of cells were glycophorin C positive (80 and 90%; Fig 6) and a subset of these had begun to express glycophorin A (42% and 61%) and band III (11% and 24%). Essentially all cells in these day-6 to -9 cultures maintained their expression of the different CD164 epitopes (>92%; Fig 6 and data not shown). It was of interest to note, however, that in these cultures only a very small proportion of these nucleated erythroid cells that were exclusively glycophorin A or band III positive either failed to express or expressed very low levels of the CD164 epitopes. The day-9 erythroid-cultured cells were also analyzed on the FACSCalibur to compare the levels of cell surface expression of the CD164 epitopes and those of the erythroid specific markers. As indicated in Fig 7, the cultured erythroid cells at day 9 could be divided into two subsets, both exhibiting low side light scatter characteristics, but with either low to medium (the R1 subset) or medium to high forward light scatter parameters (the R2 subset). Both subsets expressed glycophorin C in high amounts (histograms B and C). However, cells within the R1 or lower forward scatter gate expressed higher levels of glycophorin A (histogram E) and band III (histogram H) than those in the R2 or higher forward scatter gate (histograms F and I, respectively). The R1 gated cells were essentially all glycophorin A positive, but could be separated into two subsets based on positive or negative expression of band III (histogram H). This suggested that cells within the lower forward scatter gate were more mature, but not fully developed, nucleated erythroid components. However, all subsets exhibited essentially the same surface fluorescence intensities when stained with the 103B2/9E10, as indicated in a typical experiment in Fig 7 (histograms J to L). Thus, the 103B2/9E10 epitope is evident before the expression of specific erythroid markers and is lost only at the late stages of erythroid differentiation.
Immunofluorescence staining with CD164 and erythroid specific MoAbs of cord blood CD34+ cells cultured under erythroid conditions. CD34+ cells were isolated from cord blood and cultured for 5 to 9 days under the conditions described. Cultured cells were cytocentrifuged, fixed in acetone, and stained by dual immunofluorescence using DAPI as a nuclear counterstain. Of interest are day-5 cultured cells stained in the Golgi region with 103B2/9E10 (A) and105A5 (B) antibodies. These cells were negative with anti-glycophorin C (C). Similar staining was obtained for day-9 cultured cells using 103B2/9E10 (D). Also shown are day-9 cells labeled with an IgG3 irrelevant isotype-matched negative control (E), anti–glycophorin C (F), anti–glycophorin A (G), and band III (H); and day-6 cultured cells labeled with 103B2/9E10 plus FITC–anti-mouse IgG3 (I through K) and glycophorin C (I), glycophorin A (J), or band III (K) plus Texas Red–anti-mouse IgG1. Arrow heads indicate double-stained cells and arrows show cells stained only with 103B2/9E10 (I through K); (A through H: 40 × original magnification; I through K: 100 × original magnification).
Immunofluorescence staining with CD164 and erythroid specific MoAbs of cord blood CD34+ cells cultured under erythroid conditions. CD34+ cells were isolated from cord blood and cultured for 5 to 9 days under the conditions described. Cultured cells were cytocentrifuged, fixed in acetone, and stained by dual immunofluorescence using DAPI as a nuclear counterstain. Of interest are day-5 cultured cells stained in the Golgi region with 103B2/9E10 (A) and105A5 (B) antibodies. These cells were negative with anti-glycophorin C (C). Similar staining was obtained for day-9 cultured cells using 103B2/9E10 (D). Also shown are day-9 cells labeled with an IgG3 irrelevant isotype-matched negative control (E), anti–glycophorin C (F), anti–glycophorin A (G), and band III (H); and day-6 cultured cells labeled with 103B2/9E10 plus FITC–anti-mouse IgG3 (I through K) and glycophorin C (I), glycophorin A (J), or band III (K) plus Texas Red–anti-mouse IgG1. Arrow heads indicate double-stained cells and arrows show cells stained only with 103B2/9E10 (I through K); (A through H: 40 × original magnification; I through K: 100 × original magnification).
Flow cytometric analysis of day-9 cultured cord blood erythroid cells. Cord blood CD34+ cells were cultured for 9 days under erythroid conditions as described. These cells were labeled with MoAbs to the erythroid-specific markers, glycophorin C (A through C), glycophorin A (D through F), or band III (G through I) or with the CD164(103B2/9E10) MoAb (J through L), followed by FITC anti-mouse IgG. Cells were analyzed on a FACSCalibur on the basis of forward and side light scatter parameters. Either the whole cell population was analyzed (dark histograms A, D, G, J) or the cells were gated into two subsets based, both with low side scatter and with low to medium forward scatter (R1) or medium to high forward scatter (R2) before fluorescence analysis (dark histograms B, E, H, K for R1 and C, F, I, L for R2). Values adjacent to each histogram represent median fluorescence values. Isotype-matched negative controls for A through I were an irrelevant IgG1 mouse MoAb (light histograms in A through C) or an irrelevant mouse IgG3 MoAb (light histograms in J through L). Median fluorescence values for negative controls were A = 9; B = 9; C = 6; J = 9; K = 9; L = 8.
Flow cytometric analysis of day-9 cultured cord blood erythroid cells. Cord blood CD34+ cells were cultured for 9 days under erythroid conditions as described. These cells were labeled with MoAbs to the erythroid-specific markers, glycophorin C (A through C), glycophorin A (D through F), or band III (G through I) or with the CD164(103B2/9E10) MoAb (J through L), followed by FITC anti-mouse IgG. Cells were analyzed on a FACSCalibur on the basis of forward and side light scatter parameters. Either the whole cell population was analyzed (dark histograms A, D, G, J) or the cells were gated into two subsets based, both with low side scatter and with low to medium forward scatter (R1) or medium to high forward scatter (R2) before fluorescence analysis (dark histograms B, E, H, K for R1 and C, F, I, L for R2). Values adjacent to each histogram represent median fluorescence values. Isotype-matched negative controls for A through I were an irrelevant IgG1 mouse MoAb (light histograms in A through C) or an irrelevant mouse IgG3 MoAb (light histograms in J through L). Median fluorescence values for negative controls were A = 9; B = 9; C = 6; J = 9; K = 9; L = 8.
CD164 Is Differentially Expressed During B Lymphoid and Myelomonocytic Development
It was of interest that the CD34+CD164(103B2/9E10)lo/− cell population in bone marrow (Fig 8) consisted mainly of CD19+ B-cell precursors, suggesting that the most immature B-cell subset in bone marrow is CD164−. Developing B cells in bone marrow, however, were CD164(103B2/9E10)+ as evidenced in Fig 8, which further shows that 60% to 70% of CD34− B cells expressing CD19 or CD20 surface molecules coexpress the CD164(103B2/9E10) epitope. In contrast, the majority of CD34− T cells, in bone marrow, staining with the CD3 marker were found in the CD164(103B2/9E10)lo/− cell gate (Fig 8). To determine whether this CD164 epitope was maintained on peripheral blood lymphocyte subsets, three individual Ficoll separated peripheral blood samples were scatter gated on the lymphoid gate and analyzed by two-color FACS analysis for the B lymphoid marker, CD20, and for the T-cell markers, CD3, CD4, and CD8 (Fig 9). The majority of CD3+, CD4+, CD8+, and CD20+ cells fell within the CD164(103B2/9E10)lo/− gate. Analyses of the MFI indicated that for these three peripheral blood samples the CD20+ B cells showed lower levels of CD164(103B2/9E10) expression (MFI = 3.95 ± 0.29) than the CD3+ T cell subset (MFI = 6.69 ± 0.44). In addition, within the T-cell subset, the CD8+ cells (MFI = 4.11 ± 0.33) showed lower levels of CD164(103B2/9E10) epitope expression than did the CD4+cells (MFI = 8.47 ± 0.12). Our other studies (data not shown) examining the correlated expression of the CD164(103B2/9E10) epitope and defined lymhoid surface markers on cord blood cells indicated that the majority of cord blood CD34−CD19+ and CD20+ B cells were CD164(103B2/9E10)+, whereas the CD34−CD3+ T cells were CD164(103B2/9E10)lo/−. Together these results show that T cells in bone marrow and peripheral blood are very weakly positive for the 103B2/9E10 epitope. However, the expression of this epitope was differentially regulated on developing B cells, with the most primitive bone marrow CD34+CD19+ B cells and the most mature peripheral blood B cells being CD164(103B2/9E10)lo/−, whereas a large proportion of maturing CD34−CD19+ or CD34−CD20+ B cells in bone marrow were CD164(103B2/9E10)+.
Bone marrow lymphoid cells differentially express the CD164(103B2/9E10) epitope. Representative dot plots from one of two separate experiments showing CD34+ bone marrow cells and Ficoll separated bone marrow mononuclear cells labeled with the 103B2/9E10 MoAb or with an IgG3 negative control MoAb plus CD34-PerCP and with the PE-conjugated MoAbs indicated before counterstaining with FITC anti-mouse IgG3 and analysis on the FACSCalibur. In contrast to the more mature CD34−CD19+ and CD34−CD20+ B cells, the immature CD34+CD19+ B-cell subset failed to express the CD164(103B2/9E10) epitope. The median fluorescence intensity for CD164(103B2/9E10) staining of the CD34−CD3+cells was 9.08 after subtraction of the isotype-matched negative control value, indicating that the CD34−CD3+ T-cell subset was weakly reactive with the 103B2/9E10 MoAb.
Bone marrow lymphoid cells differentially express the CD164(103B2/9E10) epitope. Representative dot plots from one of two separate experiments showing CD34+ bone marrow cells and Ficoll separated bone marrow mononuclear cells labeled with the 103B2/9E10 MoAb or with an IgG3 negative control MoAb plus CD34-PerCP and with the PE-conjugated MoAbs indicated before counterstaining with FITC anti-mouse IgG3 and analysis on the FACSCalibur. In contrast to the more mature CD34−CD19+ and CD34−CD20+ B cells, the immature CD34+CD19+ B-cell subset failed to express the CD164(103B2/9E10) epitope. The median fluorescence intensity for CD164(103B2/9E10) staining of the CD34−CD3+cells was 9.08 after subtraction of the isotype-matched negative control value, indicating that the CD34−CD3+ T-cell subset was weakly reactive with the 103B2/9E10 MoAb.
The expression of the CD164(103B2/9E10) epitope on peripheral blood lymphoid subsets. Peripheral blood mononuclear cells were labeled with the 103B2/9E10 MoAb or with an IgG3negative control MoAb plus the FITC-conjugated MoAbs indicated before counterstaining with PE-conjugated anti-mouse IgG3. Lymphoid cells were gated on low forward and low side scatter gates before analysis on the FACSCalibur.
The expression of the CD164(103B2/9E10) epitope on peripheral blood lymphoid subsets. Peripheral blood mononuclear cells were labeled with the 103B2/9E10 MoAb or with an IgG3negative control MoAb plus the FITC-conjugated MoAbs indicated before counterstaining with PE-conjugated anti-mouse IgG3. Lymphoid cells were gated on low forward and low side scatter gates before analysis on the FACSCalibur.
In earlier experiments,7 we have shown that myeloid precursors, the pre–CFU-GM and GM-CFC, were CD164(103B2/9E10)+, whereas this epitope continues to be expressed on monocytes but is virtually absent from mature neutrophils in peripheral blood (Fig 2). We therefore used three cell-surface markers to analyze the expression of this CD164 epitope on CD34− myelomonocytic cells in bone marrow. These were CD14,48 which is expressed from the promonocyte to the monocyte stages; the myelomonocytic marker, CD6549; and CD66b,50 which exhibits high levels of expression on myelocytes and metamyelocytes and lower levels of expression on the more primitive promyelocytes and early myelocytes and on the more mature band forms and neutrophils in the neutrophilic lineage. Figure 10 confirms that essentially all CD14+ cells within the monocytic lineage and all except the more mature cells within the neutrophilic lineage are CD164(103B2/9E10)+.
The CD164(103B2/9E10) epitope is expressed on myelomonocytic cells in bone marrow. Representative dot plots from one of two separate experiments showing Ficoll separated bone marrow mononuclear cells labeled with the 103B2/9E10 MoAb or with an IgG3 negative control MoAb plus CD34-PerCP and with the FITC-conjugated MoAbs indicated before counterstaining with PE anti-mouse IgG3 and analysis on the FACSCalibur. Gates were set to exclude the CD34+ cell population before myelomonocytic cell analysis.
The CD164(103B2/9E10) epitope is expressed on myelomonocytic cells in bone marrow. Representative dot plots from one of two separate experiments showing Ficoll separated bone marrow mononuclear cells labeled with the 103B2/9E10 MoAb or with an IgG3 negative control MoAb plus CD34-PerCP and with the FITC-conjugated MoAbs indicated before counterstaining with PE anti-mouse IgG3 and analysis on the FACSCalibur. Gates were set to exclude the CD34+ cell population before myelomonocytic cell analysis.
Identification of CD164 Containing PAC Clones
A PAC library containing 120,000 recombinants was screened with full-length CD164 cDNA probes, and two positive clones (termed CD164 PAC1 and PAC5) were analyzed in detail. Restriction enzyme analyses of these clones, using EcoR1, BglII,Pst 1, HindIII, and BamH1 revealed that the pattern of bands on a 0.7% agarose gel stained with ethidium bromide was similar but not exactly identical (data not shown). When these gels were Southern blotted and hybridized with the 1.173 kbEcoRV/HindIII CD164 probe C, however, the pattern of bands observed for CD164 PAC1 was identical to that obtained with CD164 PAC5. Southern blotting of human placental genomic DNA digested with all five restricton enzymes and hybridized with CD164 cDNA probe C produced the same banding pattern (data not shown), indicating that the PAC clones had not undergone gross rearrangements within the region of the CD164 gene. A single hybridization band was observed with each enzyme, the smallest being an EcoR1 fragment of approximately 2.2 kb, whereas the largest fragment of greater than or equal to 18 kb was obtained after BamH1 restriction digestion. When the blot was reprobed with the 5′ end of the cDNA, two additonal BamH1 fragments of 2 and 3 kb were identified. These three BamH1 fragments cover the entire CD164 gene and span approximately 23 kb of DNA. Complete sequence analysis has been obtained from these BamH1 fragments derived from the CD164 PACs after subcloning into pCRScript and these contain a sequence that is identical to the CD164 cDNA from 1 to 2867 bp, but they are interspersed with intronic sequences of varying sizes.42
PCR Analysis of Somatic Cell Hybrids Localizes CD164 to Chromosome 6
PCR analysis was performed using DNA from a set of somatic cell hybrids (Tables 3 and4), from the BglII subclone of CD164 PAC1 and from three human genomic DNA samples. For the human genomic DNA, the CD164 PAC1 subclone and the hybrid containing human chromosome Xq13 translocated to 6p21, t(6,X)(p21,q13), single bands of approximately 1.1 kb and 500 bp were obtained with the MGC24-Gp- F3/B3 and F4/B4 primers, respectively, on ethidium bromide–stained gels. These bands hybridized to the CD164 cDNA probe C. An example of the PCR products generated with the MGC24-Gp-F4/B4 primers from the somatic cell hybrid panel is shown in Fig 11A and B. These PCR products were not generated with the hybrid DNA containing the X chromosome alone or in conjunction with chromosomes other than 6. This indicated that the CD164 gene was very likely to be located on chromosome 6. A sequence comparison of the products generated by PCR analysis of the hybrids revealed identical sequence for both the cDNA clones, the BglII subclones of the CD164 PACs, and genomic DNA from male or female peripheral blood leukocytes.
Monochromosomal Somatic Cell Hybrid DNA Panel From the UK HGMP Resource Centre
| Hybrid . | Chromosome . | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y | |
| GM07299 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − |
| GM10826B | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10253 | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| HHW416 | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10114 | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| MCP6BRA | − | − | − | − | − | / | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | / | − |
| CLONE21E | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| C4A | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | / | − | − |
| GM10611 | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 762-8a | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| JICL4 | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 1aA9602+ | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | + | − | + | − |
| 289 | − | − | − | − | − | − | − | / | − | − | / | / | + | − | − | − | − | − | − | − | − | − | − | − |
| GM10479 | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | / | − | − | − | − | − | − | − | − |
| HORLI | − | − | − | − | − | − | − | − | − | − | / | − | − | − | + | − | − | − | − | − | − | − | / | − |
| 2806H7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − |
| PCTBA1.8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − |
| DL18TS | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| GM10612 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − |
| GM10478 | − | − | − | / | − | − | − | / | − | − | − | − | − | − | − | − | − | − | − | + | − | / | + | − |
| THYB1.3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | − |
| PgME25NU | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | / | − |
| HORL9X | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − |
| 853 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| Hybrid . | Chromosome . | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y | |
| GM07299 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − |
| GM10826B | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10253 | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| HHW416 | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10114 | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| MCP6BRA | − | − | − | − | − | / | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | / | − |
| CLONE21E | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| C4A | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | / | − | − |
| GM10611 | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 762-8a | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| JICL4 | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 1aA9602+ | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | + | − | + | − |
| 289 | − | − | − | − | − | − | − | / | − | − | / | / | + | − | − | − | − | − | − | − | − | − | − | − |
| GM10479 | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | / | − | − | − | − | − | − | − | − |
| HORLI | − | − | − | − | − | − | − | − | − | − | / | − | − | − | + | − | − | − | − | − | − | − | / | − |
| 2806H7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − |
| PCTBA1.8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − |
| DL18TS | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| GM10612 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − |
| GM10478 | − | − | − | / | − | − | − | / | − | − | − | − | − | − | − | − | − | − | − | + | − | / | + | − |
| THYB1.3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | − |
| PgME25NU | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | / | − |
| HORL9X | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − |
| 853 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
Hybrid: GM07299, 1 and X; MCP6BRA, Xqter-Xq13:6p21-6qter; C4a, 8 and fragment of 22; 762-8a, 10 and Y; 1aA9602 + ve, 12, 21 and X; 289, 13 plus fragments of 8, 11, and 12; GM10479, 14 plus part of 16 (probably 16p13.3-16q22.1); HORLI, 15, 11q, part of Xp and proximal Xq; GM10478, 20, 4 (part), 8 (part), 22q, and X; GM10612, 9p stronger than 9q, only 10% of cells with whole 9; PGME25NU, 22, part of Xp.
+ Indicates presence of chromosome; − indicates absence of chromosome; / indicates chromosome translocation, extra chromosomes, or other modifications.
Somatic Cell Hybrid DNA Panel Positive for CD164
| Hybrid . | Chromosome . | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y | |
| GM07299 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10826B | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10253 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| HHW416 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10114 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| MCP6BRA | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| CLONE21E | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| C4A | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10611 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 762-8a | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| JICL4 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 1aA9602+ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 289 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10479 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| HORLI | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 2806H7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| PCTBA1.8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| DL18TS | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10612 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10478 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| THYB1.3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| PgME25NU | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| HORL9X | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 853 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Hybrid . | Chromosome . | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y | |
| GM07299 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10826B | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10253 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| HHW416 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10114 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| MCP6BRA | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| CLONE21E | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| C4A | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10611 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 762-8a | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| JICL4 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 1aA9602+ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 289 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10479 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| HORLI | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 2806H7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| PCTBA1.8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| DL18TS | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10612 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GM10478 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| THYB1.3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| PgME25NU | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| HORL9X | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 853 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
DNA samples were PCR amplified using CD164 primers as indicated in Materials and Methods and analyzed on 2% agarose gels. Positive bands of the correct size were obtained with the chromosome 6 DNA sample only. These products were Southern blotted and probed with CD164 cDNA Probe C or sequenced and were shown to have sequence identifical to CD164 cDNA. −, no PCR product; +, PCR product of the appropriate size. Human, mouse, and hamster genomic DNA samples were also analyzed, but only the human DNA proved positive for CD164 under the conditions used.
CD164 is located on human chromosome 6q21. Somatic cell hybrids shown in Tables 3 and 4 were analyzed by PCR for the presence of the CD164 gene. (A) Ethidium bromide–stained gel of PCR products using the MGC24-Gp-F4/B4 primer pairs. This gel was Southern blotted and probed with CD164 cDNA probe C as indicated in (B). Partial metaphase spreads (C) showing localization of the BglII subclone of CD164 PAC1 to human chromosome band 6q21 (arrowed and shown on the idiogram).
CD164 is located on human chromosome 6q21. Somatic cell hybrids shown in Tables 3 and 4 were analyzed by PCR for the presence of the CD164 gene. (A) Ethidium bromide–stained gel of PCR products using the MGC24-Gp-F4/B4 primer pairs. This gel was Southern blotted and probed with CD164 cDNA probe C as indicated in (B). Partial metaphase spreads (C) showing localization of the BglII subclone of CD164 PAC1 to human chromosome band 6q21 (arrowed and shown on the idiogram).
FISH Localizes CD164 to Chromosome 6q21
Sixty metaphase spreads were examined for signal using each of theBglII subclones of CD164 PACS 1 and 5. Probe efficiency was approximately 60%, with signal being observed on both chromatids of chromosome 6 on band 6q21 only (Fig 11C).
DISCUSSION
The interaction of hematopoietic progenitor cells with stromal/endothelial cells in their immediate microenvironment is thought to be of major importance in the regulation of hematopoietic stem cell self-renewal, quiescence, commitment, and migration. These interactions involve cooperation between adhesion receptors, their cognate ligands, and cytokines. The adhesion receptors/ligands implicated in these processes belong to several families of adhesion molecules. These include the integrins, selectins, Ig superfamily, and sialomucins. Recently, mucin-like receptors or sialomucins, in particular, have assumed some importance in hematopoiesis, with four of these receptors, CD34, PSGL-1, CD43, and CD164, having been identified on primitive hematopoietic precursor cells and/or their associated stromal/endothelial elements.5,7,19-27,51 Our own studies and those of other researchers indicate that these receptors are involved in mediating or regulating hematopoietic precursor cell adhesion in vitro and/or the regulation of hematopoietic progenitor cell proliferation.5,7,20,31 32
In this report, our studies have concentrated on the CD164 molecule. To this end, we have characterized the reactivity of the first four MoAbs, 103B2/9E10, 105A5, 67D2, and N6B6, to this sialomucin. Each of these antibodies shows distinct patterns of reactivity on CD34+hematopoietic progenitor cells, labeling on average 55% to 93% of these cells. When compared with other CD164 epitopes, the 103B2/9E10 epitope is more highly expressed on CD34+ cells from both cord blood and bone marrow, whereas the 105A5 epitope exhibits the most variable level of expression. Our recent studies suggest that this may reflect glycosylation differences in the 105A5 and 103B2/9E10 epitopes on different CD34+ subsets. We have cloned the CD164 cDNA5,7 and have shown that this gene encodes an 80 to 90 kD heavily O-glycosylated protein that is able to form homodimers with an apparent molecular weight of 160 to 180 kD. Our recent studies have also shown that the CD164 receptor mediates the adhesion of CD34+ hematopoietic progenitor cells to bone marrow stroma, since this adhesion can be partially blocked by the 103B2/9E10 MoAb.5,7 In addition, two-color phenotypic selection, based on expression of CD34 and the CD164(103B2/9E10) epitope, revealed that essentially all the committed myeloid and erythroid progenitors and their immature precursors were present in this CD164+population and correspondingly absent from the CD34+CD164− subset. In contrast, mature peripheral blood cells showed only low or negligible levels of CD164 expression. In view of these results, we have characterized, in more detail, the expression of the 103B2/9E10 epitope on CD34+and CD34− subsets of cord blood and bone marrow cells. Multiparameter flow cytometric analyses revealed that the majority (>90%) of CD34+ human bone marrow and cord blood cells that expressed CD90(Thy-1), CD117(c-kit), AC-133, and CD135(FLT3) also coexpressed the 103B2/9E10 epitope. Because human AC-133+ cells have the capacity for hematopoietic engraftment in fetal sheep,52 these results suggest that the 103B2/9E10 epitope is present on a very primitive subset of CD34+ cells.52,53 Similar analyses of the CD34lo/−CD164(103B2/9E10 epitope)+subsets have also indicated that one of the most prominent populations consists of nucleated erythroid cells, with the appearance of this CD164 epitope preceding the expression of the erythroid markers, glycophorin C, glycophorin A, and band III. This is consistent with our previous studies5 7 showing that the 103B2/9E10 and 105A5 epitopes of CD164 are present on the majority of day-14 CFU-GEMM and BFU-E. Of added interest was the finding that the expression of the 103B2/9E10 epitope was differentially regulated during B-cell development, because the majority of CD34− B cells in bone marrow expressed the 103B2/9E10 epitope, whereas both mature B cells from peripheral blood and the most immature CD34+CD19+ B-cell precursors were CD164(103B2/9E10 epitope)lo/−.
Our recent studies45 indicate that the 103B2/9E10 and 105A5 epitopes both localize to the N-terminal domain of the CD164 molecule encoded by exon 1. Although the epitope recognized by the 103B2/9E10 antibody was not affected by sialidase treatment, that detected by 105A5 was completely abrogated by CD164 desialylation. This indicates that these epitopes are spatially distinct from one another and implies that the epitope required for CD164-mediated cell adhesion is not sialylated. Further to these finding, we have shown by cross-blocking experiments that the binding of the 105A5 MoAb was partially inhibited by prelabeling the cells with the 103B2/9E10 MoAb, but not vice versa. This partial inhibition might be due to the induction, by the 103B2/9E10 antibody, of a conformational change in the CD164 molecule that results in masking the 105A5 epitope. It therefore seems possible that these two epitopes may cooperate or compete with one another in the initial adhesion of CD34+ cells to stromal cells, with expression of the 105A5 epitope serving to modulate the function of the CD164(103B2/9E10) epitope. Further studies are in progress to fully characterize the epitopes recognized by these different CD164 MoAb. However, it is possible that the different patterns of expression of the CD164 epitopes on CD34+ cells reflect differences in glycosylation patterns, carbohydrate modifications, or splice variant expression among different precursor cells within the hematopoietic progenitor cell pool.
One major unanswered question in hematopoiesis involves the mechanisms by which CD34+ hematopoietic progenitor cells identify particular microenvironmental niches. It is thought that the lodgement of such cells in these niches is required for the maintenance of their quiescent state or for their self-renewal or commitment to particular hematopoietic lineages. It is possible that the sialomucins as a family may act to regulate hematopoietic proliferation or differentiation. Evidence for this comes from both in vitro and in vivo studies. For instance, transfection of the long form of CD34 into M1 cells inhibits their differentiation, whereas transfection of the short isoform has no effect.33 In other studies, ligation of the CD43 engenders an apoptotic response in more mature cells within the CD34hiLin− subset but not in the more immature stem or severe combined immunodeficiency disease (SCID) repopulating cells.24,25 Our recent experiments suggest that ligation of CD164 with both the 103B2/9E10 and 105A5 MoAbs can inhibit the clonogenic growth of CD34+CD38lo/− cells.7 An alternative way of determining the potential importance of these mucin-like molecules is to generate knock-out mice lacking the gene of interest or to create transgenic mice overexpressing a particular sialomucin in a specified subset of hematopoietic cells and then to examine their effects on hematopoietic development. Although studies are limited, CD34-null embryonic stem (ES) cells and mice have been generated.35In the mutant ES cells, erythroid and myeloid development is delayed during embryoid body formation, but it can be restored by the introduction of either a full-length or cytoplasmically truncated CD34 gene into these cells. In the CD34-null mice, the number of clonogenic progenitor cells is reduced in the yolk sac, fetal liver, adult bone marrow, and adult spleen, and cytokine-induced expansion of bone marrow hematopoietic progenitors ex vivo is inhibited. These studies suggest that cytokines may cooperate with adhesion receptors to induce cell proliferation and/or differentiation by presenting cytokines on the CD34 molecule,54 by interaction between CD34 and cytokine receptors expressed on the same cell, or by engagement of the CD34 molecule on one cell with its cognate ligand on an opposing cell. Such an adhesion process might precede or be required for interaction with membrane associated growth factors on accessory cells. It will be of interest to see if CD164 possesses similar functions or if signaling via CD164 is required for the regulation of hematopoietic cell proliferation or differentiation.
In the final section of this report, we have identified PAC clones containing genomic fragments of CD164 and have used these to localize the CD164 gene to human chromosome 6q21. These PAC clones have now been fully sequenced and the genomic structure of CD164 determined.42 These constructs, together with the murine CD164 gene, when cloned, will provide important reagents for identifying elements that regulate CD164 gene expression in hematopoietic progenitor cells and for generating transgenic and knock-out mice for the analysis of the function of CD164 in vivo. In addition, the PAC clones containing CD164 may help to identify other genes on chromosome 6q21 that are related to CD164 or that have similar functions to CD164 in regulating hematopoietic progenitor cell proliferation and differentiation.
ACKNOWLEDGMENT
The authors thank Professor Sir D.J. Weatherall and Professor L. Kanz for their support, Dr D Buck for the AC-133 MoAb, and Heike Hanisch and Doris Schweigert for excellent technical assistance.
Supported by the Medical Research Council (S.M.W., J.L.-P.) and Leukaemia Research Fund (S.M.W.) of Great Britain; Smith-Kline-Beecham (L.H.B.); INTAS/RFBR, PECO, and E.U. Concerted Action and Biotech Grants on the Human Haematopoietic Stem Cell (S.M.W.); the Deutsche Forschungsgemeinschaft (SFB 510, project A1 to H.J.B.) and the fortune project 244 of the University of Tübingen (I.R.); grants from the Anti-Cancer Foundation of the Universities of South Australia and the National Health and Medical Research Council of Australia (P.J.S.); the Imperial Cancer Research Fund (T.J., D.S.); and the Taiwanese Government (J.Y.-H.C.).
Address reprint requests to Suzanne M. Watt, PhD, MRC Molecular Haematology Unit, Institute of Molecular Medicine, The John Radcliffe Hospital, Headington, Oxford OX3 9DS, UK.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

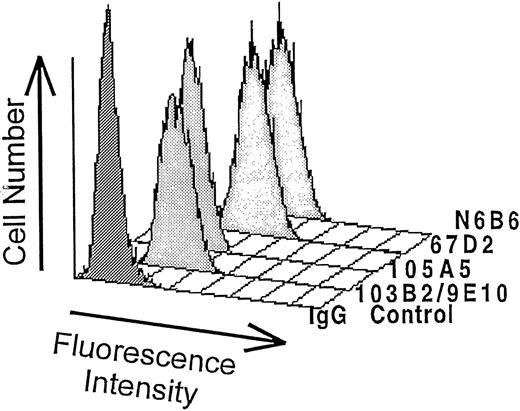
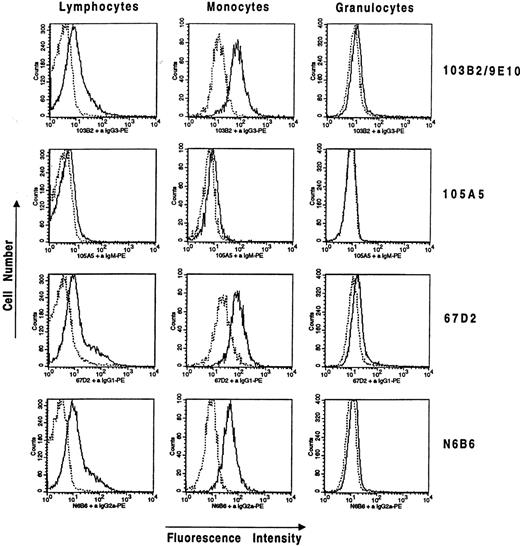
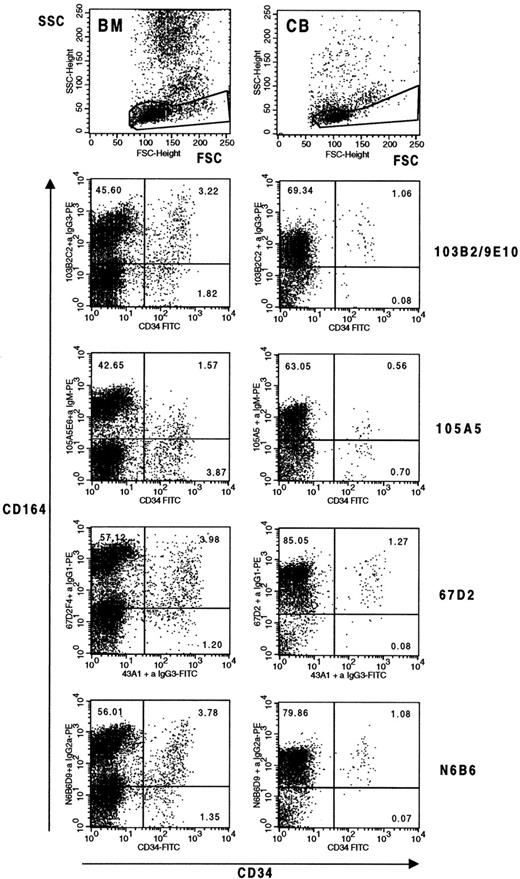
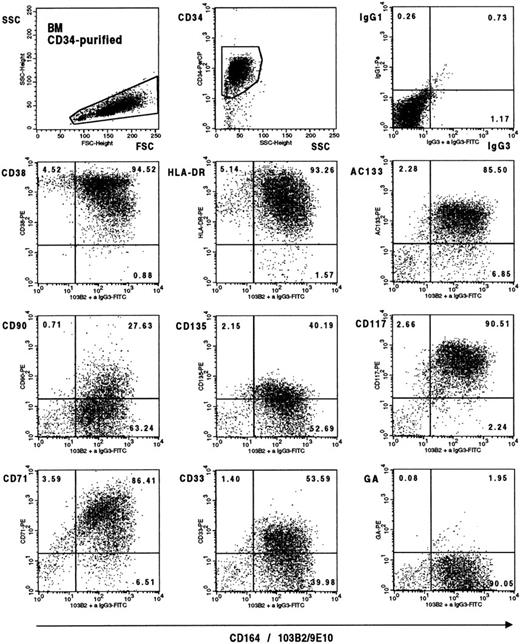
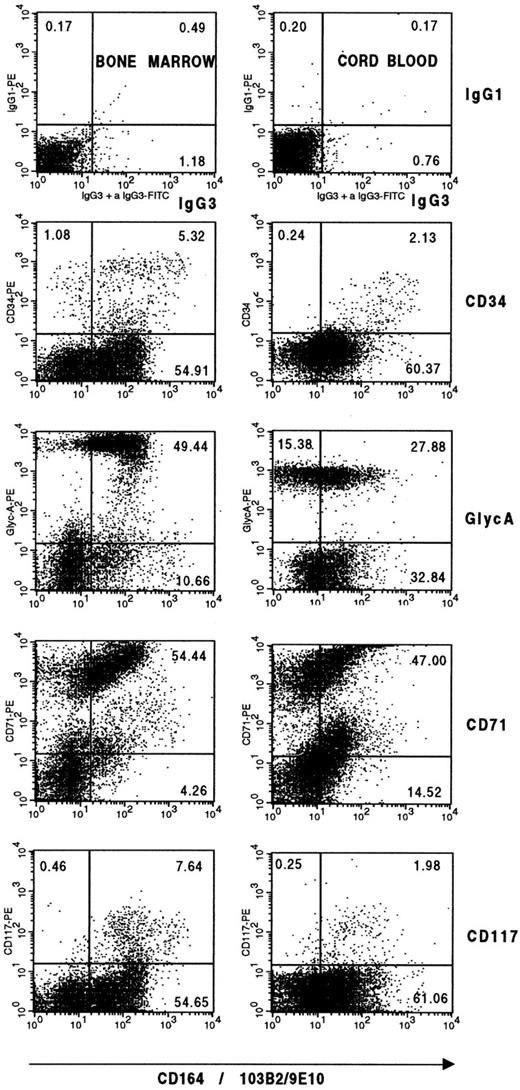
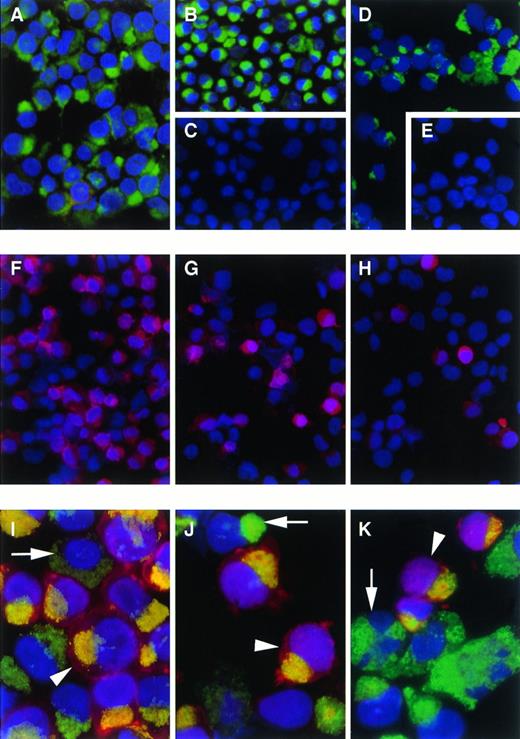
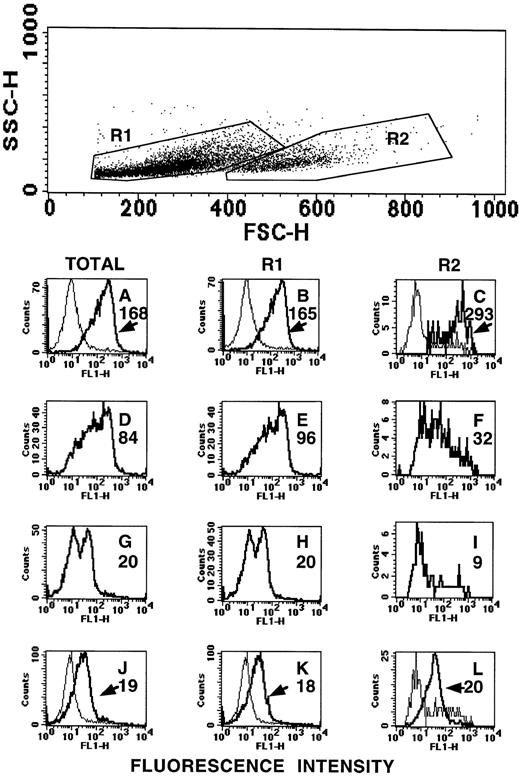
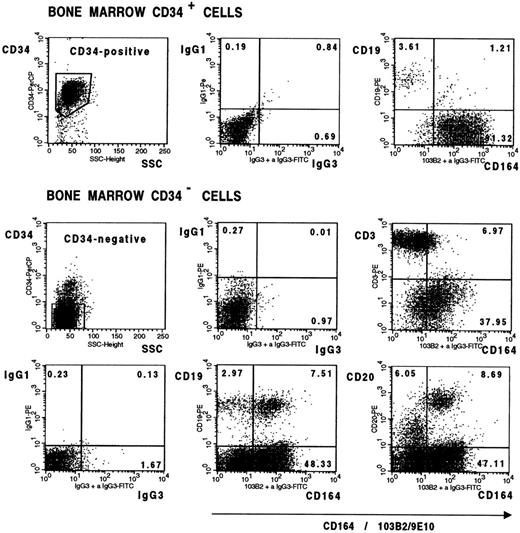
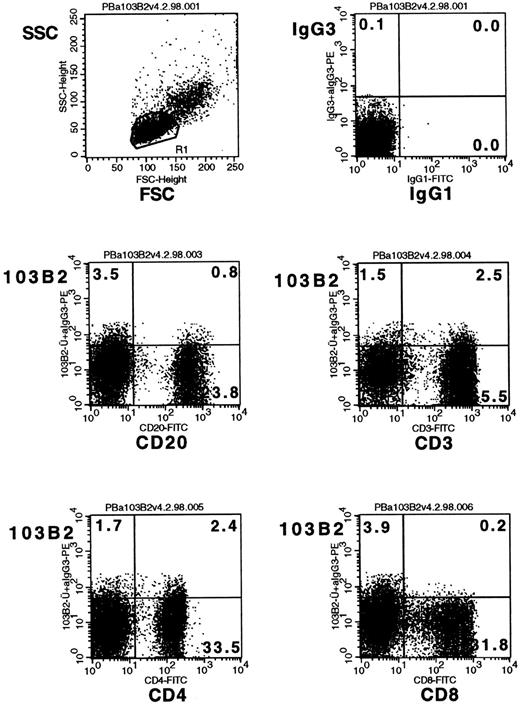
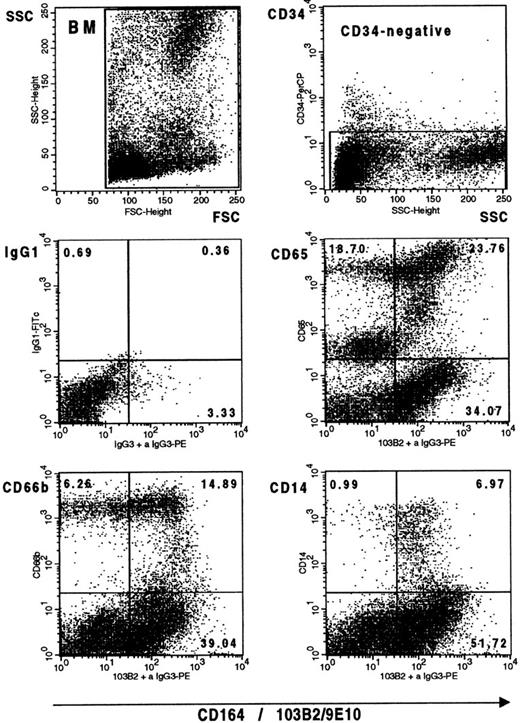
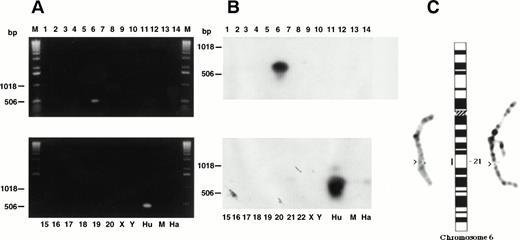
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal