Abstract
Primitive and definitive erythropoiesis represent distinct hematopoietic programs that differ with respect to stage of development, transcriptional control, and growth regulation. Although these differences have been recognized for some time, the relationship of the two erythroid lineages to each other is not well established. We have used a model system based on the hematopoietic development of embryonic stem (ES) cells in culture to investigate the origins of the earliest hematopoietic populations. Using ES cells transduced with a retrovirus that overexpresses the HOX11 gene, we have established factor-dependent hematopoietic cell lines that represent novel stages of embryonic hematopoiesis. Analysis of three of these cell lines indicates that they differ with respect to cytokine responsiveness, cell surface markers, and developmental potential. Two of the cell lines, EBHX1 and EBHX11, display the unique capacity to generate both primitive and definitive erythroid progeny as defined by morphology and expression of βH1 and βmajor globin. The third line, EBHX14, has definitive erythroid and myeloid potential, but is unable to generate cells of the primitive erythroid lineage. Analysis of the cytokine responsiveness of the two lines with primitive erythroid potential has indicated that exposure to leukemia inhibitory factor (LIF) results in the upregulation of βH1 and a change in cellular morphology to that of primitive erythrocytes. These findings are the first demonstration of a clonal cell line with primitive and definitive hematopoietic potential and support the interpretation that these lineages may arise from a common precursor in embryonic life. In addition, they suggest that LIF could play a role in the regulation of primitive erythropoiesis.
© 1998 by The American Society of Hematology.
THE MURINE EMBRYONIC hematopoietic system undergoes rapid and dynamic changes in both the lineages produced and the sites of production.1,2 The earliest stage of hematopoietic development, known as primitive hematopoiesis, occurs in the yolk sac blood islands and is restricted primarily to the generation of a unique population of erythroid cells, commonly referred to as embryonic or primitive erythrocytes.2-4 These primitive erythrocytes are large, nucleated and produce the embryonic forms of globin.3,4 Three to 4 days after the initiation of yolk sac hematopoiesis, intraembryonic hematopoiesis is established, initially in the region destined to form the aorta-gonad-mesonephros and shortly thereafter in the fetal liver.2,5-7 The onset of fetal liver hematopoiesis marks the switch from the primitive to the definitive hematopoietic program, which is characterized by the production of a broad spectrum of lineages including adult type erythrocytes. These cells differ from their embryonic counterparts in that they are smaller, enucleate, and produce adult globins.3 4 With the establishment of the definitive hematopoietic program, yolk sac hematopoiesis declines and primitive erythrocytes are no longer produced.
Although these developmental changes within the hematopoietic system have been well established for many years, the relationship of the primitive and definitive lineages to one another and the mechanisms regulating them remain poorly defined. Recent gene targeting studies, as well as naturally occurring mutations in mice, have provided some insights into the molecular events involved in the establishment of primitive and definitive hematopoiesis and have documented clear differences in the transcriptional regulation and growth control of these populations. For instance, transcription factors such as myb8 and acute myeloid leukemia-1 (AML-l; core-binding factor [CBF]α2)9,10 are essential for the establishment of definitive hematopoiesis, but appear to be dispensable for primitive erythropoiesis. The c-kit receptor tyrosine kinase and its ligand, kit ligand (KL), are essential for progression into the definitive program, but are not required for the establishment of primitive erythropoiesis in the yolk sac.1,11 Similarly, erythropoietin is essential for maturation of the definitive erythroid lineage, but not for primitive erythropoiesis.12 13 Together these studies suggest that primitive and definitive hematopoiesis represent separate developmental programs that are regulated by different molecular mechanisms and display different growth requirements.
Further understanding of the relationship between primitive and definitive hematopoiesis, including the mechanisms involved in the establishment of the two systems, will require access to early precursor populations as they develop. Whereas definitive precursors are present in the liver throughout most of fetal life and in the bone marrow for all of adult life, the generation of primitive erythroid precursors is restricted to a narrow window of embryonic development that precedes the establishment of the yolk sac blood islands.14 This limited time of development at a stage when the embryo is extremely small and not readily accessible has made the isolation and characterization of this population difficult, if not impossible. To overcome the problem of accessibility of embryonic hematopoietic precursors, a number of groups have used an in vitro model system based on the capacity of embryonic stem (ES) cells to generate hematopoietic precursors in culture.15-17 Analysis of embryoid bodies (EBs) from the differentiating ES cells has shown that hematopoietic differentiation in these cultures recapitulates many aspects of the onset of hematopoietic development in utero, including the switch from the primitive to the definitive program.18 19
The establishment of hematopoiesis in culture from ES cells not only provides access to rare, early developing populations, but also provides a unique system for addressing questions related to the effects of altered gene expression on hematopoietic precursor development, growth, and differentiation. The outstanding advantage of this approach is that genes can be introduced at the level of the starting ES population and subsequently be expressed as the primitive hematopoietic precursors develop from prehematopoietic mesoderm. In addition to providing new insights into gene function, overexpression of genes, particularly those with transforming and/or immortalizing potential, provides a unique opportunity to establish cell lines representing various stages of embryonic hematopoietic development.
Within this context, members of the HOX gene family are of interest, as recent studies have shown that overexpression of these genes can lead to selective expansion and in some instances, immortalization of different hematopoietic precursor populations.20-23 At least two different HOX genes have been shown to have potent immortalizing capacity in primary hematopoietic cells. The first,Hox-2.4 (Hoxb8), whose expression is upregulated in WEHI-3B tumor cells, is able to immortalize bone marrow-derived hematopoietic precursors when transduced and expressed in these cells in the context of a retroviral vector.21 The second,HOX11, originally identified from translocations in certain T-cell acute lymphoblastic leukemias, also displays strong immortalizing potential when transduced into primary mouse bone marrow–derived precursors.20 Given this capacity to immortalize adult marrow precursor populations, we were interested in determining if this class of genes would exhibit similar properties when overexpressed in embryonic hematopoietic precursors. In this report, we show that immortalized hematopoietic cell lines can be generated from ES cell–derived EBs that overexpress the HOX11gene. The lines generated from these EBs are factor-dependent and display unique developmental potentials including the capacity to generate both primitive and definitive erythroid cells. Using these cell lines, we have identified leukemia inhibitory factor (LIF) as a potential novel regulator of the primitive erythroid lineage.
MATERIALS AND METHODS
Retroviral-mediated transfer of the HOX11 gene to ES cells.
The construction of the MSCV-HOX11 and MSCV-NEO (MSCVv2.1) retroviral vectors and the generation of the corresponding helper-free ecotropic retroviral vector producer lines GP+E-86/MSCV-HOX11 and GP+E-86/MSCVv2.1, respectively, have been reported.20,24 25Virus producer lines were maintained in Dulbecco's modified Eagle medium (DMEM) with 4.5 g/L glucose (Life Technologies, Gaithersburg, MD) supplemented with 10% calf serum (Hyclone Laboratories, Logan, UT) in a humidified atmosphere containing 5% CO2/95% air at 37°C. Supernatants were collected from subconfluent cultures 24 hours after the medium was changed to DMEM supplemented with 1.5 × 10−4 mol/L monothioglycerol and 15% fetal calf serum (FCS), filtered through 0.45-mm filters and used immediately to transduce ES cells. Virus titers were in the range of 4 to 8 × 106 colony-forming units/mL when assayed on NIH3T3 fibroblasts in the presence of 400 μg/mL G418.
ES cell transductions were performed essentially as described for embryonal carcinoma cells.26 Briefly, 1-mL aliquots of vector supernatants were added to 4 × 105 ES cells per 60-mm tissue culture dish and the cultures were incubated in the presence of 4 μg/mL polybrene (hexadimethrine bromide; Sigma, St Louis, MO) at 37°C for 2 hours. ES cell culture medium (see below) was added (4 mL) and the incubations were continued at 37°C for 24 hours. This procedure was repeated, and 24 hours later the cell monolayers were trypsinized into single cell suspensions and transferred to 100-mm tissue culture dishes. Transduced cells were selected in 400 μg/mL G418 and the developing colonies were pooled on day 7 and used in the EB differentiation experiments.
Growth and differentiation of ES cells.
Wild-type CCE ES cells, as well as those transduced with MSCV-HOX11 and MSCV-NEO retroviral vectors, were maintained on gelatinized flasks in DMEM supplemented with 15% FCS (Hyclone), penicillin, streptomycin, 1.5 × 10−4 mol/L monothioglycerol (MTG; Sigma) and saturating amounts of a Chinese hamster ovary (CHO) LIF-containing supernatant (1% conditioned medium; CM). Two days before the initiation of differentiation, cells were transferred to Iscove's modified Dulbecco's medium (IMDM) containing the above components. After 2 days of growth in IMDM, ES cells were trypsinized into a single-cell suspension and plated into differentiation medium containing 1% methyl cellulose in IMDM supplemented with 15% FCS, 2 mmol/L glutamine (Life Technologies), and 4.5 × 10−4 mol/L MTG. The differentiation cultures were performed in Petri grade dishes and maintained in a humidified chamber in a 5% CO2/air mixture at 37°C for 7 days.
Establishment and maintenance of the EBHX cell lines.
Day 7 EBs generated from ES cells transduced with either the MSCV-HOX11 or the MSCV-NEO virus were dissociated by trypsinization and the cells cultured in liquid for 2 months. These liquid cultures were performed in 100-mm Petri grade dishes in IMDM supplemented with 5% FCS, 1.5 × 10−4 mol/L MTG and either KL (1% CM) and erythropoietin (Epo) (2 U/mL) or interleukin-3 (IL-3) (1% CM) and Epo. Duplicate cultures seeded at a density of 5 × 105 cells/mL were maintained with and without G418 for each population of cells. Vigorously growing cultures were either passaged to new dishes or depleted of cells as deemed necessary to maintain appropriate cell densities. Cells from both the MSCV-HOX11 and MSCV-NEO containing EBs grew in the presence of G418. Four and 8 weeks after the initiation of the liquid cultures, G418R cells from each set of conditions were cultured in methylcellulose in the presence of appropriate cytokines. Seven to 10 days later, individual colonies from the methylcellulose cultures were transferred to microtiter wells containing the above described medium. Rapidly growing populations were passaged to larger cultures and established as immortalized cell lines. The immortalized lines could be maintained indefinitely in the presence IMDM, 5% FCS, 1.5 × 10−4 mol/L MTG and the appropriate cytokines (line maintenance conditions). Subclones of the EBHX11 line were generated by single cell micromanipulation, whereas those of EBHX1 were obtained from colonies generated in methylcellulose under sparse conditions. These subclones were maintained in the same conditions as the parental populations.
Differentiation cultures for globin expression analysis.
For analysis of globin expression in colonies, EBHX cells were cultured in methylcellulose in IMDM supplemented with 5% plasma-derived serum (PDS; Antech, Tyler, TX), 5% protein-free medium (PFMHII; Life Technologies), glutamine (2 mmol/L), ascorbic acid (50 μg/mL), and appropriate cytokines. Liquid differentiation cultures consisted of the same components with the exception of the methylcellulose.
Polymerase chain reaction (PCR) globin analysis.
Globin expression patterns in the cell lines were determined using the global amplification strategy of Brady et al.27 Either individual colonies or small numbers (<1,000) of cells from liquid cultures were lysed in 4 μL of first strand buffer. Reverse transcription, tailing, and PCR procedures were performed as previously described,27 with the exception that the (dT)-x oligonucleotide was shortened to CATCTCGAGCGGCCGC(T)24. Amplified products from the PCR reaction were separated on an agarose gel and transferred to a Z-probe GT membrane (Bio-Rad, Richmond, CA). For analysis of βmajor globin and L32 expression, the resulting blots were hybridized with 32P randomly primed cDNA probes corresponding to the 3′ region of these genes. To analyze βH1 expression, a probe was prepared by annealing two oligonucleotides, (5′TGGAGTCAAAGAGGGCATCATAGACACATGGG3′, 5′CAGTACACTGGCAATCCCATGTG3′), which share an 8 base homology at their 3′ termini. This βH1 specific oligonucleotide was labeled with 32P using a Klenow fill-in reaction. Hybridizations were performed using the method of Church and Gilbert.28 βmajor and L32 cDNA probes were hybridized and washed at 65°C, while the βH1 oligonucleotide probe was hybridized and washed at 42°C.
RNA preparation and Northern blot analysis.
Poly(A)+ RNA was isolated from the EBHX cell lines using an oligo(dT)-cellulose column. For expression analysis, 3 to 5 μg of poly(A)+ RNA from each sample was run in a 1.0% agarose-formaldehyde gel and subsequently transferred to Z-probe GT membrane (Bio-Rad). The membranes were hybridized with appropriate 32P-labeled cDNA probes or with the βH1 specific oligonucleotide.
Growth factors.
IL-11, IL-6, macrophage colony-stimulating (M-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and vascular endothelial growth factor (VEGF) were purchased from R & D Systems, Minneapolis, MN. LIF and c-kit ligand were derived from medium conditioned by CHO cells transfected with LIF and KL expression vectors (kindly provided by Genetics Institute, Cambridge, MA). For some experiments, recombinant LIF and KL (R & D Systems) were used in place of the conditioned medium. IL-3 was obtained from medium conditioned by X63 AG8-653 myeloma cells transfected with a vector expressing IL-329 or purchased from R & D Systems.
Fluorescence-activated cell sorting (FACS) analysis.
Immunofluorescence flow cytometric analysis was performed essentially as described20 30 with saturating concentrations of affinity-purified rat monoclonal antibodies recognizing murine hematopoietic cell-surface antigens. Fluorescein isothiocyanate-conjugated reagents included: M1/70HL, anti-CD11b/Mac-1 (Boehringer Mannheim, Laval, Quebec); C71/16, anti-CD18/CD11abc (PharMingen, San Diego, CA); 2.4G2, anti-CD32/CD16(FcγRII/RIII) (PharMingen); IM7.8.1, anti-CD44/Ly-24 (obtained from the American Type Culture Collection, Rockville, MD); 30-F11.1, anti-CD45/Ly-5 (ATCC); RA3-6B2, anti-CD45R/B220 (PharMingen); 30-H12, anti-CD90.2/Thy-1.2 (PharMingen); and R6B-8C5, anti-Ly-6G/Gr-1 (PharMingen). Biotinylated reagents included: anti-AA4.1 (generously provided by John McKearn, Searle Research and Development, Monsanto Co, St Louis, MO); M1/69.16.11.HL, anti-CD24/HSA (ATCC); PS/2, anti-CD49d/VLA-4 α4 (kindly supplied by Paul Kincade, Oklahoma Medical Research Foundation, Oklahoma City, OK); and E13-161-7, anti–Sca-1/Ly-6A (a generous gift from Dr Irving Weissman, Stanford University, Stanford, CA). To prevent nonspecific Fc receptor binding, cells (106) were first incubated with 1 mL of culture supernatant from the 2.4G2 anti-CD32/CD16(FcγRII/RIII) hybridoma (ATCC). Biotinylated monoclonal antibodies were developed with R-phycoerythrin streptavidin (Molecular Probes, Eugene, OR) as a second step. Viable cells were gated by a combination of forward and orthogonal light scatter and were analyzed on an Epics Elite flow cytometer (Coulter Corp, Miami, FL).
Radioimmunoprecipitation analysis of HOX11.
Radiolabeling of proteins and immunoprecipitations were performed essentially as described.20 30 Briefly, cells (2 × 107) were labeled for 6 hours at 37°C with 300 μCi [35S]methionine (Amersham Canada Ltd, Oakville, Ontario) in 1 mL of methionine-deficient DMEM (ICN Biomedicals, Costa Mesa, CA) supplemented with 5% FCS and the appropriate cytokines. Cells were washed with cold phosphate-buffered saline and lysed in 1 mL of RIPA buffer (50 mmol/L Tris pH 7.5, 150 mmol/L NaCl, 1% Nonidet P-40, 1% deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 2 mmol/L phenylmethylsulfonyl fluoride) for 30 minutes on ice. The cell extract was clarified by centrifugation and the lysate was precleared by incubation with 40 μg of whole mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) at 4°C overnight and addition of 100 μL formalin-treated S aureus cells (Pansorbin, Calbiochem-Novabiochem Corp, San Diego, CA). The HOX11 proteins were precipitated with 5 μL rabbit HOX11 antiserum (kindly provided by Ming Lu, University of California, San Diego, CA) for 1 hour at 4°C and the immune complexes were collected with 75 μL of 100 mg/mL protein A sepharose CL-4B (Pharmacia Biotech, Baie D'Urfé, Quebec) in phosphate-buffered saline. The immunoprecipitates were washed four times (once in RIPA buffer, once in RIPA buffer containing 0.5 mol/L NaCl, followed by two additional washes in RIPA buffer alone) and resuspended in 30 μL of Laemmli sample buffer (2% SDS, 10% glycerol, 60 mmol/L Tris pH 6.8, 0.001% bromphenol blue) containing 100 mmol/L dithiothreitol. Samples were boiled for 3 minutes and electrophoresed through 12% SDS-polyacrylamide gels. Dried gels were exposed to Kodak XAR-5 film (Eastman Kodak company, Rochester, NY).
RESULTS
Establishment of EBHX cell lines.
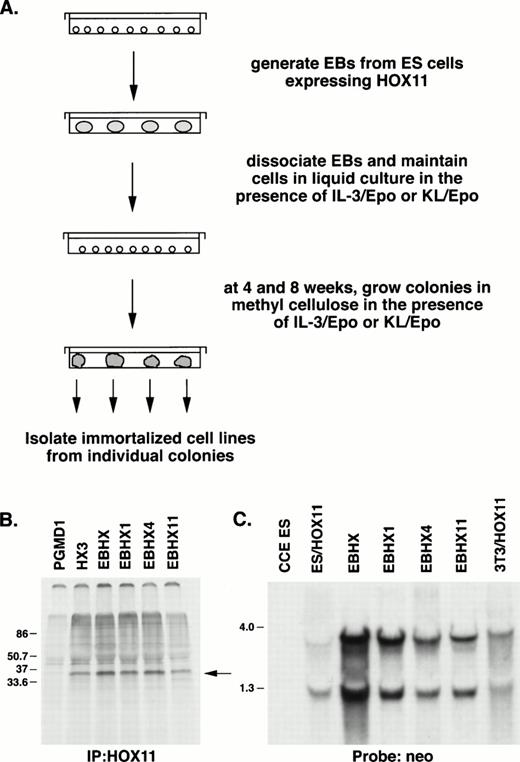
To overexpress HOX11 in early embryonic hematopoietic populations, ES cells were transduced with the MSCV-HOX11 retrovirus, which expresses the HOX11 gene from the LTR promoter and the selectableneo gene from a downstream internal pgk promoter.20 24 Transduced ES cells were selected in G418 for 7 days and then used for the generation of EBs as outlined in Fig 1A. Control ES cells were transduced with MSCV-NEO expressing only the neo gene. Analysis of EBs generated from MSCV-HOX11-transduced ES cells demonstrated the presence of HOX11 mRNA and protein, as well as neo transcripts (Fig 1B and C), indicating that the vector maintained expression of both theHOX11 and neo genes throughout the ES differentiation period.
(A) Protocol used to generate EBHX cell lines. (B) Immunodetection of HOX11 protein (37 kD, arrow) in EBHX cell lines by immunoprecipitation of radiolabeled proteins with HOX11-specific antiserum. PGMD1, an IL-3–dependent myeloid progenitor line used as negative control; HX3, a bone marrow-derived HOX11-immortalized line used as positive control; EBHX, day 6 EBs generated from MSCV-HOX11 transduced ES cells; EBHX1, EBHX4, EBHX11, three EB-derived HOX11-immortalized cell lines. (C) Northern blot analysis with a neo probe demonstrating LTR-directed (4.0 kb) HOX11 mRNA (which contains downstream neo sequences) and pgk-directed (1.3 kb) neo mRNA in EBHX cell lines. CCE ES, wild-type ES cells; ES/HOX11, MSCV-HOX11 transduced ES cells; 3T3/HOX11, NIH3T3 fibroblasts transduced with the MSCV-HOX11 vector.
(A) Protocol used to generate EBHX cell lines. (B) Immunodetection of HOX11 protein (37 kD, arrow) in EBHX cell lines by immunoprecipitation of radiolabeled proteins with HOX11-specific antiserum. PGMD1, an IL-3–dependent myeloid progenitor line used as negative control; HX3, a bone marrow-derived HOX11-immortalized line used as positive control; EBHX, day 6 EBs generated from MSCV-HOX11 transduced ES cells; EBHX1, EBHX4, EBHX11, three EB-derived HOX11-immortalized cell lines. (C) Northern blot analysis with a neo probe demonstrating LTR-directed (4.0 kb) HOX11 mRNA (which contains downstream neo sequences) and pgk-directed (1.3 kb) neo mRNA in EBHX cell lines. CCE ES, wild-type ES cells; ES/HOX11, MSCV-HOX11 transduced ES cells; 3T3/HOX11, NIH3T3 fibroblasts transduced with the MSCV-HOX11 vector.
Seven days after the initiation of ES differentiation, the developing EBs were dissociated and the cells cultured in liquid, either in the presence of IL-3 and Epo or in the presence of KL and Epo. Precursor analysis did not show any significant difference in the hematopoietic potential of the HOX11 expressing and control EBs at this stage of development. Four and 8 weeks later, cells from both sets of conditions were assayed in methylcellulose in the presence of the same cytokines used in the liquid cultures. At both time points, there were clear differences in the types of colonies that grew from the HOX11 expressing cells compared with the NEO control cells. All colonies in the control cultures had similar morphologies and were found to contain only mast cells. In contrast, HOX11-expressing cells generated colonies that displayed significant heterogeneity with respect to cell size and the degree of disperseness of the cells within the colonies. In addition, a significant number of the colonies in these cultures developed hemoglobinized erythroid cells. Many of the colonies derived from the HOX11 expressing EBs consisted of a mixture of maturing hematopoietic cells together with cells with an immature morphology. Individual colonies from cultures at both time points were transferred to microtiter wells in their respective conditions (IL-3/Epo or KL/Epo) in the presence of G418. At this stage, immortalized cell lines could be established from approximately 10% of these colonies. Medium to large colonies with an obvious undifferentiated component were most efficient at generating cell lines. A total of 16 embryoid body-derived HOX11 (EBHX) lines were established in the initial experiment, 10 from the IL-3/Epo cultures and 6 from the KL/Epo cultures.
Based on differences in cell morphology, 3 lines were selected for further characterization. These included EBHX11 isolated at 8 weeks from the KL/Epo cultures and EBHX14 and EBHX1 isolated at 4 and 8 weeks, respectively, from the IL-3/Epo cultures. Two additional lines, EBHX15 and EBHX4, established from the IL-3/Epo cultures at 4 and 8 weeks, as well as subclones of EBHX1 (EBHX1-C7) and EBHX11 (EBHX11-15), were included in some of the analyses. To determine if HOX11 was involved in the immortalization event, three of the lines were analyzed for the presence of HOX11 protein. As shown in Fig 1B, HOX11 protein was detected in EBHX1, EBHX4, and EBHX11. In addition, all three lines also expressed neo mRNA (Fig 1C). These findings demonstrate that the MSCV retroviral vector is able to maintain expression of both transduced genes in the EB-derived cell lines and is consistent with the interpretation that HOX11 is essential for their generation.
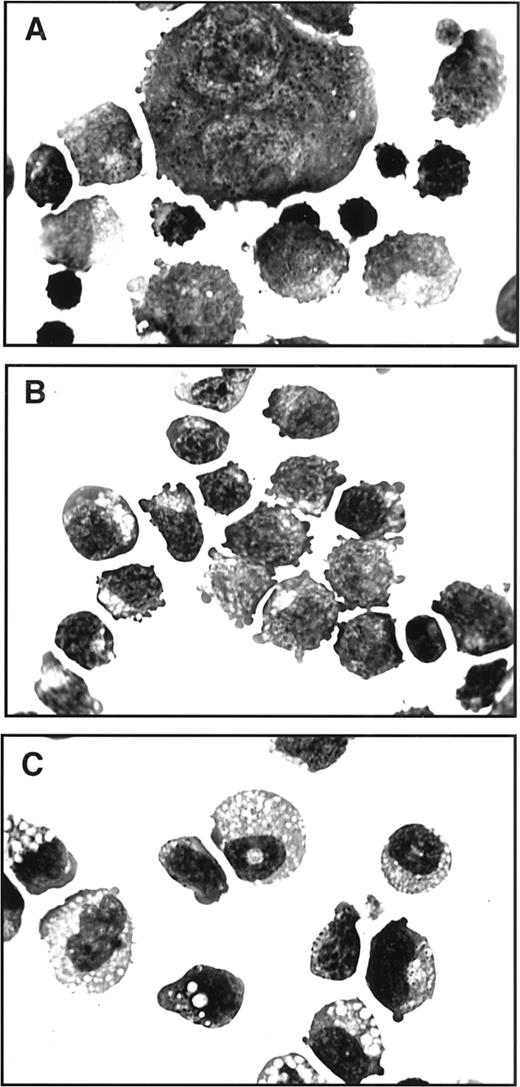
EBHX1, EBHX11, and EBHX14 cells display strikingly different morphologies. EBHX1 consists of two cell types, medium to small cells with erythroid characteristics and large megakaryocyte-like cells (Fig 2A). EBHX11 cells are relatively homogeneous in size and have an erythroblastic morphology (Fig 2B), whereas EBHX14 cells show signs of both erythroid and myeloid differentiation potential (Fig 2C). EBHX4 cells were almost identical to those of the EBHX1 line, whereas EBHX15 cells display erythroid and myeloid morphology, similar to EBHX14 cells (not shown).
Morphology of cells in three different EBHX cell lines. EBHX1 (A), EBHX11 (B), and EBHX14 (C). Cells were stained with May-Grünwald and Giemsa. Original magnification × 1,000.
Morphology of cells in three different EBHX cell lines. EBHX1 (A), EBHX11 (B), and EBHX14 (C). Cells were stained with May-Grünwald and Giemsa. Original magnification × 1,000.
Growth factor responsiveness of EBHX cell lines.
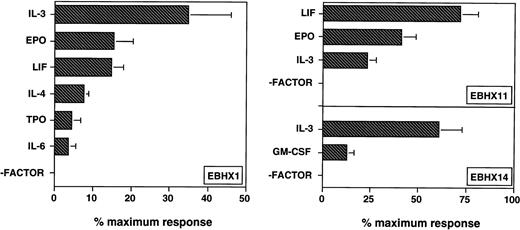
To further define the developmental potential of the EBHX lines, their growth responsiveness to different cytokines in methylcellulose cultures was next analyzed. The cytokines tested included IL-2, IL-3, IL-4, IL-6, IL-7, IL-11, Epo, thrombopoietin (TPO), KL, G-CSF, GM-CSF, M-CSF, LIF, insulin-like growth factor-1 (IGF-1), VEGF, and basic fibroblast growth factor (FGF). Only data for cytokines found to have a growth promoting activity are shown (Fig 3). None of the lines generated colonies in the absence of growth factors, demonstrating a strict dependence on cytokines for growth in methylcellulose. Of the three lines analyzed, EBHX1 cells responded to the broadest spectrum of cytokines, showing the highest response to IL-3. These cells also responded to Epo, LIF, IL-4, TPO, and IL-6. EBHX11 cells responded best to LIF, but did show significant responses to Epo and IL-3. Although the EBHX11 line was generated in the presence of KL and Epo, the cells showed no significant growth response to KL, either alone or together with Epo. Nevertheless, the cells are routinely maintained in KL and Epo, as they do express the c-kit encoded receptor (see below) and the interaction with KL may be important to preserve the potential of the line. EBHX14 cells respond to IL-3 and GM-CSF, consistent with the interpretation that they have myeloid potential. Colony growth by this line was somewhat cell density-dependent and was best at concentrations of 1 × 104 to 5 × 104 cells per mL. None of the lines showed a significant growth response to M-CSF, G-CSF, IL-2, IL-7, IL-11, KL, VEGF, basic FGF, or IGF-1. Combinations of factors were not extensively tested, with the exception of the large mixture used to measure the maximal growth response and the combination of IL-3/Epo or KL/Epo used for cell line maintenance. The cytokine responsiveness of the subclones, EBHX1-C7 and EBHX1-15, was identical to that of the parental lines (not shown).
Growth factor responsiveness of EBHX cell lines. EBHX cells were cultured in methylcellulose in the presence of the indicated cytokines and the developing colonies scored 7 to 8 days later. EBHX1 cells were cultured at 1 × 104 to 5 × 104cells/mL, EBHX11 at 250 to 750 cells/mL, and EBHX 14 at 1 × 104 to 5 × 104 cells/mL. Maximum response was determined by the number of colonies that grew in response to a broad mixture of cytokines, which included IL-3, IL-6, IL-11, Epo, KL, LIF, GM-CSF, G-CSF, and M-CSF. Colony-forming cell frequencies in this mixture were approximately 14% for EBHX1, 50% for EBHX11, and 20% for EBHX14. Cytokines were used at the following concentrations: IL-2, 1% conditioned medium; IL-3, 1 ng/mL; IL-6, 5 ng/mL; IL-7, 16 ng/mL; IL-11, 25 ng/mL; Epo, 2 U/mL; KL, 100 ng/mL; LIF, 1 ng/mL; IL-4, 1% conditioned medium; TPO, 5 ng/mL; G-CSF, 30 ng/mL; GM-CSF, 15 ng/mL; M-CSF, 5 ng/mL; IGF-1, 5 ng/mL; and basic FGF, 10 ng/mL. Bars represent standard error of the mean (SEM) from three independent experiments.
Growth factor responsiveness of EBHX cell lines. EBHX cells were cultured in methylcellulose in the presence of the indicated cytokines and the developing colonies scored 7 to 8 days later. EBHX1 cells were cultured at 1 × 104 to 5 × 104cells/mL, EBHX11 at 250 to 750 cells/mL, and EBHX 14 at 1 × 104 to 5 × 104 cells/mL. Maximum response was determined by the number of colonies that grew in response to a broad mixture of cytokines, which included IL-3, IL-6, IL-11, Epo, KL, LIF, GM-CSF, G-CSF, and M-CSF. Colony-forming cell frequencies in this mixture were approximately 14% for EBHX1, 50% for EBHX11, and 20% for EBHX14. Cytokines were used at the following concentrations: IL-2, 1% conditioned medium; IL-3, 1 ng/mL; IL-6, 5 ng/mL; IL-7, 16 ng/mL; IL-11, 25 ng/mL; Epo, 2 U/mL; KL, 100 ng/mL; LIF, 1 ng/mL; IL-4, 1% conditioned medium; TPO, 5 ng/mL; G-CSF, 30 ng/mL; GM-CSF, 15 ng/mL; M-CSF, 5 ng/mL; IGF-1, 5 ng/mL; and basic FGF, 10 ng/mL. Bars represent standard error of the mean (SEM) from three independent experiments.
The strong response of EBHX11 cells to LIF and Epo suggested that cytokines other than the combination of KL/Epo might be able to support their growth for an extended period of time. To address this question, EBHX11 cells were passaged from KL/Epo to cultures containing either LIF or only Epo. In both conditions, the cells survived, grew, and could be easily maintained over a 6-month period without any apparent signs of senescence. Although there were no obvious significant differences in the growth rate of these populations, the cells maintained in LIF, EBHX11L, were more adhesive than the parental cells and tended to grow in clusters, particularly at high cell density. The cells grown in Epo, EBHX11E, were similar to those maintained in KL/Epo. In addition to these adhesive differences, EBHX11L cells showed different patterns of globin gene expression compared to EBHX11 and EBHX11E cells (see below).
Surface phenotype of EBHX cell lines.
To better define the lineage relationship of the EBHX lines, their cell-surface phenotype was determined by immunofluorescence flow cytometric analysis using a panel of monoclonal antibodies directed against hematopoietic cell surface antigens. A summary of the analysis of five different lines is presented in Table 1. EBHX1 and EBHX4 exhibited similar expression patterns that included low levels of Thy-1 and high levels of the low-affinity Fc receptor, FcγRII/III, found on myeloid, B-cell, T-cell, and natural killer cell precursors.31,32 In addition, cells from these two lines expressed the hyaluronan receptor CD44 found on most hematopoietic precursors,33 the integrin very late antigen (VLA)-4 present on precursors and differentiated myeloid and lymphoid cells,34 and lymphocyte function-associated antigen (LFA)-1/CD18 found on most leukocytes.35 A subpopulation of EBHX1, but not of EBHX4, also expressed TER-119, an erythroid lineage-restricted antigen.36 The cells from these two lines did not express AA4.1, a marker found on fetal liver stem cells and precursors,37 Sca-1 (Ly-6A) found on fetal liver and adult marrow stem cells,38-40 heat stable antigen (HSA), and the panleukocyte antigen CD45, markers found on most hematopoietic precursors,35 Gr-1, a marker of the granulocyte lineage,41 Mac-1, a myeloid differentiation antigen,35 and B220, a marker of the B-cell lineage.42 EBHX14 and EBHX15 also showed similar expression patterns and displayed the broadest spectrum of surface markers of all lines tested. Both lines contain subpopulations of cells that expressed AA4.1, TER-119, Mac-1, and Gr-1. In addition, these lines also expressed Thy-1, FcγRII/III, CD44, VLA-4, and CD18. The expression pattern of these two lines did differ in that EBHX15 expressed CD45, intercellular adhesion molecule (ICAM)-1, HSA, and no detectable Sca-1, whereas EBHX14 expressed very low levels of CD45 and HSA and no detectable ICAM-1. In addition, a small subpopulation of EBHX14 did express Sca-1. EBHX11 cells showed the most restricted pattern of the lines tested and were found to express only CD44, VLA-4, HSA, and ICAM. The results from the surface marker analysis further documents differences between these lines and suggest that EBHX14 and EBHX15 contain the most immature cell types, as defined by AA4.1 expression, EBHX-1 and EBHX4 represent intermediate stage cells, and EBHX11 represents a relatively late stage population.
Surface Marker Analysis of EBHX Lines
| Marker . | EBHX1 . | EBHX4 . | EBHX11 . | EBHX14 . | EBHX15 . |
|---|---|---|---|---|---|
| Precursors | |||||
| AA4.1-150 | 0.2 (0.8) | 0.5 (0.9) | 0.1 (0.6) | 7.6 (1.1) | 18.3 (3.2) |
| Sca-1 (Ly-6A)-150 | 0.4 (0.9) | 0.7 (0.8) | 0.1 (0.6) | 5.2 (1.4) | 1.1 (2.1) |
| Thy-1 | 6.8 (4.7) | 3.1 (4.3) | 0.3 (0.5) | 86.4 (30.7) | 99.4 (62.5) |
| FcγRII/III (CD32/CD16) | 78.2 (3.7) | 85.0 (5.4) | 0.1 (0.6) | 91.4 (5.7) | 99.7 (39.9) |
| CD44 | 91.5 (8.9) | 94.3 (10.2) | 98.1 (9.2) | 92.2 (7.7) | 16.9 (23.9) |
| VLA-4 (CD49d)-150 | 77.1 (3.4) | 98.0 (4.0) | 73.6 (1.2) | 99.5 (5.3) | 99.5 (20.4) |
| HSA (CD24)-150 | 0.1 (0.9) | 0.3 (0.8) | 99.7 (6.6) | 5.1 (2.3) | 98.5 (12.5) |
| Lineage-restricted | |||||
| TER-119-151 | 6.5 (1.8) | 0.8 (1.8) | 0.1 (0.7) | 13.4 (4.4) | 28.0 (8.6) |
| Mac-1 (CD11b) | 0.8 (1.8) | 0.2 (1.4) | 0.3 (0.6) | 4.1 (2.6) | 11.8 (12.1) |
| Gr-1 (Ly-6G) | 1.0 (1.7) | 0.4 (2.2) | 0.3 (0.5) | 23.2 (20.3) | 6.3 (31.5) |
| B220 (CD45R) | 1.2 (4.6) | 0.6 (2.4) | 0.3 (0.5) | 0.6 (1.6) | 0.4 (7.8) |
| Panleukocyte | |||||
| CD45 (Ly-5) | 1.2 (2.1) | 0.3 (2.1) | 0.2 (0.6) | 3.4 (2.3) | 84.8 (6.5) |
| CD18/CD11abc (LFA-1β2) | 96.8 (6.0) | 98.4 (7.2) | 0.3 (0.5) | 98.7 (10.1) | 98.5 (23.7) |
| ICAM-1 (CD54)-151 | 1.8 (2.2) | 0.4 (1.6) | 27.8 (1.7) | 0.7 (1.6) | 30.9 (9.2) |
| Marker . | EBHX1 . | EBHX4 . | EBHX11 . | EBHX14 . | EBHX15 . |
|---|---|---|---|---|---|
| Precursors | |||||
| AA4.1-150 | 0.2 (0.8) | 0.5 (0.9) | 0.1 (0.6) | 7.6 (1.1) | 18.3 (3.2) |
| Sca-1 (Ly-6A)-150 | 0.4 (0.9) | 0.7 (0.8) | 0.1 (0.6) | 5.2 (1.4) | 1.1 (2.1) |
| Thy-1 | 6.8 (4.7) | 3.1 (4.3) | 0.3 (0.5) | 86.4 (30.7) | 99.4 (62.5) |
| FcγRII/III (CD32/CD16) | 78.2 (3.7) | 85.0 (5.4) | 0.1 (0.6) | 91.4 (5.7) | 99.7 (39.9) |
| CD44 | 91.5 (8.9) | 94.3 (10.2) | 98.1 (9.2) | 92.2 (7.7) | 16.9 (23.9) |
| VLA-4 (CD49d)-150 | 77.1 (3.4) | 98.0 (4.0) | 73.6 (1.2) | 99.5 (5.3) | 99.5 (20.4) |
| HSA (CD24)-150 | 0.1 (0.9) | 0.3 (0.8) | 99.7 (6.6) | 5.1 (2.3) | 98.5 (12.5) |
| Lineage-restricted | |||||
| TER-119-151 | 6.5 (1.8) | 0.8 (1.8) | 0.1 (0.7) | 13.4 (4.4) | 28.0 (8.6) |
| Mac-1 (CD11b) | 0.8 (1.8) | 0.2 (1.4) | 0.3 (0.6) | 4.1 (2.6) | 11.8 (12.1) |
| Gr-1 (Ly-6G) | 1.0 (1.7) | 0.4 (2.2) | 0.3 (0.5) | 23.2 (20.3) | 6.3 (31.5) |
| B220 (CD45R) | 1.2 (4.6) | 0.6 (2.4) | 0.3 (0.5) | 0.6 (1.6) | 0.4 (7.8) |
| Panleukocyte | |||||
| CD45 (Ly-5) | 1.2 (2.1) | 0.3 (2.1) | 0.2 (0.6) | 3.4 (2.3) | 84.8 (6.5) |
| CD18/CD11abc (LFA-1β2) | 96.8 (6.0) | 98.4 (7.2) | 0.3 (0.5) | 98.7 (10.1) | 98.5 (23.7) |
| ICAM-1 (CD54)-151 | 1.8 (2.2) | 0.4 (1.6) | 27.8 (1.7) | 0.7 (1.6) | 30.9 (9.2) |
Results are the percentages of labeled cells (log mean fluorescence intensity) determined by direct staining of FcγRII/III-blocked cells with fluorescein isothiocyanate-conjugated antibodies.
Abbreviations: LFA, lymphocyte function-associated antigen; HSA, heat stable antigen; ICAM, intercellular adhesion molecule; Sca, stem cell antigen; VLA, very late antigen.
Direct staining of FcγRII/III-blocked cells with biotinylated antibodies and R-phycoerythrin-streptavidin.
Indirect staining with fluorescein isothiocyanate-conjugated secondary antibody (unblocked FcγRII/III).
Gene expression analysis of EBHX cell lines.
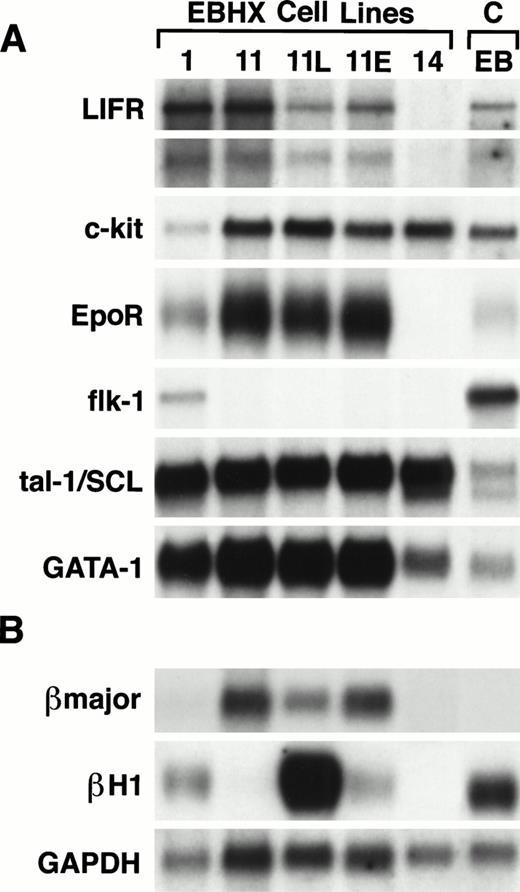
Given these observed differences in the lines, we were next interested in determining their patterns of specific receptor, transcription factor, and globin gene expression. Included in this analysis were EBHX1, EBHX11, and EBHX14, as well as the subpopulations of EBHX11 that were maintained in either LIF (EBHX11L) or Epo (EBHX11E). All of the lines, which responded to LIF, expressed two isoforms of the LIF receptor gene,43 a large message of approximately 10 kb and a smaller message of approximately 4.5 kb (Fig 4). Similarly, all of the lines that showed a response to Epo alone expressed the Epo receptor. c-Kit mRNA was detected in all of the lines consistent with the interpretation that they represent early hematopoietic precursor populations. EBHX1 was the only cell line that expressed flk-1, suggesting that it could represent an early stage of embryonic hematopoietic development, a stage characterized by expression of this receptor.19With respect to transcription factors, all lines expressed tal-1/SCL a member of the helix-loop-helix family of factors that is found in early hematopoietic cells and required for the establishment of the embryonic hematopoietic system.44-46 In addition, all of the lines expressed GATA-1, a transcription factor normally found in cells of the erythroid, mast cell, and megakaryocytic lineages.47However, in this case, EBHX1 and EBHX11, which contain Epo-responsive erythroid precursors, showed higher levels of GATA-1 expression than EBHX14, which contains less mature precursors that require both IL-3 and Epo for growth.
Expression analysis of EBHX cell lines. (A) Northern blot analysis of receptor and transcription factor gene expression. (B) Northern blot analysis of globin gene expression. The relative amounts of RNA loaded are indicated by hybridization to the probe specific for glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Cells from day 4.5 EBs were included as a control (C).
Expression analysis of EBHX cell lines. (A) Northern blot analysis of receptor and transcription factor gene expression. (B) Northern blot analysis of globin gene expression. The relative amounts of RNA loaded are indicated by hybridization to the probe specific for glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Cells from day 4.5 EBs were included as a control (C).
In contrast to the above expression patterns, the globin analysis showed some unexpected and important differences among the lines. EBHX11, EBHX11L, and EBHX11E all expressed βmajor globin, consistent with the interpretation that they represent erythroid precursors at a late stage of maturation. EBHX1 and EBHX14 did not express detectable amounts of βmajor globin, suggesting that the erythroid precursors in these lines represent an intermediate stage of development. Although EBHX11, EBHX11L, and EBHX11E expressed comparable levels of adult goblin, they displayed striking differences in the levels of βH1 expression. The parental line grown in the presence of KL/Epo showed little βH1 expression, whereas the subpopulation maintained in LIF expressed significant levels of this embryonic globin. The line maintained in Epo showed low, but detectable, levels of βH1. In addition to EBHX11L, EBHX1 also expressed some βH1. βH1 expression was not detected in EBHX14 cells. The presence of cells that express βH1 suggests that lines EBHX1 and EBHX11 have the potential to generate cells of the primitive erythroid lineage. Furthermore, the high levels of βH1 in EBHX11L cells indicates that LIF could be an important mediator in the upregulation of this embryonic globin and possibly in establishing the primitive erythroid program.
Primitive and definitive erythroid potential of the EBHX cell lines.
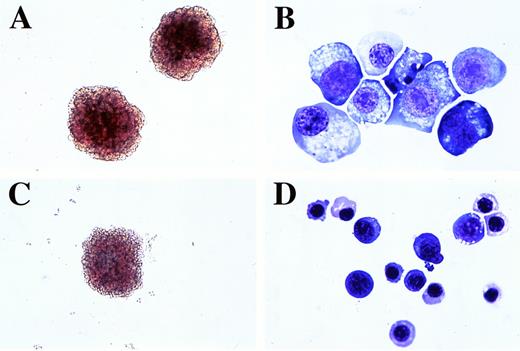
To further explore the primitive and definitive potential of these lines, EBHX11 cells were cultured in methylcellulose in the presence of LIF or Epo and the cells analyzed for globin expression patterns and morphologic characteristics of these lineages. Epo was used in place of KL/Epo, as studies subsequent to the Northern blot analysis indicated that EBHX11 cells do not express significant levels of βH1 when cultured in the presence of this cytokine. Colonies generated in both sets of conditions showed visible signs of hemoglobinization (Fig 5A and C), but did display distinct differences in morphology. In the presence of LIF, the colonies had a tighter, more compact appearance than those generated in response to Epo. More than 50% of the cells within the LIF-stimulated colonies were large and displayed characteristics of primitive erythrocytes (Fig5B). The remainder of these colonies typically consisted of undifferentiated cells and cells with a definitive erythroid morphology. In contrast, the majority of the cells in the Epo-stimulated colonies were significantly smaller than those in the LIF-stimulated colonies and had the morphology of cells of the definitive erythroid lineage (Fig 5D). The Epo-stimulated colonies also contained some cells with an undifferentiated morphology. Similar differences were observed in colonies generated from the EBHX11-15 subclone and from EBHX1 cells in these two sets of conditions (not shown).
EBHX11-derived colonies and cells. (A) Colonies grown in the presence of LIF for 7 days. (B) Cells from LIF-stimulated colonies. (C) Colony grown in the presence of Epo for 7 days. (D) Cells from Epo-stimulated colonies. Original magnification for (A and C), ×200 and for (B and D), ×1,000.
EBHX11-derived colonies and cells. (A) Colonies grown in the presence of LIF for 7 days. (B) Cells from LIF-stimulated colonies. (C) Colony grown in the presence of Epo for 7 days. (D) Cells from Epo-stimulated colonies. Original magnification for (A and C), ×200 and for (B and D), ×1,000.
Globin analysis of the colony-derived cells from these two conditions confirmed the morphologic differences. Colonies generated from EBHX11, EBHX11-15, or EBHX1 in the presence of LIF or LIF/Epo expressed readily detectable levels of βH1 (Fig 6). In contrast, those generated in the presence of Epo, KL/Epo, or IL-3/Epo showed little, if any, embryonic globin expression. The lack of significant βH1 expression in EBHX11-derived colonies generated in the presence of Epo or in EBHX1-derived colonies grown in the presence of IL-3 and Epo differs from the previous Northern blot analysis, which demonstrated low levels of embryonic globin in these populations. These differences could reflect differences in the culture conditions used for cell line maintenance compared with those used for differentiation (see Materials and Methods). βmajor was expressed in colonies generated in all conditions, although in many experiments EBHX11 colonies or cells grown in the presence of LIF expressed lower levels of the adult globin than those grown in the presence of Epo or KL/Epo (see also Fig 7). Erythroid colonies generated from EBHX14 cells in the presence of IL-3/Epo or in the presence of IL-3/LIF/Epo did not express detectable levels of βH1. Together, these findings strongly suggest that lines EBHX11, the subclone EBHX11-15 and EBHX1, have the potential to generate cells of the primitive and definitive erythroid lineages and that LIF is an important regulator of the primitive erythroid program.
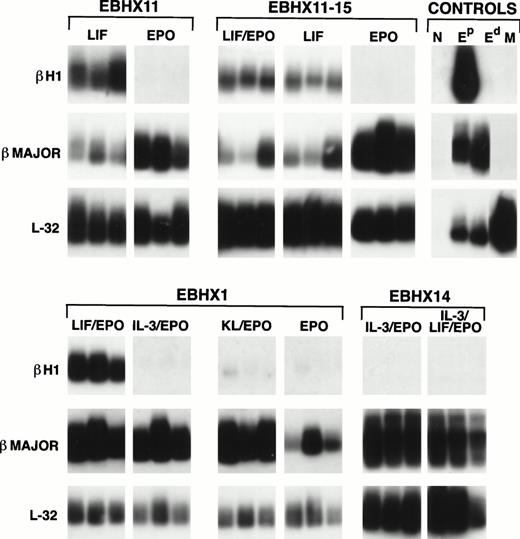
Globin expression analysis of EBHX-derived colonies. Colonies were grown in the indicated cytokines for 7 days and then picked and analyzed for βH1 and βmajor expression by Poly(A) PCR. The analysis of three individual colonies is shown for each set of conditions. EBHX11-15 is a subclone of EBHX11. Control Eprepresents primitive erythroid colonies grown from day 6 EB precursors, control Ed are definitive erythroid colonies generated from fetal liver precursors, and control M are macrophage colonies grown from day 6 EB-derived precursors. N represents PCR reagents with no cells added. Expression of the ribosomal L32 gene was used as an indication of the amount of material in each lane. Cytokines were used at the concentrations indicated in the legend to Fig 3.
Globin expression analysis of EBHX-derived colonies. Colonies were grown in the indicated cytokines for 7 days and then picked and analyzed for βH1 and βmajor expression by Poly(A) PCR. The analysis of three individual colonies is shown for each set of conditions. EBHX11-15 is a subclone of EBHX11. Control Eprepresents primitive erythroid colonies grown from day 6 EB precursors, control Ed are definitive erythroid colonies generated from fetal liver precursors, and control M are macrophage colonies grown from day 6 EB-derived precursors. N represents PCR reagents with no cells added. Expression of the ribosomal L32 gene was used as an indication of the amount of material in each lane. Cytokines were used at the concentrations indicated in the legend to Fig 3.
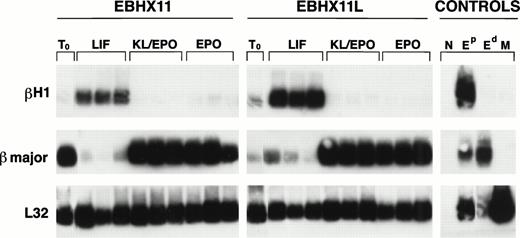
Globin expression analysis of EBHX11 and EBHX11L cells grown under different conditions. Cells were seeded into microtiter wells at a concentration of 30 or 100 cells per well, maintained in the indicated cytokines for 7 days, and then analyzed for βH1 and βmajor expression by Poly(A) PCR. Three samples from each set of conditions are shown. T0 represents the starting populations. Controls are the same as those in Fig 6. Expression of the ribosomal L32 gene was used as an indication of the amount of material in each lane.
Globin expression analysis of EBHX11 and EBHX11L cells grown under different conditions. Cells were seeded into microtiter wells at a concentration of 30 or 100 cells per well, maintained in the indicated cytokines for 7 days, and then analyzed for βH1 and βmajor expression by Poly(A) PCR. Three samples from each set of conditions are shown. T0 represents the starting populations. Controls are the same as those in Fig 6. Expression of the ribosomal L32 gene was used as an indication of the amount of material in each lane.
The previous data indicated that switching EBHX11 cells from KL/Epo to LIF results in the onset of a primitive erythroid program characterized by the generation of large cells with the morphology of primitive erythrocytes, which express βH1 globin. We were next interested in determining if EBHX cells exposed to LIF were still able to generate cells with a definitive phenotype when removed from LIF and cultured in KL/Epo. To address this question, EBHX11L cells that were maintained in LIF for 6 months were cultured in microtiter wells in the presence of LIF, KL/Epo, or Epo. As a control, parental EBHX11 cells were cultured under identical conditions. After 7 days of culture, both lines showed similar changes in goblin gene expression in response to the different cytokines (Fig 7). In the presence of LIF, both EBHX11L and EBHX11 cells expressed readily detectable levels of βH1 as expected. However, when cultured in KL/Epo or Epo alone, EBHX11L cells downregulated βH1 expression, indicating that the line maintained in LIF retained the capacity to generate cells with a definitive erythroid phenotype.
DISCUSSION
In this report we describe the generation and characterization of novel hematopoietic cell lines from EBs that were genetically modified to overexpress the HOX11 gene. All of the established EBHX lines analyzed expressed the HOX11 gene originally transduced into the starting ES population, all retained factor dependency, and all showed some capacity to differentiate. Several features of these cell lines distinguish them from virtually all other hematopoietic cell lines established to date. First, they were generated from day-7 EBs, a population representative of the embryonic or yolk sac phase of hematopoiesis.18 19 Most other hematopoietic cell lines have been derived from fetal or adult precursors. Second, at least two of the cell lines display the unique capacity to generate both primitive and definitive erythroid progeny. To our knowledge, this is the first demonstration of a clonal cell line with both primitive and definitive hematopoietic potential.
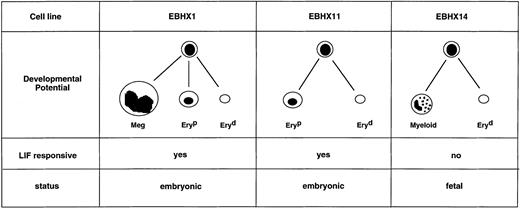
The developmental potential of the EBHX cell lines, summarized in Fig 8, raises some important questions with respect to the origin of the primitive and definitive erythroid lineages. Studies in both the avian and murine systems have provided evidence that primitive erythropoiesis and definitive hematopoiesis arise at distinct times and sites during embryogenesis and as such suggest that these populations derive from separate precursors.48-50 Further support for this concept was provided by findings, which demonstrated that the primitive and definitive erythroid lineages can arise from separate precursors in developing EBs.51 While these observations are consistent with the interpretation that these lineages have separate origins, several studies have challenged this notion and provided evidence indicating they arise from a common precursor early in development.52 53 The restricted potential of EBHX11 and its subclone, EBHX11-15, further supports the concept of a common ancestor for primitive and definitive hematopoiesis and suggests that, at early stages of development, the primitive and definitive erythroid lineages develop from an erythroid committed precursor. One concern with these interpretations is that they are based on the potential of immortalized cell lines, which may not accurately reflect potential of normal embryonic precursors. Our current studies are focused on the identification of comparable precursor populations in EBs and developing embryos.
Summary of the developmental potential of three EBHX cell lines. Meg, megakaryocyte; Eryp, primitive erythroid; Eryd, definitive erythroid.
Summary of the developmental potential of three EBHX cell lines. Meg, megakaryocyte; Eryp, primitive erythroid; Eryd, definitive erythroid.
The capacity to respond to LIF defines two separate populations of erythroid precursors represented by the different EBHX lines. LIF-responsive cells found in the EBHX11 line have both primitive and definitive erythroid potential and may be representative of precursors found in the yolk sac blood islands. The LIF nonresponsive population present in the EBHX14 line is unable to generate primitive erythroid progeny and could be the equivalent of the earliest definitive precursors found in the fetal liver. The correlation of LIF responsiveness with primitive erythroid potential suggests that this molecule could be a regulator of yolk sac erythropoiesis. This is an important observation as regulation of the growth and maturation of this early erythroid lineage is not well defined. The only known cytokine that stimulates primitive erythroid precursors in culture is Epo and, as with the definitive erythroid lineage, it appears to act at the equivalent of the CFU-E stage of development.15Although Epo can stimulate these precursors in culture, studies on Epo and Epo receptor knockout mice demonstrate that this cytokine is not essential for development of the lineage in vivo and suggest that other growth regulators are involved.12 13
Although our data implicate LIF as a possible regulator of primitive erythropoiesis, analysis of knockout mice indicates that this factor is not essential for development of this lineage in the embryonic yolk sac. Mice lacking LIF or gp130, the signaling component of the LIF receptor, survive through embryonic development, suggesting that neither the ligand nor the normal receptor complex is absolutely required for primitive erythropoiesis.54-56 However, as the early erythroid population in these animals was not characterized, it is not known if this lineage was negatively affected by these mutations. The fact that the LIF receptor is expressed in the EBHX lines would suggest that LIF or a LIF-like molecule that can bind this receptor does play some role in the regulation of the primitive erythroid lineage. Preliminary studies indicate that other cytokines that signal through gp130 including IL-6, IL-11, and oncostatin M are unable to mediate the switch to the primitive phenotype, suggesting that the effect could be specific to LIF (unpublished observation, 1997). Future studies will determine whether other molecules, either known or novel, have effects similar to those of LIF.
Immortalization of EB-derived embryonic precursors by HOX11 confirms and extends our earlier findings that constitutive expression of HOX11 immortalized bone marrow–derived precursors,20,30 and further demonstrates the potential of this class of genes to dramatically alter growth regulation within the hematopoietic system. In particular, our current studies document that HOX11 can affect the growth and development of embryonic hematopoietic precursors. In an earlier study, Helgason et al23 showed that deregulated expression of HOXB4 resulted in an expansion of the multipotential and erythroid precursor pool in EBs. However, overexpression of this particular HOX gene did not lead to the immortalization of these precursors. The strong immortalizing potential of HOX11 in the different hematopoietic populations raises important questions as to mechanisms of action. A recent study suggests that HOX11 can disrupt cell cycle checkpoints through interaction with specific protein phosphatases.57 Whether or not this is its primary mode of action in the EBHX lines remains to be determined.
In summary, we have shown that ectopic HOX11 expression in EBs leads to the immortalization of embryonic hematopoietic precursors. The EBHX cell lines derived from these precursors are representative of different stages of embryonic hematopoietic development and are the first to demonstrate both primitive and definitive potential. Using these lines, we have obtained evidence suggesting that LIF could play a role in the regulation of the primitive erythroid program. The availability of these lines provides a unique opportunity to further characterize the growth regulation of the primitive erythroid lineage, as well as to define the molecular events involved in its establishment.
ACKNOWLEDGMENT
We thank Ming Lu for the HOX11 antiserum, Scott Robertson, Georges Lacaud, Mitch Weiss, and Suzanne Kirby for critically reading the manuscript, and Leigh Landskroner for expert assistance with graphics and photography.
Supported in part by Grant No. R01 HL48834A from the National Institutes of Health, Bethesda, MD, and by the National Cancer Institute of Canada, Toronto, Canada, (8398) with funds from the Canadian Cancer Society (to R.G.H.).
Address reprint requests to Gordon Keller, PhD, National Jewish Medical and Research Center, 1400 Jackson St, Denver, CO 80206; e-mail: kellerg@njc.org.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal