Abstract
The most upstream hypersensitive site (HS) of the β-globin locus control region (LCR) in humans (5′ HS 5) and chickens (5′ HS 4) can act as an insulating element in some gain of function assays and may demarcate a β-globin domain. We have mapped the most upstream HSs of the mouse β-globin LCR and sequenced this region. We find that mice have a region homologous to human 5′ HS 5 that is associated with a minor HS. In addition we map a unique HS upstream of 5′ HS 5 and refer to this novel site as mouse 5′ HS 6. We have also generated mice containing a targeted deletion of the region containing 5′ HS 5 and 6. We find that after excision of the selectable marker in vivo, deletion of 5′ HS 5 and 6 has a minimal effect on transcription and does not prevent formation of the remaining LCR HSs. Taken together these findings suggest that the most upstream HSs of the mouse β-globin LCR are not necessary for maintaining the β-globin locus in an active configuration or to protect it from a surrounding repressive chromatin environment.
THE MULTIGENE β-globin locus is subject to tissue-specific and developmental regulation. Transcription of the locus is restricted to erythroid cells, and each gene is expressed during specific developmental stages.1 Initial evidence that regions far upstream of the β-like globin genes are important for regulation of the locus was derived from the analysis of naturally occurring deletions in the human locus that result in the transcriptional silencing of the cis-linked genes.2-4 These regions encompass 5 DNase I hypersensitive sites (5′ HS 1-5) located 6-22 kilobases (kb) 5′ of the human ε-globin gene. When analyzed in human tissues or after passage of human transgenes through the mouse germline, the formation of 5′ HS 1-5 is restricted to erythroid cells; however, 5′ HS 2 and 5′ HS 5 have been observed in nonerythroid cell lines.5-9
Linkage of large restriction fragments containing these 5′ HSs to a human β-globin gene leads to high level, position-independent expression in transgenic mice.10,11 This observation is in contrast with earlier studies in which isolated globin transgenes showed extensive variation in expression with the site of integration. This ability to overcome integration site effects of linked genes defines the locus control region (LCR). Analysis of a naturally occurring deletion of 5′ HS 2-5 and ∼20 kb upstream of this region (Hispanic thalassemia) suggests that the LCR has an important role in regulation of chromatin structure and replication, as well as transcription.3,12,13 The Hispanic thalassemia deletion leads to an alteration of the generalized DNase I sensitivity of the locus, alterations in the timing of replication and origin used, and the transcriptional silencing of the locus.12 13
Extensive analysis has attempted to determine which sequences within the LCR are necessary for activity and how these sequences may interact. Each HS is associated with a several hundred base pair (bp) core region of homology that is highly conserved in evolution, suggesting that these HS cores may be important for LCR function.14 Individual HSs and combinations of HSs have been assayed for the ability to direct high-level expression of linked genes in erythroid cells in transient and stable expression assays in tissue culture and in transgenic mice. The results can be summarized as (1) individually, 5′ HS 2 and 3 have enhancer activity in both transient and stable assays, whereas sites 2, 3, and 4 lead to high-level expression in transgenic mice. 5′ HS 1 and 5 have no demonstrable effects on expression in these assays; (2) combinations or arrays of individual HSs or the whole LCR have more activity than single HSs; and (3) activity of LCR fragments is restricted to erythroid cells.14
Although these studies have yielded valuable information concerning LCR structure and function, these results may be difficult to extrapolate to the intact LCR at its endogenous location due to differences in (1) the spatial organization of the LCR fragments and reporter gene(s) used, (2) whether the construct is integrated or not, (3) integration site position effects, (4) the copy number, (5) the species used, and (6) whether the construct has been passed through the germline. Several groups have attempted to overcome these potential limitations by generating transgenic mice containing low copy number yeast or P1-derived artificial chromosomes. However, it is now clear that this approach is also vulnerable to integration site effects, as well as potential cross species effects that complicate the analysis of regulatory elements.15 16
To circumvent these potential problems, we have analyzed the endogenous mouse β-globin locus by generation of specific LCR mutations using homologous recombination (HR) in ES cells followed by the generation of mutant mice. Previously this approach has been used to generate mice lacking 5′ HS 2 and 5′ HS 3, two sites shown to activate transcription in a variety of systems.17 18 Developmental analysis of these mice showed that the presence of a selectable marker gene in the LCR (an unavoidable consequence of HR) leads to dramatic effects on the transcriptional phenotype. Thus, our strategy includes recombinase-mediated removal of the selectable marker after HR. When mice and embryos lacking either 5′ HS 2 or 3 and the selectable marker were analyzed, minor decreases in expression were detected, but no change in the timing or tissue specificity of expression were noted.
The role that 5′ HS 5, the most 5′ HS defined in the human locus, plays in regulation of the β-globin locus is still unknown. Gain of function assays have not revealed a role for 5′ HS 5 in the direct activation of expression.19-22 Human 5′ HS 5 and the corresponding region of galago have been sequenced and are notable for two regions of homology.23 The first is an ∼500-bp region to which human 5′ HS 5 maps and that contains several conserved phylogenetic blocks and two consensus CACC binding protein (CACC BP) motifs. The second region is ∼200 bp and contains one of the two Drosophila topoisomerase II consensus binding sites that map to the region. Both regions map to a 2.6-kb restriction fragment that has been shown to be a scaffold attachment region (SAR).24 Because this region contains the most 5′ HS of the locus, is an SAR, is conserved in evolution, yet has no direct effect on expression, it has been suggested that this region may be important as a boundary or insulator element and might be important for defining the 5′ extent of a β-globin locus “domain.”8,14,19,21,22,24 25
Two groups have shown that the most 5′ HS of the chicken β-globin locus, chicken 5′ HS 4, marks a transition in chromatin structure of the region in erythroid cells.26,27In contrast to the region upstream of chicken 5′ HS 4, the chick β-globin domain downstream of 5′ HS 4 is uniformly sensitive to DNase I, and histone H4 in this region is highly acetylated. In addition, chicken 5′ HS 4 has been shown to have insulator activity in several stable transformation systems and has more activity when present in high copy number.19,28 In contrast to the experimental evidence for chicken 5′ HS 4, analysis of human 5′ HS 5 has not revealed a consistent biologic role. For example, it has been reported that human 5′ HS 5 has a weak ability to insulate a reporter gene from activation by mouse 5′ HS 2 in a stable transformant assay, and that single copies of human 5′ HS 5 are capable of insulating a reporter gene from the activation effects of human 5′ HS 3 in an expression level assay.19,21Similarly, two groups report that single copies of human 5′ HS 5 flanking reporter genes lead to less variability of expression amongst clones, suggesting that the reporter gene may be insulated from integration site effects.20,22 In contrast, a single copy of human 5′ HS 5 did not insulate a human β-globin gene from activation by 5′ HS 1-4 in single copy transgenic lines.9 Thus, the function of human 5′ HS 5 remains highly speculative.
To determine the function of the most 5′ HS of the murine β-globin locus at its endogenous location, we have mapped and sequenced the region upstream of mouse 5′ HS 4 and find the homologue to human 5′ HS 5 as well as a unique HS further upstream. We have generated mice lacking this region and show that deletion of this region has no significant effect on erythroid or nonerythroid expression of the locus.
MATERIALS AND METHODS
Cloning and sequencing of mouse 5′ HS 5 and 6.
Clones containing mouse 5′ HS 4, 5, and 6 were isolated from a 129-mouse library derived from AK-7 ES cells (gift of A. Imamoto and P. Soriano, Fred Hutchinson Cancer Research Center) using 5′ HS 4 as a probe (−21,221 to −20,509 relative to the Ey cap). A clone with a 15-kb insert was isolated and subcloned using standard methods. Sequencing was done using dye terminators and universal or custom synthesized primers on an ABI 377XL automated sequencer. At a minimum, both strands of all regions were sequenced but for most regions the sequence was determined from at least three overlapping sequence runs (GenBank Accession No. AF071080). Homologies were generated using the computer program L-FASTA (W.R. Pearson, University of Virginia). Pairwise alignments and multiple alignments were generated using the computer programs sim and yama2, respectively.29-31 The generation of percent identity plots (PIPs) was as described.32
DNase I hypersensitive site mapping.
Cells were isolated from the spleens of 129 mice 4 to 6 days after a phenylhydrazine induced hemolytic anemia, at which time at least 50% of the cells are erythroid.26 Hypersensitive site mapping was confirmed using mouse erythroleukemia (MEL) cells. Isolation of nuclei, digestion with DNase I, restriction enzyme digestion, and blotting were done as previously described.13 Mapping was done using probes hybridizing to both upstream and downstream extents of the appropriate restriction fragments. Probes used were restriction enzyme fragments or obtained by polymerase chain reaction (PCR). Probes used map to the following nucleotides with respect to the Ey cap: −33,780 to −33,145 (KH end), −29,645 to −28,991 (LCR probe 1), −28,979 to −27,981 (KP middle), −27,974 to − 27,558 (LCR probe 2), −22,193 to −21,416 (LCR probe 3), −14,684 to −14,118 (LCR probe 4), and −3,468 − 2,868 (Nhe Hpa).
Targeted deletion of mouse 5′ HS 5 and 6.
The 3.5-kb region from a Hpa I site at −28,560 to aEcoRV site at −25,062 relative to the Ey cap was replaced with a PGK-hygro selectable marker flanked by loxP sites, destroying both restriction enzyme sites in the process. The targeting construct had 5.2 kb and 3.7 kb of flanking 5′ and 3′ homology, respectively, as well as a MC HSV-TK gene to allow negative selection for nonhomologous recombinants. Linearized targeting vector (30 to 60 μg) was electroporated into AK7 ES cells (gift of A. Imamoto and P. Soriano) and grown on mitomycin C treated feeders that are hygromycin resistant and produce leukemia inhibitory factor (gift of B. Zambrowicz [Fred Hutchinson Cancer Research Center] and P. Soriano). Cells were selected in 150 μg/mL hygromycin B (Calbiochem, San Diego, CA) and 2 μg/mL gancyclovir. Gancyclovir gave a 2.5- to 5-fold enrichment. Resistant colonies were expanded and screened by Southern blotting, and clones with the correct structure were injected into C57 blastocysts using standard techniques. To faithfully excise the selectable marker, mice carrying the Δ5,6 Hygro mutation were bred to mice containing a cytomegalovirus (CMV)-Cre transgene (TgN[CMV-Cre]1AN).33 All offspring that inherited Cre and a Δ5,6 Hygro allele showed evidence of a site specific recombination event by Southern blotting. To assure that the presence of the Cre protein was not a factor, these mice were bred again and only animals with a correct Δ5,6 ΔHygro structure and lacking the Cre transgene were used for further study. Cre was detected by PCR using the following primers: Cre F ACCTGATGGACATGTTCAGG, and Cre R CTACACCTGCGGTGCTAAC.
Reverse-transcription polymerase chain reaction (RT-PCR) assays.
RNA isolation and RT-PCR was done as described previously except that RT reactions were done at 37°C.17 Gels were quantitated with a PhosphoImager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The value for each band was normalized to background and adjusted for the number of cytosine residues. In addition to wild-type controls, all gels contained at least duplicate wild-type S/D samples from the same RT sample. The D to S ratio from these controls was averaged and adjusted to 1.0. All other D to S ratios from the gel were adjusted by the same factor. Primers were as described.17
RESULTS
Mapping and sequencing of mouse 5′ HS 5 and 6.
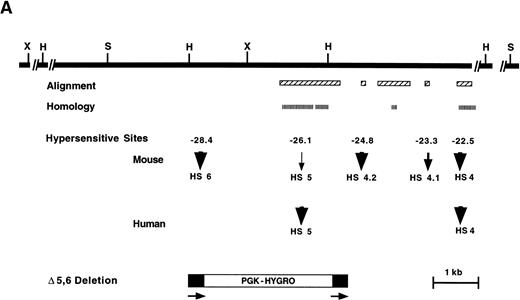
5′ HS 5 is the most 5′ HS described in the human β-globin locus and has been mapped and sequenced.5,8,22 Until now, the most 5′ mapped HS in mouse was 5′ HS 4, which is highly conserved in evolution.34 To determine if the mouse locus contains an additional 5′ HS analogous to human, a nonrepetitive probe immediately 3′ of mouse 5′ HS 4 was used to clone 11.5-kb 5′ to 5′ HS 4 from an ES cell library. Probes downstream of 5′ HS 4 and from the 5′ end of this clone were used to map DNase I HSs in MEL cells and erythroid cells isolated from the spleens of anemic 129 strain mice. In addition to 5′ HS 4, which maps at −22.5 kb relative to the cap site of the Ey gene, four HSs of varying intensity map to approximately −23.3, −24.8, −26.1, and −28.4 kb (Figs 1A and B, 2, and 3). No HSs were detectable from −28.5 to −34.3 kb (Fig 3 and data not shown).
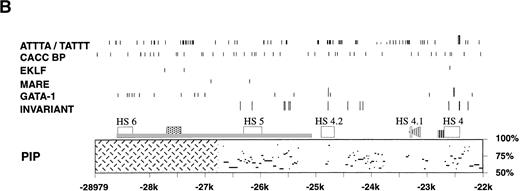
Analysis of the upstream region of the mouse β-globin LCR. (A) Map of the 5′ HS 4, 5 and 6 region. Upper solid black line represents the 5′ HS 4, 5, and 6 region and restriction enzyme sites used for HS mapping. Alignment: boxes with diagonal cross hatch represent regions that can be aligned with the human locus using the program yama2. Homology: boxes with vertical hatch represent regions with >40% identity with human and are a more stringent gauge of homology. HSs: arrows show HSs and their relative intensity. Mouse HSs are placed relative to the mouse map, whereas human HSs are placed relative to the homologous regions of the mouse sequence. The numbers above the arrows are the map positions relative to the Ey cap of the mouse HSs. Bottom diagram represents the targeting construct that deletes HS 5, HS 6 and the region of homology to human HS 5. Arrows and black stripes represent loxP sites that flank the construct. Restriction enzyme sites: S- SphI, H- HpaI, X- XbaI. (B) Sequence features in the 5′ HS 4, 5, and 6 region. Regions that each HS maps to are represented by open rectangles. L1 and B2 repeat elements are represented by a gray rectangle and triangle, respectively. The black bar represents the region deleted by homologous recombination. The striped line represents the thymidine-rich tract. Below the map is a PIP of the mouse-human comparison of the region. The percent identity in each gap-free aligning segment is plotted using coordinates of the mouse sequence with the cap site of the Ey gene being +1. Cross hatching indicates the region for which no human sequence is available for comparison. The lines above the map show sequence features as noted below. In each case short lines mark any occurrence of the specified sequence and tall lines mark sites that are conserved between mouse, human, and galago. Invariant: blocks of at least 7 bp that are invariant in mouse, human, and galago. GATA-1: sites matching the consensus WGATAR. MARE: sites matching the Maf-associated response element consensus TGASTCA. Several basic leucine zipper proteins contain this motif in their consensus binding sequence including NFE-2, AP-1, and LCRF1/Nrf1. EKLF: sites matching the consensus CCNCNCCC. BKLF also has been shown to bind this consensus. CACC BP: sites matching the consensus CACC, a sequence motif common to Krüppel-like Zn finger proteins such as Sp1 and EKLF. ATTTA/TATTT: sites matching the specified sequences.
Analysis of the upstream region of the mouse β-globin LCR. (A) Map of the 5′ HS 4, 5 and 6 region. Upper solid black line represents the 5′ HS 4, 5, and 6 region and restriction enzyme sites used for HS mapping. Alignment: boxes with diagonal cross hatch represent regions that can be aligned with the human locus using the program yama2. Homology: boxes with vertical hatch represent regions with >40% identity with human and are a more stringent gauge of homology. HSs: arrows show HSs and their relative intensity. Mouse HSs are placed relative to the mouse map, whereas human HSs are placed relative to the homologous regions of the mouse sequence. The numbers above the arrows are the map positions relative to the Ey cap of the mouse HSs. Bottom diagram represents the targeting construct that deletes HS 5, HS 6 and the region of homology to human HS 5. Arrows and black stripes represent loxP sites that flank the construct. Restriction enzyme sites: S- SphI, H- HpaI, X- XbaI. (B) Sequence features in the 5′ HS 4, 5, and 6 region. Regions that each HS maps to are represented by open rectangles. L1 and B2 repeat elements are represented by a gray rectangle and triangle, respectively. The black bar represents the region deleted by homologous recombination. The striped line represents the thymidine-rich tract. Below the map is a PIP of the mouse-human comparison of the region. The percent identity in each gap-free aligning segment is plotted using coordinates of the mouse sequence with the cap site of the Ey gene being +1. Cross hatching indicates the region for which no human sequence is available for comparison. The lines above the map show sequence features as noted below. In each case short lines mark any occurrence of the specified sequence and tall lines mark sites that are conserved between mouse, human, and galago. Invariant: blocks of at least 7 bp that are invariant in mouse, human, and galago. GATA-1: sites matching the consensus WGATAR. MARE: sites matching the Maf-associated response element consensus TGASTCA. Several basic leucine zipper proteins contain this motif in their consensus binding sequence including NFE-2, AP-1, and LCRF1/Nrf1. EKLF: sites matching the consensus CCNCNCCC. BKLF also has been shown to bind this consensus. CACC BP: sites matching the consensus CACC, a sequence motif common to Krüppel-like Zn finger proteins such as Sp1 and EKLF. ATTTA/TATTT: sites matching the specified sequences.
Mapping of mouse 5′ HS 4 and 5. Wild-type 129 mice were treated with phenylhydrazine as described, and nuclei were isolated from spleen cells on day 4. A DNase I series was generated, digested with Sph I (S) to completion, Southern blotted, and hybridized with a 5′ probe. The “0” lane shows the expected parent band in untreated nuclei followed by samples having undergone increasing DNase I digestion. Degradation bands map to −22.5 kb, −23.3 kb, −24.8 kb, and −26.1 kb relative to the Ey cap (in parentheses), which we refer to as 5′ HS 4, 4.1, 4.2 and 5, respectively (see text). An additional band mapping to −28.4 kb maps to this region but is outside the range of resolution of this gel. Molecular size standards are marked on the left. The size of the parent band, placement of the probe used (KP middle), and the size of the degradation bands are diagrammed below. Sph I cuts at −14,121 and −30,281 relative to the Ey cap. Mapping of the sites was confirmed by hybridizing with a probe from the 3′ end of the same restriction fragment as well as mapping with additional restriction enzymes (data not shown).
Mapping of mouse 5′ HS 4 and 5. Wild-type 129 mice were treated with phenylhydrazine as described, and nuclei were isolated from spleen cells on day 4. A DNase I series was generated, digested with Sph I (S) to completion, Southern blotted, and hybridized with a 5′ probe. The “0” lane shows the expected parent band in untreated nuclei followed by samples having undergone increasing DNase I digestion. Degradation bands map to −22.5 kb, −23.3 kb, −24.8 kb, and −26.1 kb relative to the Ey cap (in parentheses), which we refer to as 5′ HS 4, 4.1, 4.2 and 5, respectively (see text). An additional band mapping to −28.4 kb maps to this region but is outside the range of resolution of this gel. Molecular size standards are marked on the left. The size of the parent band, placement of the probe used (KP middle), and the size of the degradation bands are diagrammed below. Sph I cuts at −14,121 and −30,281 relative to the Ey cap. Mapping of the sites was confirmed by hybridizing with a probe from the 3′ end of the same restriction fragment as well as mapping with additional restriction enzymes (data not shown).
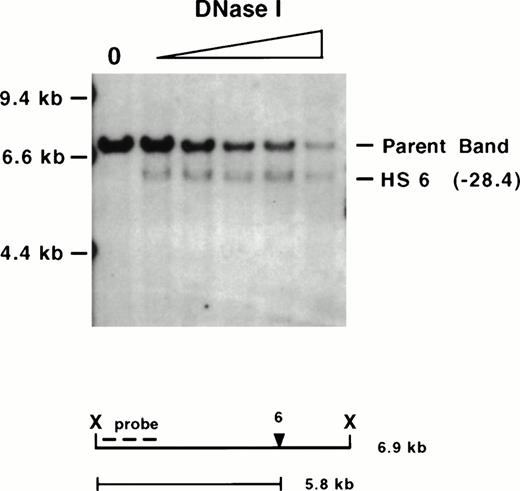
Mapping of mouse 5′ HS 6. The same wild-type 129-mouse erythroid DNase I series displayed in Fig 2 was digested withXba I (X), Southern blotted, and hybridized with a probe on the 5′ end. Molecular size standards are marked on the left. The “0” lane shows the expected parent band in untreated nuclei followed by samples having undergone increasing DNase I digestion. The expected 6.9-kb parent band and a degradation band that maps to −28.4 kb (in parentheses), which we refer to as 5′ HS 6 (see text), are observed. The size of the parent band, placement of the probe used (KH end), and the size of the degradation band are diagrammed below.Xba I cuts at −27,346 and −34,263 relative to the Ey cap. Mapping of the site was confirmed by hybridizing with a probe from the 3′ end of the same restriction fragment as well as mapping with additional restriction enzymes (data not shown).
Mapping of mouse 5′ HS 6. The same wild-type 129-mouse erythroid DNase I series displayed in Fig 2 was digested withXba I (X), Southern blotted, and hybridized with a probe on the 5′ end. Molecular size standards are marked on the left. The “0” lane shows the expected parent band in untreated nuclei followed by samples having undergone increasing DNase I digestion. The expected 6.9-kb parent band and a degradation band that maps to −28.4 kb (in parentheses), which we refer to as 5′ HS 6 (see text), are observed. The size of the parent band, placement of the probe used (KH end), and the size of the degradation band are diagrammed below.Xba I cuts at −27,346 and −34,263 relative to the Ey cap. Mapping of the site was confirmed by hybridizing with a probe from the 3′ end of the same restriction fragment as well as mapping with additional restriction enzymes (data not shown).
To identify and compare the murine sequence with human and galago, a segment of ∼7 kb upstream of mouse 5′ HS 4 was sequenced. Sequence was analyzed to identify (1) regions of homology (regions with greater than 40% identity to the human sequence), (2) regions of alignment (regions with lesser homology that nonetheless could be aligned with human and galago sequences under less stringent conditions), (3) invariant blocks (blocks of at least 7 bp that are invariant in mouse, human, and galago), (4) putative transcription factor binding sites present in other LCR HS regions, and (5) motifs that are repeated elsewhere in the LCR (Fig 1A and B).
Four regions of the mouse sequence that align with human and galago can be distinguished, and notably, HSs map to three of these. We have compared sequence and HS patterns in the mouse with human and galago to name these HSs. The longest region of alignment is 1.4 kb, is centered at −26.0 kb, and contains six invariant blocks. The least intense HS, present at the limit of detection, maps to the center of this region. This region has ∼60% identity to the region that human 5′ HS 5 has been mapped to (Fig 1A and B). Despite this being a minor HS in the mouse, its coincidence with the region of homology with human 5′ HS 5 identifies this site as mouse 5′ HS 5. A short 59-bp region at −23.3 kb maps near a relatively weak HS. Within the precision of the HS mapping, this block of homology falls within the mouse HS. No equivalent HS is seen in the human β-globin LCR. Because this is the first HS upstream of 5′ HS 4, and 5′ HS 5 has been defined, we call this site 5′ HS 4.1. This nomenclature is advantageous in that HSs that are conserved evolutionarily maintain identical numbering regardless of whether additional sites are present in some species. A short 78-bp region at −24.8 kb has a string of potential GATA-1 binding sites; this string is shorter in mouse compared with human and galago. One GATA site is conserved in all three species and is part of an invariant 10-bp block (Fig 1B). A moderately intense HS maps to this region. No comparable HS has been mapped to the homologous region in humans. Because this is the second HS upstream of 5′ HS 4 that is present in mouse but not human, we call this 5′ HS 4.2. Finally, a long 709-bp region, centered at −24.1 kb, contains three invariant blocks and a 130-bp subregion with 68% identity with human (Fig 1A and B). Of note, no HS maps within this region.
The region containing mouse 5′ HS 5 differs from the homologous region of human 5′ HS 5 in several ways in addition to those noted above. The consensus sequences of binding sites for Krüppel-like Zn finger proteins such as Sp1 and EKLF can contain a CACC motif; thus, they have been designated CACC BPs (CACC binding proteins). This motif is conserved in several LCR HSs.14,25,35 Human 5′ HS 5 is centered on a pair of CACC motifs that are part of a 20-bp dyad that is conserved in galago.14 These CACC BP sites are not present in mouse and may explain the disparity in the intensity of the HSs amongst species. In addition, several HSs from β-globin LCRs contain consensus sequences for Maf recognition elements (MAREs).14,36,37Several basic leucine zipper-containing proteins, including NFE-2 and LCRF1/Nfr1, recognize this core consensus (TGASTCA), and it has been suggested that the homo- and heterodimeric proteins that recognize MAREs play an important regulatory role in differentiation.36 In addition to the invariant blocks noted above, the region encompassing mouse 5′ HS 5 maps contains one of only two MAREs in this 7-kb region (Fig 1B). This motif is not conserved in the primate sequences. Of note, the second MARE is at −26.9 kb, just 5′ to the beginning of the available sequence from human and galago. This MARE is within a 157-bp region of mouse sequence that also contains two CACC motifs and a GATA site. Finally, a 2.6-kb HindIII fragment that contains human 5′ HS 5 is notable for being an SAR, a region that has been associated with topoisomerase II binding sites.22,24 In addition topoisomerase II cleavage sites in the chicken β-globin locus map to DNase I HSs.26 The human 5′ HS 5 region contains two blocks that contain single-base differences from the Drosophila topoisomerase II consensus sequence,22 but these are not conserved in the mouse.
The mouse β-globin LCR contains an additional HS upstream of 5′ HS 5.
An additional major HS maps 2.3 kb 5′ to mouse 5′ HS 5 at −28.4 kb (Fig 3). No additional HSs are detectable in the 5.8 kb upstream of this site in erythroid tissues. The absence of available DNA sequence from homologous region of other species precludes an assessment of sequence conservation. We refer to this HS as mouse 5′ HS 6. The region encompassing 5′ HS 6 contains a high density of potential binding sites for the erythroid transcription factor GATA-1, (Fig 1B), consistent with several other β-globin LCR HSs.
The sequence upstream of 5′ HS 4 was analyzed for the presence of other potential transcription factor binding sites and sequence motifs noted to occur in β-globin LCRs of several species.14,25,35 As above, in contrast to the rest of the LCR, the region upstream of mouse 5′ HS 4 shows little similarity with the primate sequences. EKLF binding has been shown to be important for the expression of adult β-globin genes, and it has been suggested that it may mediate LCR-promoter interactions.38,39 Three potential EKLF sites are in this segment of the mouse sequence, one in HS 4 and two in the region with no sequence from other species (Fig1B). Several CACC motifs are in the mouse sequence, but none are conserved in human and galago (Fig 1B). Previously it was noted that the motifs ATTTA and TATTT are conserved in several regions of the LCR,14 as exemplified by the HS 4 region, but despite the presence of many such sites in the mouse 5′ HS 4 through 5′ HS 5 region, none are conserved (Fig 1B). Finally, a thymidine-rich track, with thymidine residues comprising 106 of 132 nucleotides, is immediately upstream of HS 4 but is not conserved in the primate sequences.
Targeted deletion of mouse 5′ HS 5 and 6.
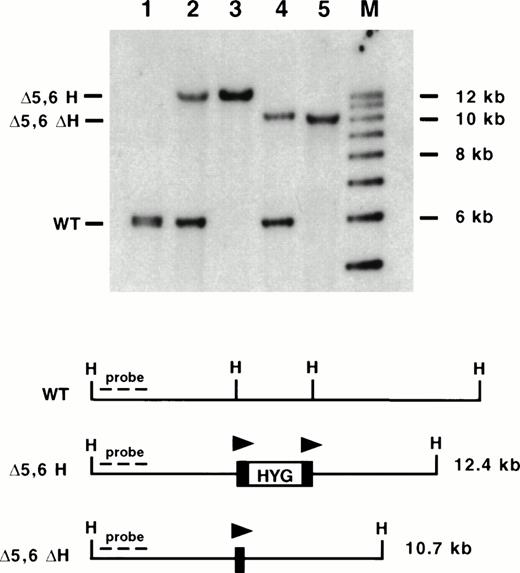
To delete the most 5′ HSs, a targeting vector was designed that would delete a 3.5-kb region containing mouse 5′ HS 6, as well as mouse 5′ HS 5 and the region of homology with human 5′ HS 5 (Fig 1A). Previously we determined that the presence of a selectable marker within the LCR dramatically affects expression of the linked globin genes.17,18,40,41 Thus, we used a selectable marker (PGK-hygro) flanked by loxP sites, the recognition sequence for the Cre recombinase. We have previously shown the utility of using site-specific recombinases to excise selectable markers after HR events in several systems.17,18,40 42 In addition, our targeting construct contained a HSV-TK gene for negative selection. AK7 ES cells were electroporated, and clones were grown in hygromycin and gancyclovir for positive and negative selection respectively. Five hundred and ninety-one colonies were screened by Southern blot analysis. Six clones had the correct structure spanning both 5′ and 3′ junctions of the targeting construct and having a single copy of PGK-hygro present for a targeting frequency of 1%. Three clones were chosen for blastocyst injection, and all showed germline transmission (Fig 4, lanes 2 and 3).
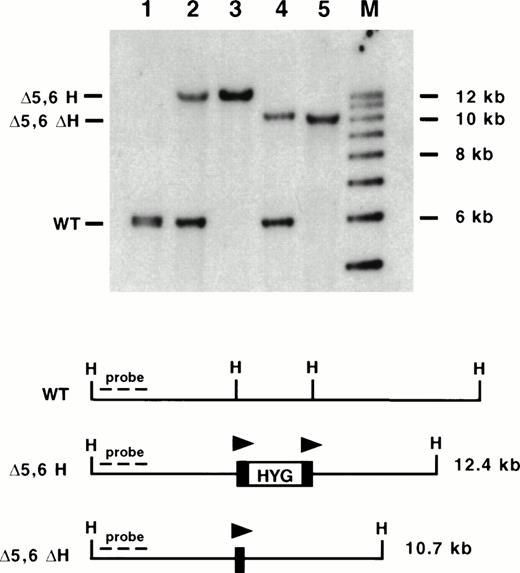
Southern blot analysis of mice containing targeted deletions of 5′ HS 5 and 6. DNA was isolated from mouse tails and digested with HpaI (H), blotted, and hybridized to an upstream probe (KH end). Lane 1: wild-type (WT) mouse; lane 2: ▵5,6 H/WT mouse; lane 3: ▵5,6 H/▵5,6 H mouse; lane 4: ▵5,6 ▵H/WT mouse; lane 5: ▵5,6 ▵H/▵5,6 ▵H mouse; M: molecular size standards. The targeted mutation replaced a 3.5-kb Hpa I to EcoRV fragment with a 2.0-kb PGK-hygro marker, destroying both restriction enzyme sites and leading to a larger Hpa I restriction fragment (▵5,6 H; lanes 2 and 3). Excision of PGK-hygro decreases the restriction fragment size by 2.0 kb (▵5,6 ▵H; lanes 4 and 5). Triangles represent lox P sites.
Southern blot analysis of mice containing targeted deletions of 5′ HS 5 and 6. DNA was isolated from mouse tails and digested with HpaI (H), blotted, and hybridized to an upstream probe (KH end). Lane 1: wild-type (WT) mouse; lane 2: ▵5,6 H/WT mouse; lane 3: ▵5,6 H/▵5,6 H mouse; lane 4: ▵5,6 ▵H/WT mouse; lane 5: ▵5,6 ▵H/▵5,6 ▵H mouse; M: molecular size standards. The targeted mutation replaced a 3.5-kb Hpa I to EcoRV fragment with a 2.0-kb PGK-hygro marker, destroying both restriction enzyme sites and leading to a larger Hpa I restriction fragment (▵5,6 H; lanes 2 and 3). Excision of PGK-hygro decreases the restriction fragment size by 2.0 kb (▵5,6 ▵H; lanes 4 and 5). Triangles represent lox P sites.
In vivo excision of the selectable marker.
To analyze mice with 5′ HS 5 and 6 deleted and free of any effects of the selectable marker, the mutant mice containing the marker (Δ5,6H) were bred with mice containing a Cre recombinase transgene driven by a CMV promoter (TgN[CMV-Cre]1AN).33 Southern blot analysis of tail DNA revealed that all seven mice that inherited the Cre transgene showed an accurate Cre-mediated excision of the selectable marker (Δ5,6ΔH; data not shown). Two of the seven mice contained a population of cells in which the selectable marker had not been excised; thus, although efficient, excision did not take place in every cell in the first generation. The degree of chimerism in tail may not reflect chimerism in the hematopoietic system, and Southern blotting is not sufficiently sensitive to detect a small population of marker containing cells that could complicate phenotypic analysis. To generate mice homogeneous for the mutation, these first generation Δ5,6ΔH mice were bred and Δ5,6ΔH pups lacking the Cre transgene were isolated (Fig 4, lanes 4 and 5). As the Cre recombinase is not present, the excision event must have occurred in the parent; thus, the gamete must have carried the Δ5,6ΔH mutation, and the resultant mouse must be homogeneous. In addition, this strategy avoids the theoretical concern of Cre protein binding to a loxP site and altering the phenotype. Mice homozygous for the Δ5,6ΔH mutation were generated and revealed no decreased viability.
Erythroid tissue from these homozygotes was used for DNase I mapping and revealed that although 5′ HS 5 and 6 do not form, all other sites form normally (Fig 5 and data not shown). Thus, we have deleted mouse 5′ HS 5 and 6, no new HSs form in their place, and this deletion does not result in a change in the chromatin conformation of the locus that extinguishes the ability of other HSs to form.
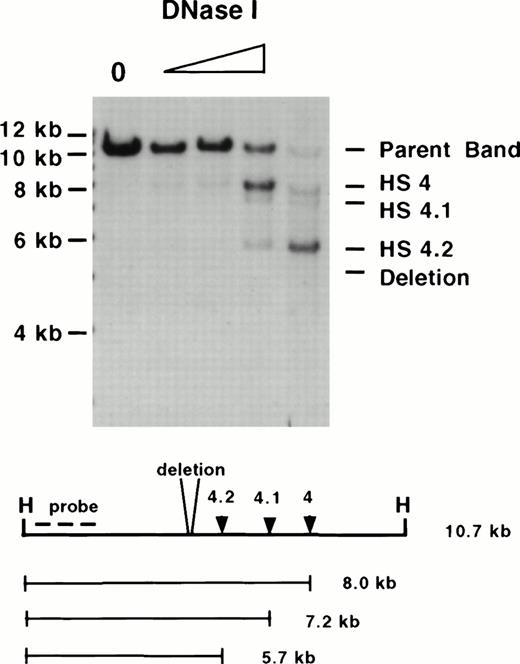
Mapping of HSs in homozygous ▵5,6 ▵H/▵5,6 ▵H mice. A DNase I series of erythroid tissue from a homozygous mutant mouse was digested with Hpa I (H) and hybridized with a 5′ probe (the same enzyme and probe combination as used in Fig4). Although 5′ HS 4, 4.1, and 4.2 form, no HS forms at the site of the deletion. Molecular size standards are marked on the left. The size of the parent band, placement of the probe used, and the size of the degradation bands are diagrammed below.
Mapping of HSs in homozygous ▵5,6 ▵H/▵5,6 ▵H mice. A DNase I series of erythroid tissue from a homozygous mutant mouse was digested with Hpa I (H) and hybridized with a 5′ probe (the same enzyme and probe combination as used in Fig4). Although 5′ HS 4, 4.1, and 4.2 form, no HS forms at the site of the deletion. Molecular size standards are marked on the left. The size of the parent band, placement of the probe used, and the size of the degradation bands are diagrammed below.
Deletion of 5′ HS 5 and 6 does not have a major effect on the level or pattern of expression.
To determine if the deletion of mouse 5′ HS 5 and 6 has an effect on the level, timing, or tissue specificity of β-globin gene expression, mice and embryos were analyzed by internally controlled quantitative RT-PCR assays. These assays exploit RFLPs between the two major β-globin alleles in mice, HbbS and HbbD. Previously we showed that primer pairs specific for the Ey, βh1, and the adult β-globin genes coamplify mRNA from HbbS and HbbD homologues equally and that cleavage at polymorphic restriction enzyme sites result in the quantitative determination of expression from one allele compared to the other.17 Targeted mutations were made on a HbbD allele in ES cells derived from 129 mice. Mutant mice were bred to mice carrying a wild-type HbbS allele allowing comparison of expression from the mutant HbbD allele to that of the internal control HbbS allele in these heterozygotic animals. These assays accurately quantitate ratios of D-specific to S-specific transcription from 0.1 to 0.9.17For developmental analysis all three sets of primers were used to examine RNA from yolk sac from day postconception (dpc) 10.5 embryos, liver from dpc 15.5 fetuses, and peripheral blood in adults.
Analysis of expression in heterozygotes carrying the Δ5,6H mutation is summarized in Table 1. A moderate decrease was observed in βh1 expression, with a lesser effect on adult β-globin expression, but no effect on Ey or fetal expression of β-globin was detectable. Analysis of mice lacking 5′ HS 5 and 6 without the selectable marker (Δ5,6ΔH) is shown in Fig 6 and is quantitated in Table 1. Minor decreases in embryonic expression of βh1 and fetal expression of β-globin expression were noted, whereas embryonic expression of Ey and adult expression of β-globin expression were unchanged. No abnormalities in globin gene switching were detectable (Fig 6). In addition, no globin gene expression was observed in day 3.5 blastocysts; in day 6.5 or 7.5 yolk sacs; or in adult thymus, brain, muscle, gut, testes, or kidney (data not shown).
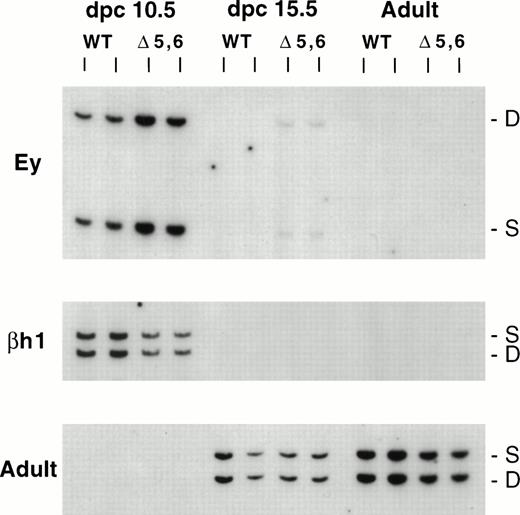
Expression of β-like globin genes in ▵5,6 ▵H/WT heterozygous mice. The HbbD allele (D) carries the targeted mutation, whereas the HbbS allele (S) is wild-type. RNA was analyzed from dpc 10.5 yolk sac, dpc 15.5 fetal liver, and adult peripheral blood using RT-PCR assays for Ey, bh1, and adult primers (top, middle, and bottom panels). WT, wildtype D/S animals; ▵5,6, mutant ▵5,6 ▵H/S heterozygous mice. S and D mark the RT-PCR products from the S and D alleles, respectively.
Expression of β-like globin genes in ▵5,6 ▵H/WT heterozygous mice. The HbbD allele (D) carries the targeted mutation, whereas the HbbS allele (S) is wild-type. RNA was analyzed from dpc 10.5 yolk sac, dpc 15.5 fetal liver, and adult peripheral blood using RT-PCR assays for Ey, bh1, and adult primers (top, middle, and bottom panels). WT, wildtype D/S animals; ▵5,6, mutant ▵5,6 ▵H/S heterozygous mice. S and D mark the RT-PCR products from the S and D alleles, respectively.
DISCUSSION
The mouse β-globin LCR has a complex pattern of HSs upstream of 5′ HS 4.
The 12-kb region upstream of 5′ HS 4 has four DNase HSs in mouse, whereas only one, 5′ HS 5, has been described in human to date. In the region where sequences are available from human and galago, several aligning segments are found. The largest segment has an average of ∼60% identity with the primate sequences and spans the area to which human 5′ HS 5 maps. In contrast to 5′ HS 1,2,3, and 4, which are major HSs in both human and mouse, 5′ HS 5 is a major HS in human but is at the limit of detection in the mouse. The human 5′ HS 5 contains two CACC motifs in a region of substantial dyad symmetry that is not conserved in the mouse. Assuming that these sites are occupied in the human, their absence in the mouse could explain the difference in HS intensity. If this is the case we would expect that when mapped, galago 5′ HS 5 would be a relatively intense site as these CACC BP sequences are conserved in the galago.14 Similarly, mouse 5′ HS 4.2 maps to a region with multiple consensus GATA-1 sites. It also contains a GGGCAG motif, which is a single mismatch from the Sp1 consensus binding site and has been suggested to bind porcine Sp1.43 This sequence is not conserved in the human, which may explain why no HS is observed.
Mouse 5′ HS 6 is currently the most 5′ HS mapped to the β-globin LCR, with no further HSs present for at least 5.8 kb upstream. Previous mapping in the human would have detected 5′ HS 6 if the spatial organization of 5′ HS 6 and 5 was conserved (2.3 kb apart in the mouse).5,8 Thus, either this HS is unique to the mouse, or it is present in the human locus, but spacing is not maintained. This region is striking in the density of potential GATA-1 binding sites, six in total with four within 120 bp. β-Globin LCR core regions from several species contain adjacent and opposing GATA-1 sites as well as NFE-2 and AP-1 sites that are bound in vitro and are thought to play a role in the regions’ enhancer activity and ability to lead to position-independent expression.14,25 35Although the opposing GATA-1 sites in 5′ HS 6 are separated by ∼30 bp and no AP-1/NFE-2 consensus site is present, it will be interesting to determine if 5′ HS 6 contains erythroid-specific enhancer activity.
Previously a 2.6-kb region containing human 5′ HS 5 was found to act as an SAR and to have two potential Drosophila topoisomerase II consensus binding sites, leading to suggestions that this region demarcated the extent of the β-globin domain.22 Although these topoisomerase II sites are not conserved in mouse 5′ HS 5, two such sites map to mouse 5′ HS 6; however, the significance of this observation is not known. In addition, a region upstream of mouse 5′ HS 6 is AT rich and contains multiple potential topoisomerase II sites, raising the possibility that these regions may act as SARs in the mouse (M. Bulger and M.A. Bender, unpublished observations, November 1997).
Deletion of the most upstream HSs does not lead to repression of the locus and does not have major effects on transcription.
Deletion of the most distal HSs of the mouse β-globin LCR has no substantial effect on globin gene expression. Several studies previously suggested that human 5′ HS 5 can act as an insulator in the appropriate context, while in other contexts no effect was seen.9,19-22 The studies presented here address the effects of deleting the most distal HSs of the murine β-globin LCR and the homologous sequences to human 5′ HS 5. If insulation of the locus were the primary function of this element, deletion of 5′ HS 5 in human or 5′ HS 5 and 6 in mouse would be expected to lead to an alteration of the chromatin environment of the locus. Manifestations of this could include repression of globin gene expression in erythroid cells, premature activation of the globin locus in development, and/or ectopic expression in nonerythroid tissues. We have obtained no evidence that deletion of 5′ HS 5 and 6 leads to the shutdown of the locus in erythroid cells. Expression is near normal and the remaining LCR HSs form normally. In addition, when mouse 5′ HS 1-6 are deleted in ES cells, the chromatin of the β-globin locus remains resistant to digestion with DNase I; however, after transfer into a human erythroid background the locus becomes sensitive, suggesting that 5′ HS 1-6 are not required to insulate the locus from any repressive upstream influences in an erythroid environment.44 Furthermore, we have obtained no evidence that deletion of 5′ HS 5 and 6 leads to the inappropriate activation of the locus. No ectopic expression is detectable in the nonerythroid hematopoietic tissues or nonhematopoietic tissues we analyzed. Although this is consistent with a lack of activation of the locus in nonerythroid cells, we cannot exclude the possibility that the locus is in an active conformation, but the absence of an appropriate erythroid cellular milieu prevents detectable transcription. Because the regions flanking the β-globin locus have not been characterized, we can not fully address whether deletion of 5′ HS 5 and 6 leads to inappropriate repression or silencing of upstream regions in nonerythroid cells or activation in erythroid cells. We can state that any such effects, if present, do not lead to any gross phenotype or decrease in viability. Thus, deletion of the most distal HS in the mouse, 5′ HS 6 and 5′ HS 5, has not provided any evidence that this region functions as a boundary element in vivo.
Recently, it has been shown that sequences from the human β-globin LCR (Garrick et al, submitted) and the HS-40 enhancer from the human α-globin locus can decrease position effect variegation in transgenic mice.45 In addition, inactivation of the transgenes increases with age of the animal.46 If 5′ HS 5-6 were to act as an insulator at its endogenous location, the region upstream of the locus might exert its effect in a stochastic and time-dependent manner, possibly revealing a transcriptional phenotype with increasing age. In the experiments reported here, only young adult mice were analyzed. Thus, it will be interesting to analyze β-like globin gene expression in these mice as they age.
The most upstream extent of the β-globin LCR differs between the mouse and human.
The region at the 5′ end of the mouse β-globin LCR has a distinctive (complex) structure that contrasts with the human LCR. Sequence comparisons and DNase I HS mapping reveal that the mouse does have a homologue of human 5′ HS 5. Although in the human this lone site upstream of 5′ HS 4 is a major site, in the mouse it is barely detectable and is overshadowed by three more intense sites (Fig1B). In addition, in contrast to the high degree of conserved motifs seen between mouse and human in the region of 5′ HS 1 through 4, the region upstream of mouse 5′ HS 4 lacks this degree of conservation of motifs for MAREs, GATA-1, CACC BPs, TATTT, and ATTTA and has a thymidine-rich tract, suggesting that its organization may be different. This suggests that the role that the most 5′ region of the LCR plays in regulating the transcription, chromatin structure, and replication of the β-globin locus may vary amongst species.
The selectable marker has a minor effect on gene level of expression.
Previously we showed that the presence of a selectable marker within the LCR had dramatic effects on transcription in vitro and in vivo, regardless of whether it is associated with a deletion within the locus17,18,40,41; thus, it is essential to remove markers to assure accurate assessment of the transcriptional phenotype with targeted deletions.42 Although several strategies have been used to accomplish this, we have chosen to flank selectable markers with a recognition sequence for the Cre site-specific recombinase. While the marker can be excised by transient expression of the Cre recombinase in vitro, this requires additional time in culture, thus reducing the potential for germline transmission of the mutation. By using mice expressing a CMV-Cre transgene, we and others find efficient but not complete excision of markers in F1 Cre containing pups,33 similar to Cre transgenes used previously.47 48 This lack of complete excision shows the necessity of analyzing F2 animals and embryos that lack the Cre transgene to assure complete excision while also avoiding the potential for the Cre enzyme to bind a residual loxP site and affect expression in vivo.
Comparison of several strains of mutant mice with selectable markers inserted into the β-globin LCR reveals dramatic differences in the transcriptional phenotypes. Targeted replacement of 5′ HS 2 with a PGK-neo gene in the same transcriptional orientation as the globin genes leads to homozygous lethality, whereas replacement of 5′ HS 3 in the opposite orientation reduces viability by 50%.17,18 In contrast, replacement of 5′ HS 5 and 6 with a PGK-hygro gene in the same transcriptional orientation as the globin genes did not affect viability and had relatively minor effects on transcription, even when normalizing for the effect of the deletion without the marker present. There are several factors that may contribute to the decreased effect of the marker gene seen here including (1) the marker is further from the endogenous genes, (2) the marker does not lie between the HSs that affect transcription most significantly (5′ HS 2, 3, and 4) and the endogenous genes, (3) the marker does not disrupt the organization of 5′ HS 2, 3, and 4, (4) the transcriptional orientation of the marker gene, and (5) the specific selectable marker promoter and gene used. These factors could affect LCR activation of globin gene expression regardless of whether tracking, looping, “holocomplex,” or other models are correct.49 Analysis of mice with PGK-neo replacing 5′ HS 1 and 4 and mice with the identical marker in both orientations at the same insertion site will be useful in evaluating the contribution of these factors and in understanding how the LCR influences gene expression.
ACKNOWLEDGMENT
We are grateful to Webb Miller for generating sequence analysis graphics; A. Imamoto, B. Zambrowicz, and P. Soriano for sharing cell lines and ES cell advice; A. Nagy for sharing TgN(CMV-Cre)1AN mice; and the FHCRC Biotechnology Center for oligonucleotide synthesis and automated sequencing. We thank M. Bulger, D. Cimbora, and H. Blanton for critical reading of the manuscript.
Supported by the National Institutes of Health (NIH), Grant No. P30 HD28834, through the University of Washington Child Health Research Center (M.A.B.), NIH Grant Nos. DK52854 and DK44746 (M.G.), National Library of Medicine grants RO1LM05110 and RO1LM05773 (R.H.), a Burroughs-Wellcome Fund Career Development Award (S.F.), and a core grant to the FHCRC Biotechnology Center. M.A.B. is a Howard Hughes Medical Institute Physician Postdoctoral Fellow.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Mark Groudine, MD, PhD, Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave North–Mailstop A3-025, Seattle, WA 98109-1024.