Abstract
Using an RNase protection assay, globin mRNA species expressed in clones derived from Ba/F3 and B6SUtA cells transfected with the erythropoietin receptor (EpoR) and selected with erythropoietin (Epo) were compared with globin mRNA species induced in corresponding parental cells by sodium butyrate (SB) and trichostatin A (TSA). βMajor/βminor- and -1/-2–globin mRNAs were the major species, with trace amounts of ɛ-globin mRNA, formed in Epo-stimulated EpoR+ Ba/F3 clones, whereas SB and TSA allowed expression of all species of globin mRNAs, ie, ɛ, βh1, βmajor/βminor, ζ, and -1/-2, in parental Ba/F3 cells. In contrast, ɛ- and -1/-2–globin mRNAs were the major species present in Epo-stimulated EpoR+ B6SUtA clones, whereas SB and TSA activated ɛ-, βh1-, βS/βT-, and -1/-2–globin genes in parental B6SUtA cells; ζ-globin mRNA was not detected in SB- and TSA-treated B6SUtA cells. Because TSA is a specific inhibitor of histone deacetylase, the mimicry of action exhibited by SB and TSA suggests that the effects of SB are mediated through its ability to inhibit histone deacetylase and that histone deacetylase is an integral part of the repression of globin genes in these interleukin-3–dependent cells. Efficient coinduction of embryonic and adult types of globin mRNA in bone marrow cell lines derived from adult mice indicates that adult hematopoietic precursors possess an embryonic nature. These cell lines are useful models to study the mechanism(s) of developmental globin gene switching.
MURINE INTERLEUKIN-3 (IL-3)–dependent bone marrow–derived cell lines are considered to be immature because of their dependency on IL-3 and to be “untransformed” because of their dependency on a cytokine for survival and growth.1These cells differ from cell lines established from hematopoietic malignancies, which proliferate in the absence of cytokines and whose movement along the pathway to maturity is impaired by alterations in regulatory mechanisms. Two murine IL-3–dependent cell lines, Ba/F32,3 and B6SUtA,4 acquire responsiveness to the mitogenic and differentiation-inducing properties of erythropoietin (Epo) on introduction of the Epo receptor (EpoR) by transfection. Epo-induced differentiation is shown by accumulation of βmajor-globin mRNA in EpoR+ Ba/F3 cells2,3 and by accumulation of hemoglobin in B6SUtA cells.4 Whereas some leukemia cell lines have the ability to terminally differentiate in vitro in response to a variety of nonphysiological chemical agents,5,6 the capacity of factor-dependent cells to respond to chemical inducers of differentiation has not been well studied. We have been interested in establishing model systems in which signal molecules involved in the differentiation pathways used by both Epo and chemical inducers can be dissected. To this end, we have evaluated a wide range of potential chemical inducers in B6SUtA cells using benzidine positivity to measure hemoglobin accumulation and have identified sodium butyrate, diazepam, and hemin as positive initiators of maturation.4 In this report, we have analyzed in depth the globin mRNA species expressed in Epo-stimulated EpoR+ clones of Ba/F3 and B6SUtA cells and have compared these with the globin mRNA species induced in parental cells by chemical inducers. Furthermore, to determine whether sodium butyrate (SB) exerts its action through an ability to inhibit histone deacetylase, comparative studies were conducted with trichostatin A (TSA), a specific inhibitor of histone deacetylase.7 These investigations collectively revealed unique features characteristic of these IL-3–dependent cell lines.
The murine diffuse haplotype β-globin locus (in strains such as BALB/c and DBA/2) contains five genes, including εy, βh0, βh1, βmajor, and βminor, whereas the murine single haplotype β-globin locus (in strains such as C57BL/6) contains εy, βh1, βS, and βT.8 The majority of the βh0 gene is deleted in the single haplotype β-globin cluster.9 The βh0 gene in the diffuse haplotype has been proposed to be a pseudogene.9 The murine α-globin locus contains three functional genes (ζ, α-1, and α-2).10,11 Whereas the human exhibits embryonal to fetal and fetal to adult hemoglobin switches, the mouse shows only one switch, from embryonic to adult hemoglobin.12 In the mouse, the embryonic to adult switch takes place at 10.5 days of gestation. The primitive nucleated erythroid cells in the yolk sac contain two embryonic β-globins, βh1 and εy, and α-globins, ζ and α-1/α-2. The definitive erythrocytes from fetal liver, spleen, and bone marrow contain adult-type β-globins-βmajor/βminor(βS/βT), although there is a report which shows that εy-globin mRNA is produced not only in the yolk sac but also in the fetal liver during midgestation, suggesting that εy gene expression is analogous to that of the fetal globin genes in the human.13 The α-1/α-2 genes are expressed throughout development and are the only genes expressed during adult life.12
The mechanism by which the developmental switching of globin genes is executed and the mechanism by which the induction of globin genes by the physiological cytokine Epo, as well as by chemical inducers, occurs is far from complete. One of the obstacles to the understanding of these phenomena is the limitations of in vitro model systems for molecular analysis. Thus, the availability of tissue culture cell lines in which expression of both embryonic and adult-type globins becomes permissive on exposure to inhibitors of histone deacetylase would provide a first step toward the identification of factors involved in repression and derepression of globin genes. The results of our previous2-4 and current studies point to the utility of IL-3–dependent bone marrow cell lines as model systems for (1) the dissection of growth versus differentiation signals transduced by Epo, and (2) the dissection of the differentiation pathways used by the physiological cytokine Epo and inhibitors of histone deacetylase.
MATERIALS AND METHODS
Cells.
Murine IL-3–dependent Ba/F3 cells were obtained from Dr H. F. Lodish of the Whitehead Institute for Biomedical Research (Cambridge, MA). They were established from the bone marrow of an adult BALB/c mouse (β-globin locus diffuse haplotype),9 and were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 10% conditioned medium from WEHI-3B murine myelomonocytic cells (WEHI-CM) as a source of IL-3.14 Clones of Ba/F3 cells expressing the EpoR were obtained by transfection of the eukaryotic expression plasmid for the EpoR, p75/15-EpoR,4 which contained the EpoR cDNA under the control of the human metallothionein IIA promoter (clones Ba-ER-1 to -4). These clones were developed in 0.8% methylcellulose medium containing 15% FBS and 1 U/mL of Epo (human recombinant Epo generously donated by the R. W. Johnson Pharmaceutical Research Institute, Raritan, NJ). The E-7 clone was obtained from Ba/F3 cells by transfection of p75/15-EpoR, followed by selection with 0.8 mg/mL of G418 in medium containing 0.8% methylcellulose, 15% FBS, and 10% WEHI-CM. The E-7 clone shows Epo-dependent accumulation of βmaj/min globin mRNA on exposure to the cytokine.2
Murine IL-3–dependent B6SUtA cells,15 kindly provided by Dr J. S. Greenberger of the University of Massachusetts Medical Center (Worcester, MA), were adapted to replicate in RPMI 1640 medium containing 10% FBS and 10% WEHI-CM in this laboratory.4 B6SUtA cells were established from the bone marrow of an adult B6.S mouse15 (B6.S has the genetic background of C57BL/6 mice which belong to the β-locus single haplotype).8 9 Clones of B6SUtA cells expressing the EpoR (B6-ER-1 to -4) were obtained by transfection of p75/15-EpoR, followed by selection with 1 U/mL of Epo and 0.7 mg/mL of G418 in medium containing 0.8% methylcellulose and 15% FBS. Because of the overgrowth of nontransfected and/or transiently transfected cells, which interfered with the formation of discrete colonies of stable EpoR transfected cells when clones were selected with Epo as the sole selecting agent, it was necessary to select EpoR+B6SUtA clones in the presence of both Epo and G418.
Friend murine erythroleukemia (MEL) 745-PC4 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 15% FBS. MEL745-PC4 was derived from a DBA/2 mouse which belongs to the β-globin locus diffuse haplotype.16 Eleven and one-half–day-old DBA/2 mouse embryonal blood was prepared as described by Kovach et al.17
MEL-PC4 and Ba/F3 cells at an initial cell density of 4 × 104 cells/mL, and B6SUtA cells at an initial density of 6 × 104 cells/mL were treated with various chemical agents for 3 days before RNA extraction. The percent of inhibition of cell growth was calculated according to the following equation: [log(final cell density of the control culture) − log(final cell density of the treated culture)]/[log(final cell density of the control culture) − log(initial cell density of the control culture)] × 100.
RNA extraction.
Total cellular RNA was isolated by a guanidinium/CsCl gradient sedimentation procedure,18 or extracted using TRIZOL reagent (GIBCO-BRL, Rockville, MD) from 1 to 4 × 107 cells according to the instructions of the manufacturer.
Northern hybridization.
Total cellular RNA (10 μg) was separated by electrophoresis, transferred to a nitrocellulose membrane, and hybridized with cDNA probes as described previously.2 To probe for different types of globin mRNA, duplicate blots were made rather than stripping and reprobing the same blot to avoid incomplete stripping. The nitrocellulose filters were washed at 55°C in 0.2 × SSC (0.15 mol/L sodium chloride/0.015 mol/L sodium citrate, pH 7.0). To obtain a murine βmajor-globin cDNA probe, a 457-bp fragment containing the entire murine βmajor-globin cDNA19 was amplified by reverse transcription polymerase chain reaction (RT-PCR) from dimethyl sulfoxide (DMSO)-treated MEL 745-PC4 cell RNA and cloned into pRc/CMV (Invitrogen, San Diego, CA). pBluescript KS containing a part of embryonic εy2 cDNA20 was supplied by Dr Ajay Bhargava of Yale University (New Haven, CT) and a 0.25-kb ε cDNA insert was excised by SpeI/EcoRV digestion. The actin probe was a 2-kb PstI fragment of chicken β-actin from pA1.21
RNase protection assays.
In vitro transcription kits (MAXIscript) and ribonuclease protection assay kits (RPA II) were purchased from Ambion, Inc (Austin, TX). Detailed procedures provided by the manufacturer were followed. Antisense probes were made by labeling 1 μg of linearized plasmid DNA with 50 μCi (3.125 μmol/L) of [α-32P]UTP (800 Ci/mmol) in a volume of 20 μL. The transcription mixture was separated on a 5% polyacrylamide/8 mol/L urea gel, and the antisense probe corresponding to a full-length transcript was eluted from the gel. Three kinds of antisense probes (ε, βh1, and βmaj/min or ζ, α-1/α-2, and actin), each of which contained 0.9 to 1.8 × 105 cpm, were combined and hybridized with 10 μg of total cellular RNA according to the streamlined procedure provided by the manufacturer. The RNA-RNA hybrids were treated with a mixture of RNase A and RNase T1, and the digested fragments were analyzed on a 5% polyacrylamide/8 mol/L urea gel.
Murine globin probes for RNase protection assays.
A 17-base difference exists between βmajor- and βminor-globin coding regions.22 The antisense βmaj/min-globin probe was constructed by amplifying a 113-bp fragment encompassing parts of exon l and exon 2 by PCR using pRc/CMV-βmajor-globin containing the entire βmajor-globin cDNA described above as a template and subcloning the fragment into pCR2.1 by TA cloning (Invitrogen). This portion was selected because of identity in the nucleotide sequence between these two homologous globin genes.22pBluescript-εy2 was described above. PBSmαT96 containing an embryonic/adult-type α-globin DNA fragment, and pSP64Mζ10,23 were gifts of Drs Murat Arcasoy and Bernard Forget of Yale University. pSP65βh124,25 was provided by Dr Thalia Papayannopoulou (University of Washington, Seattle, WA). All globin probes were subjected to DNA sequencing. Three bases were different in the 96-base protected region between the α-1-26 and α-2–globin genes (the murine α-2–globin DNA sequence was kindly provided by Dr Aya Leder of Harvard Medical School, Boston, MA). The α probe in PBSmαT96 contained 1 base corresponding to the α-1 sequence, 1 base corresponding to the α-2 sequence, and 1 base unrelated to either sequence.
RESULTS
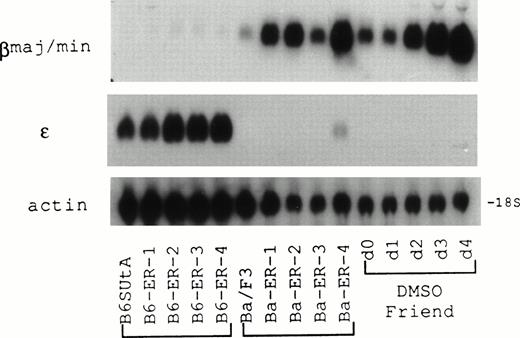
Murine IL-3–dependent Ba/F3 and B6SUtA cells produced subsets of clones with different phenotypes on transfection of an EpoR expression plasmid followed by selection with a drug resistance marker, one subtype being sensitive only to the mitogenic activity of Epo, and the other sensitive to both the mitogenic and differentiation inducing properties of the cytokine.2,4 In contrast, Ba/F3 clones transfected with the EpoR expression plasmid and selected with Epo were uniformly positive for differentiation, expressing βmajor-globin mRNA; furthermore, an inverse relationship existed between the content of β-globin mRNA and the level of EpoR mRNA.3 RNA prepared from EpoR+ Ba/F3 clones (Ba-ER-1 to -4) and EpoR+ B6SUtA clones (B6-ER-1 to -4) that were selected by Epo-dependent growth was hybridized with βmajor- and ε-globin cDNA probes (Fig 1). Although the Ba/F314and B6SUtA15 cell lines were established from adult BALB/c and B6.S mouse bone marrow, respectively, with IL-3 as a stimulant for cell growth, developmentally different types of globin mRNA were expressed in their respective EpoR+ clones. The levels of βmajor-globin mRNA were markedly increased in EpoR+ Ba/F3 clones compared with parental Ba/F3 cells, with the content of βmajor-globin in EpoR+ Ba/F3 clones being comparable with the levels observed in DMSO-treated Friend MEL cells. In contrast, significant amounts of embryonic type ε-globin mRNA were present in parental B6SUtA cells, and ε-globin gene expression seemed to be enhanced in EpoR+ B6SUtA clones. Despite a 74% nucleotide sequence similarity between the coding regions of βmajor- and ε-globin genes,20 cross-hybridization between the heterologous globins appeared to be minimal under the conditions used for the northern hybridizations.
Expression of developmentally different β-globin mRNA species in Epo-stimulated EpoR+ clones derived from B6SUtA and Ba/F3 cells analyzed by northern hybridization. B6-ER-1 to -4 and Ba-ER-1 to -4 were EpoR+ clones derived from B6SUtA and Ba/F3 cells, respectively. Friend MEL cells exposed to 1.5% DMSO for 0 to 4 days served as a control. RNA was probed with βmajor-globin, ɛ-globin, and β-actin cDNAs.
Expression of developmentally different β-globin mRNA species in Epo-stimulated EpoR+ clones derived from B6SUtA and Ba/F3 cells analyzed by northern hybridization. B6-ER-1 to -4 and Ba-ER-1 to -4 were EpoR+ clones derived from B6SUtA and Ba/F3 cells, respectively. Friend MEL cells exposed to 1.5% DMSO for 0 to 4 days served as a control. RNA was probed with βmajor-globin, ɛ-globin, and β-actin cDNAs.
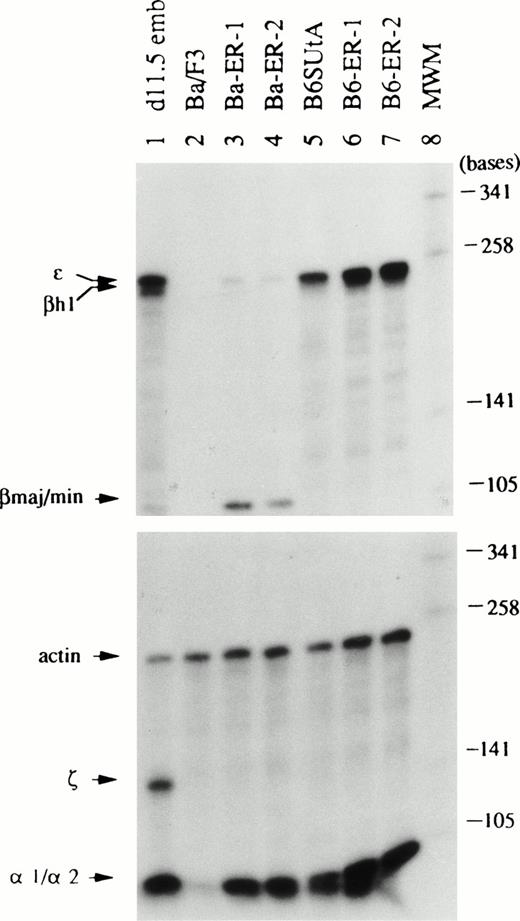
To clarify further the type of globin mRNA expressed in EpoR+ clones of Ba/F3 and B6SUtA cells, RNase protection assays capable of distinguishing between different mRNA species were used. The lengths of the probe and protected fragment for each globin mRNA are summarized in Table 1. Total cellular RNA was hybridized separately with a mixture of ε, βh1, and βmaj/min antisense probes or with a mixture of ζ, α-1/α-2, and actin antisense probes. The globin mRNA species expressed in Epo-stimulated EpoR+ Ba/F3 and B6SUtA clones, corresponding parental cells, and day 11.5 DBA/2 mouse embryonal blood are shown in Fig 2. In accord with the results of northern hybridizations, the levels of βmaj/βmin-globin mRNA in EpoR+Ba/F3 clones (Ba-ER-1 and -2) were elevated relative to that of parental cells. The levels of α-1/α-2–globin mRNAs were also increased in these clones. Trace amounts of ε-globin mRNA were also detected. However, βh1- and ζ-globin mRNAs were not detectable. Also consistent with the results of northern hybridizations, parental B6SUtA cells contained significant amounts of ε-globin mRNA. In addition, α-1/α-2–globin mRNAs were also present in parental B6SUtA cells. The levels of both ε- and α-1/α-2–globin mRNAs were elevated in EpoR+ B6SUtA clones (B6-ER-1 and -2) compared with that of parental cells. However, neither parental B6SUtA cells nor EpoR+ B6SUtA clones contained mRNAs for adult-type βS/T-globins and embryonic βh1- and ζ-globins. It should be noted that the size of the protected fragment of βh1-globin mRNA in B6SUtA cells was 180 bases instead of 245 bases in day 11.5 DBA/2 embryonal blood as discussed below. Although Epo has been reported to induce both embryonic and adult-type β-globin mRNAs in parental B6SUtA cells in the absence of cell growth, with a preferential augmentation of adult β-globin mRNA as a function of time,24 adult-type β-globin mRNAs were not detected in our clones of B6SUtA cells transfected with the EpoR and selected with Epo.
Description of the Probes for Each Globin Type Used in the RNase Protection Assays
| Globin Type . | Transcription Vector . | RE/Promoter . | Probe Length (bases) . | Protected Fragment (bases) . | Region Protected . |
|---|---|---|---|---|---|
| βmaj/min βS/T | pCR2.1 | HindIII/T7 | 241 | 113 | Part of exon I Part of exon II |
| ε | pBluescript KS | XbaI/T3 | 348 | 253 | Part of exon II Entire exon III Part of 3′UTR |
| βh1 | pSP65 | HindIII/Sp6 | 379 | 245 or 180 | Part of exon III Part of 3′UTR |
| α-1/α-2 | pBluescript SK | XbaI/T7 | 193 | 96 | Part of exon II |
| ζ | pSP64 | EcoRI/Sp6 | 239 | 157 | Part of exon I |
| Globin Type . | Transcription Vector . | RE/Promoter . | Probe Length (bases) . | Protected Fragment (bases) . | Region Protected . |
|---|---|---|---|---|---|
| βmaj/min βS/T | pCR2.1 | HindIII/T7 | 241 | 113 | Part of exon I Part of exon II |
| ε | pBluescript KS | XbaI/T3 | 348 | 253 | Part of exon II Entire exon III Part of 3′UTR |
| βh1 | pSP65 | HindIII/Sp6 | 379 | 245 or 180 | Part of exon III Part of 3′UTR |
| α-1/α-2 | pBluescript SK | XbaI/T7 | 193 | 96 | Part of exon II |
| ζ | pSP64 | EcoRI/Sp6 | 239 | 157 | Part of exon I |
After each transcription vector containing the globin DNA fragment was digested with the RE, an antisense RNA probe was transcribed with the specified promoter. The length (base number) of the probe and the protected fragment, and the region protected are summarized. The length of protected fragment of βh1-globin mRNA differed depending on the haplotype, consisting of 245 bases in the diffuse haplotype (Ba/F3 cells and 11.5-day-old DBA/2 embryonic blood) and 180 bases in the single haplotype (B6SUtA cells), as discussed in the text.
Abbreviation: RE, restriction endonuclease.
Types of globin mRNA expressed in Epo-stimulated EpoR+ clones of Ba/F3 and B6SUtA cells analyzed by RNase protection assays. Total cellular RNA (10 μg) was hybridized separately with a mixture of ɛ, βh1, and βmaj/minprobes and with a mixture of ζ, -1/-2, and actin probes. Ba-ER-1 to -2 and B6-ER-1 to -2 are described in Fig 1. RNA from DBA/2 mouse day 11.5 embryonal blood (d11.5 emb, lane 1) served as controls for embryonal globin mRNAs. Molecular weight markers (MWM, lane 8) are Sau3A-digested 32P-labeled pUC19 DNA.
Types of globin mRNA expressed in Epo-stimulated EpoR+ clones of Ba/F3 and B6SUtA cells analyzed by RNase protection assays. Total cellular RNA (10 μg) was hybridized separately with a mixture of ɛ, βh1, and βmaj/minprobes and with a mixture of ζ, -1/-2, and actin probes. Ba-ER-1 to -2 and B6-ER-1 to -2 are described in Fig 1. RNA from DBA/2 mouse day 11.5 embryonal blood (d11.5 emb, lane 1) served as controls for embryonal globin mRNAs. Molecular weight markers (MWM, lane 8) are Sau3A-digested 32P-labeled pUC19 DNA.
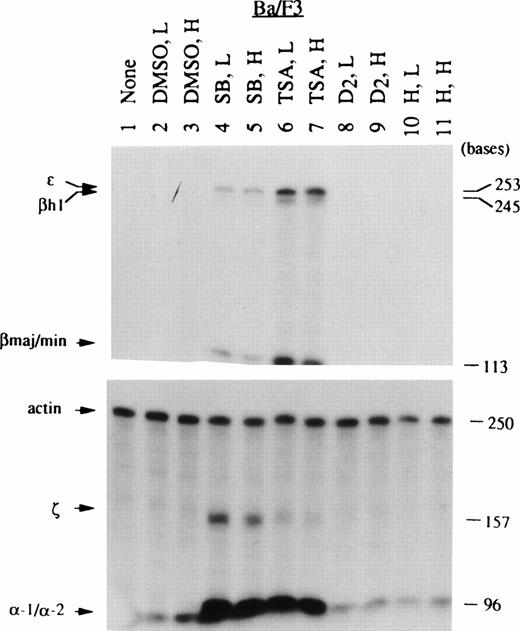
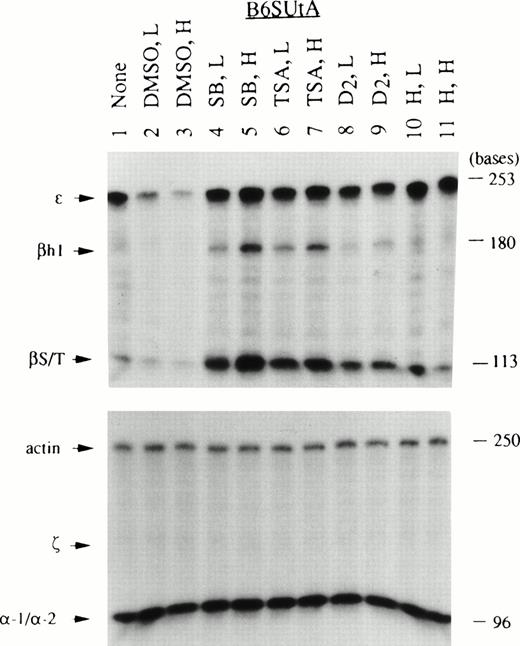
Given the specificity of globin mRNAs stimulated by Epo in EpoR+ Ba/F3 and B6SUtA clones, it was of interest to determine the types of globin mRNA induced by the chemical initiators of maturation. Although Ba/F3 cells have been reported to express erythroid-specific transcription factors such as GATA-1, NF-E2, and EKLF,27,28 and Epo-stimulated EpoR+ clones express both β- and α-globin mRNAs, these cells, unlike B6SUtA cells, do not readily become benzidine positive,2,3suggesting a defect(s) in the hemoglobin synthesizing machinery. Thus, it was necessary to monitor the sensitivity of Ba/F3 cells to chemical inducers at the globin mRNA level. To determine whether SB exerts its activity through a solvent effect, inhibition of histone deacetylase, or a combination of these actions, both DMSO and the specific inhibitor of histone deacetylase, TSA,7 were evaluated for their capacity to induce globin mRNA. Thus, the chemical inducers used included DMSO, SB, TSA, diazepam (D2), and hemin. Although Friend MEL 745 cells are known to respond to all of these agents by attaining benzidine positivity, information on the type of globins induced at the mRNA level by different initiators of differentiation is limited. Therefore, MEL 745-PC4 cells were used to compare the profiles of globin mRNAs produced by these cells with those by IL-3–dependent cell lines. Total cellular RNA was prepared from the three cell lines after exposure to two concentrations (relatively low and high) of chemical inducers which gave comparable degrees of cell growth inhibition within the same cell type. A summary of the degree of cell growth inhibition calculated from the initial and final (day 3) log cell densities and the percent benzidine positive cells is shown in Table 2. The globin mRNA species induced in MEL-PC4, Ba/F3, and B6SUtA cells by these inducers are shown in Figs 3, 4, and5, respectively. In MEL cells, DMSO and D2 were stronger inducers than SB and TSA, as measured by both benzidine assays (Table 2) and RNase protection assays (Fig 3). The globin mRNAs found in MEL cells exposed to DMSO and D2were only of the adult type (ie, βmaj/min and α-1/α-2). In contrast, benzidine-positive cells were produced in Ba/F3 cells by SB and TSA, but not by DMSO and D2 (Table2). Remarkably, treatment of Ba/F3 cells with SB and TSA resulted in the expression of all species of globin mRNAs (embryonic types: ε, βh1, and ζ; and adult types: βmaj/min and α-1/α-2), although the relative concentrations of the mRNAs induced by the two agents differed slightly (Fig 4). Thus, SB caused more prominent induction of ζ and α-1/α-2 than TSA, whereas TSA caused preferential induction of ε, βh1, and βmaj/min. In a manner analogous to Ba/F3 cells, exposure to SB and TSA caused a significant increase in the percent of benzidine-positive B6SUtA cells (Table 2). Furthermore, these agents caused accumulation of significant amounts of βS/T- and βh1-globin mRNAs in B6SUtA cells (Fig 5). However, ζ-globin mRNA, which was noticeably present in SB-treated Ba/F3 cells, was not induced in B6SUtA cells exposed to either SB or TSA.
Inhibition of Cellular Growth and Induction of Benzidine-Positive Cells by Chemical Inducers in the Cell Lines Used as Sources of RNA for RNase Protection Assays
| Inducer . | Concentration . | Friend MEL-PC4 . | Ba/F3 . | B6SUtA . | |||
|---|---|---|---|---|---|---|---|
| CD × 10−6 (% GI) . | % BP . | CD × 10−6 (% GI) . | % BP . | CD × 10−6 (% GI) . | % BP . | ||
| None | — | 3.60 (0) | 0 | 1.95 (0) | 0 | 1.04 (0) | 4 |
| DMSO | 128 mmol/L | — | — | 0.39 (34) | 4 | ||
| 192 mmol/L | 2.12 (12) | 41 | 0.60 (30) | 0 | 0.16 (65) | 4 | |
| 256 mmol/L | 0.81 (33) | 65 | 0.12 (71) | 0 | — | ||
| SB | 0.5 mmol/L | — | — | 0.45 (29) | 25 | ||
| 1.0 mmol/L | — | 0.62 (30) | 30 | 0.12 (77) | 71 | ||
| 1.5 mmol/L | 1.05 (26) | 5 | 0.17 (62) | 34 | — | ||
| 2.0 mmol/L | 0.40 (49) | 15 | — | — | |||
| TSA | 10 nmol/L | — | — | 0.44 (30) | 36 | ||
| 15 nmol/L | — | 0.43 (39) | 32 | — | |||
| 20 nmol/L | 1.43 (22) | 4 | 0.17 (62) | 30 | 0.19 (59) | 75 | |
| 25 nmol/L | 0.70 (37) | 13 | — | — | |||
| D2 | 0.05 mmol/L | — | — | 0.51 (25) | 5 | ||
| 0.10 mmol/L | 1.89 (14) | 39 | 0.87 (21) | 3 | 0.16 (65) | 37 | |
| 0.15 mmol/L | 0.71 (36) | 72 | 0.11 (75) | 0 | — | ||
| H | 0.05 mmol/L | 3.61 (0) | 14 | 1.95 (0) | 17 | 1.04 (0) | 48 |
| 0.10 mmol/L | 3.44 (1) | 18 | 1.83 (2) | 24 | 0.93 (4) | 59 | |
| Inducer . | Concentration . | Friend MEL-PC4 . | Ba/F3 . | B6SUtA . | |||
|---|---|---|---|---|---|---|---|
| CD × 10−6 (% GI) . | % BP . | CD × 10−6 (% GI) . | % BP . | CD × 10−6 (% GI) . | % BP . | ||
| None | — | 3.60 (0) | 0 | 1.95 (0) | 0 | 1.04 (0) | 4 |
| DMSO | 128 mmol/L | — | — | 0.39 (34) | 4 | ||
| 192 mmol/L | 2.12 (12) | 41 | 0.60 (30) | 0 | 0.16 (65) | 4 | |
| 256 mmol/L | 0.81 (33) | 65 | 0.12 (71) | 0 | — | ||
| SB | 0.5 mmol/L | — | — | 0.45 (29) | 25 | ||
| 1.0 mmol/L | — | 0.62 (30) | 30 | 0.12 (77) | 71 | ||
| 1.5 mmol/L | 1.05 (26) | 5 | 0.17 (62) | 34 | — | ||
| 2.0 mmol/L | 0.40 (49) | 15 | — | — | |||
| TSA | 10 nmol/L | — | — | 0.44 (30) | 36 | ||
| 15 nmol/L | — | 0.43 (39) | 32 | — | |||
| 20 nmol/L | 1.43 (22) | 4 | 0.17 (62) | 30 | 0.19 (59) | 75 | |
| 25 nmol/L | 0.70 (37) | 13 | — | — | |||
| D2 | 0.05 mmol/L | — | — | 0.51 (25) | 5 | ||
| 0.10 mmol/L | 1.89 (14) | 39 | 0.87 (21) | 3 | 0.16 (65) | 37 | |
| 0.15 mmol/L | 0.71 (36) | 72 | 0.11 (75) | 0 | — | ||
| H | 0.05 mmol/L | 3.61 (0) | 14 | 1.95 (0) | 17 | 1.04 (0) | 48 |
| 0.10 mmol/L | 3.44 (1) | 18 | 1.83 (2) | 24 | 0.93 (4) | 59 | |
Cells were exposed to chemical inducers at the indicated concentrations. At day 3, CD was measured and the percent GI was calculated by methodology described in Materials and Methods. The percent BP was determined as previously described.4
Abbreviations: DMSO, dimethylsulfoxide; SB, sodium butyrate; TSA, trichostatin A; D2, diazepam; H, hemin; CD, cell density; GI, growth inhibition; BP, benzidine-positive cells.
Globin mRNA species induced in Friend MEL745-PC4 cells by various chemical inducers analyzed by RNase protection assays. Treatment of cells with chemical inducers, RNA preparation, and RNase protection assays are described in Materials and Methods. The concentrations of agents used (L for low and H for high) are as listed in Table 2. The abbreviations of the inducers used are given in the legend of Table 2. Each RNA sample (10 μg) was hybridized separately with a mixture of ɛ, βh1, and βmaj/min probes and with a mixture of ζ, -1/-2, and actin probes as in Fig 2.
Globin mRNA species induced in Friend MEL745-PC4 cells by various chemical inducers analyzed by RNase protection assays. Treatment of cells with chemical inducers, RNA preparation, and RNase protection assays are described in Materials and Methods. The concentrations of agents used (L for low and H for high) are as listed in Table 2. The abbreviations of the inducers used are given in the legend of Table 2. Each RNA sample (10 μg) was hybridized separately with a mixture of ɛ, βh1, and βmaj/min probes and with a mixture of ζ, -1/-2, and actin probes as in Fig 2.
Globin mRNA species induced in Ba/F3 cells by various chemical inducers analyzed by RNase protection assays. RNA samples prepared from nontreated and chemically induced Ba/F3 cells were analyzed as described in Fig 3. Note that Ba/F3 cells belonging to the diffuse haplotype produced the 245-base–protected fragment of βh1 mRNA, which is longer than the protected fragment derived from B6SUtA cells, as discussed in detail in the text.
Globin mRNA species induced in Ba/F3 cells by various chemical inducers analyzed by RNase protection assays. RNA samples prepared from nontreated and chemically induced Ba/F3 cells were analyzed as described in Fig 3. Note that Ba/F3 cells belonging to the diffuse haplotype produced the 245-base–protected fragment of βh1 mRNA, which is longer than the protected fragment derived from B6SUtA cells, as discussed in detail in the text.
Globin mRNA species induced in B6SUtA cells by various chemical inducers analyzed by RNase protection assays. RNA samples prepared from nontreated and chemically induced B6SUtA cells were analyzed as described in Fig 3. Note that B6SUtA cells belonging to the single haplotype produced a 180-base–protected fragment of βh1 mRNA, shorter than that produced by Ba/F3 cells and 11.5-day-old DBA/2 mouse embryo blood as discussed in the text.
Globin mRNA species induced in B6SUtA cells by various chemical inducers analyzed by RNase protection assays. RNA samples prepared from nontreated and chemically induced B6SUtA cells were analyzed as described in Fig 3. Note that B6SUtA cells belonging to the single haplotype produced a 180-base–protected fragment of βh1 mRNA, shorter than that produced by Ba/F3 cells and 11.5-day-old DBA/2 mouse embryo blood as discussed in the text.
Because the concentrations of SB and TSA were chosen for their equivalence to DMSO in the extent of cell growth inhibition, and the induced levels of βmaj/min-globin mRNA in MEL PC4 cells by SB and TSA at these concentrations were low (Fig 3), a question remained as to whether these agents at higher concentrations had the ability to induce higher amounts of βmaj/min-globin mRNA, as well as to induce the embryonic species of globin mRNAs. SB at concentrations of 2.5 and 3.0 mmol/L caused 51% and 69% of cell growth inhibition, respectively, and induced βmaj/min-globin mRNA at a level comparable with that produced by 2.0% DMSO. Under these conditions, SB induced trace amounts of embryonic species of globin mRNAs in MEL 745-PC4 cells. Quantification of the autoradiogram by densitometry, with conversion based on the specific activity of the probes and the length of the protected fragments, showed that embryonic species of globin mRNAs were less than 1% of the βmaj/min-globin mRNA on a molar basis (data not shown). None of the globin mRNA species were detected in murine WEHI-3B myelomonocytic leukemia cells exposed to SB or hemin (data not shown). All of the cell lines, including WEHI-3B cells, treated with hemin for more than 3 days became benzidine positive. The absence of globin mRNAs in hemin-treated Ba/F3 and WEHI-3B cells suggests that the positive reaction with benzidine is caused by the intracellular accumulation of hemin.
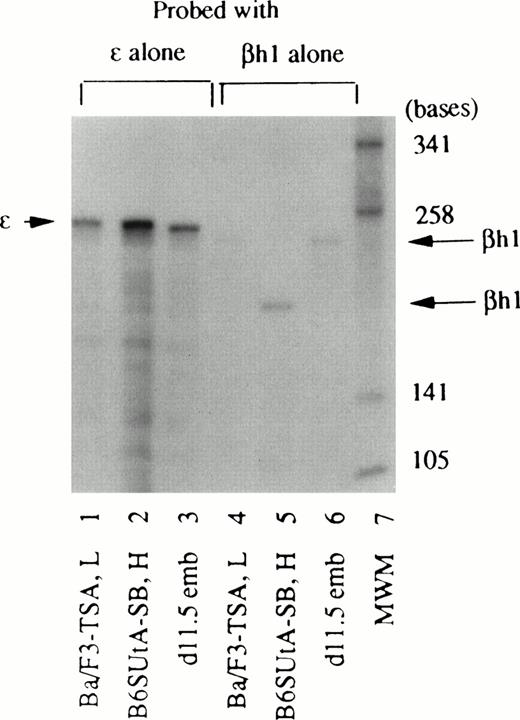
The βh1-globin antisense probe included parts of exon III and 3′UTR constructed from BALB/c (diffuse haplotype) mouse DNA. To show that the length of the protected fragment (180 bases) produced by cells that belong to the single haplotype is shorter than that (245 bases) produced by cells that belong to the diffuse haplotype, RNA from TSA-treated Ba/F3 cells (diffuse haplotype), SB-treated B6SUtA cells (single haplotype), and day 11.5–old DBA/2 embryonal blood (diffuse haplotype) were hybridized with a βh1 probe alone (Fig 6). The same set of RNA samples were also hybridized with an ε probe alone, to show that the protected fragment of ε-globin mRNA (253 bases) is equal in size among different haplotypes and is distinguishable from the longer βh1 protected fragment (245 bases). The difference in the protected fragment of βh1-globin mRNA in the two haplotypes is presumably because of the reported diversion in the nucleotide sequence in the 3′UTR region of the different mouse strains.25Although Figs 2 through 5 were derived from RNase protection assays using multiple antisense probes, RNA samples from different cell types were analyzed by hybridization with a single antisense probe to ensure that the protected fragments of all globin mRNAs, with the exception of βh1-globin mRNA, were equal in size regardless of the cell type, as well as to rule out the possibility that the absence of detection of a particular mRNA was due to overlapped fragments in the hybridization with multiple probes.
Different sizes of the protected fragment of βh1-globin mRNA produced by cells with different haplotype origins. RNA samples from Ba/F3 cells treated with 15 nmol/L TSA (Ba/F3-TSA, L), B6SUtA cells treated with 1.0 mmol/L SB (B6SUtA-SB, H), and 11.5-day-old DBA/2 mouse embryonal blood (d11.5 emb) were hybridized with the βh1-globin probe alone (lanes 4, 5, and 6). The same set of RNA samples was hybridized with the ɛ-globin probe alone (lanes 1, 2, and 3).
Different sizes of the protected fragment of βh1-globin mRNA produced by cells with different haplotype origins. RNA samples from Ba/F3 cells treated with 15 nmol/L TSA (Ba/F3-TSA, L), B6SUtA cells treated with 1.0 mmol/L SB (B6SUtA-SB, H), and 11.5-day-old DBA/2 mouse embryonal blood (d11.5 emb) were hybridized with the βh1-globin probe alone (lanes 4, 5, and 6). The same set of RNA samples was hybridized with the ɛ-globin probe alone (lanes 1, 2, and 3).
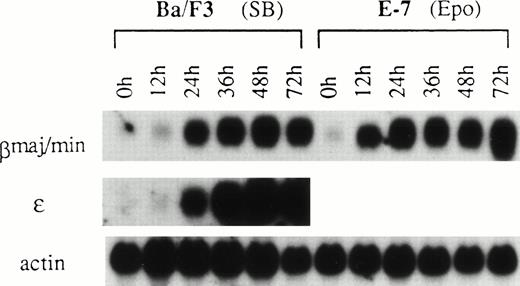
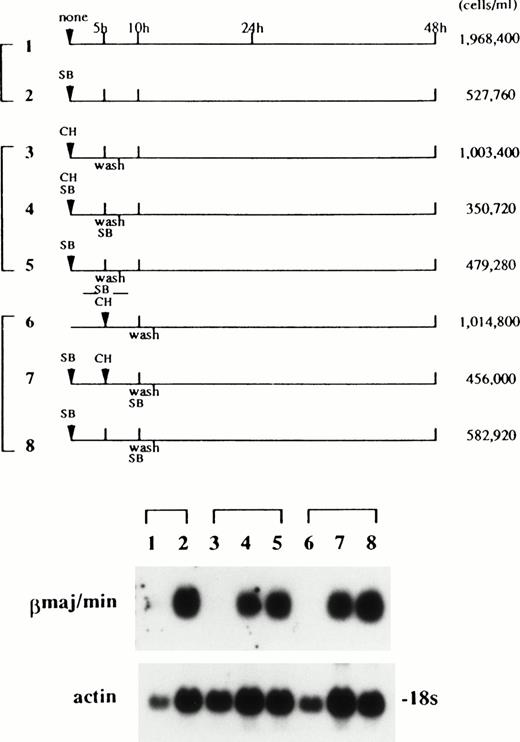
Ba/F3 cells and EpoR+ Ba/F3 clones grew in IL-3–containing medium with a doubling time of 8.9 hours. Treatment of BaF3 cells with SB, or switching of the growth factor from IL-3 to Epo in EpoR+ Ba/F3 clones, caused retardation of cell growth.3 The time course of induction of βmaj/min-globin mRNA in Ba/F3 cells by SB was compared with that by Epo in the EpoR+ Ba/F3 clone E-7,2in which the induction of βmaj/min-globin mRNA was dependent on Epo. A significant increase in β-globin mRNA content was detected in Ba/F3 cells treated with SB and in clone E-7 treated with Epo at 24 and 12 hours, respectively (Fig7). The time course of induction of ε-globin mRNA in Ba/F3 cells by SB was similar to that of βmaj/min-globin mRNA. One of the possible mechanisms responsible for the 12-hour delay in the induction of the globin gene by SB relative to that by Epo was that SB induction required newly synthesized protein(s). To evaluate this possibility, cycloheximide was added to the medium for the first or second 5 hours of the incubation period together with SB according to the schedule shown in Fig 8. Quantification of the autoradiogram by densitometry, with a correction based on the actin control, showed that the copresence of cycloheximide for the first (lane 4) and second (lane 7) 5 hours inhibited the accumulation of βmaj/min-globin mRNA by 27% and 39%, respectively, compared with respective controls (lanes 5 and 8). The moderate degrees of inhibition manifested by cycloheximide suggest that protein synthesis in the early part of the incubation period did not have a major role in the induction of βmaj/min-globin mRNA by SB. With respect to the requirement for protein synthesis in the induction of gene expression by SB, opposite results have been reported.29,30 In the J2E erythroid cell line, which is also known to be responsive to both Epo and SB, the time course of the appearance of βmajor-globin mRNA has been shown to be faster in SB-treated cells than in Epo-treated cells.31
Time course of the induction of β-globin mRNAs by SB and Epo. Parental Ba/F3 cells were treated with 1 mmol/L SB. E-7, a clone of Ba/F3 cells transfected with the EpoR and selected with G418, responds to both the mitogenic and differentiation-inducing effects of Epo.2 E-7 cells in IL-3–containing medium were washed three times with RPMI 1640 medium containing 10% FBS and exposed to 0.5 U/mL of Epo. RNA was subjected to northern hybridization with βmajor-globin, ɛ-globin, and actin probes.
Time course of the induction of β-globin mRNAs by SB and Epo. Parental Ba/F3 cells were treated with 1 mmol/L SB. E-7, a clone of Ba/F3 cells transfected with the EpoR and selected with G418, responds to both the mitogenic and differentiation-inducing effects of Epo.2 E-7 cells in IL-3–containing medium were washed three times with RPMI 1640 medium containing 10% FBS and exposed to 0.5 U/mL of Epo. RNA was subjected to northern hybridization with βmajor-globin, ɛ-globin, and actin probes.
The effects of cycloheximide (CH) on the induction of βmajor-globin mRNA by SB. Ba/F3 cells were exposed to SB and/or CH for the first or second 5-hour period according to the treatment schedules shown in the top of the figure. In panels 4 and 5, and in panels 7 and 8, SB was added back to the medium after cells were washed at 5 and 10 hours of the incubation period, respectively. RNA was extracted at 48 hours and subjected to northern hybridization (bottom of the figure). The cell densities at 48 hours are shown. The concentrations of SB and CH used were 1 mmol and 10 μmol/L, respectively. CH at this concentration inhibited protein synthesis, measured by incorporation of 35S-methionine into the acid-insoluble fraction, by more than 95% (data not shown).
The effects of cycloheximide (CH) on the induction of βmajor-globin mRNA by SB. Ba/F3 cells were exposed to SB and/or CH for the first or second 5-hour period according to the treatment schedules shown in the top of the figure. In panels 4 and 5, and in panels 7 and 8, SB was added back to the medium after cells were washed at 5 and 10 hours of the incubation period, respectively. RNA was extracted at 48 hours and subjected to northern hybridization (bottom of the figure). The cell densities at 48 hours are shown. The concentrations of SB and CH used were 1 mmol and 10 μmol/L, respectively. CH at this concentration inhibited protein synthesis, measured by incorporation of 35S-methionine into the acid-insoluble fraction, by more than 95% (data not shown).
DISCUSSION
The IL-3–dependent Ba/F3 and B6SUtA cells used in this study were derived from the adult bone marrow of BALB/c14 and B6.S15 mice, respectively. Whereas basal levels of globin mRNA of all types were negligible in Ba/F3 cells, the constitutive expression of ε- and α-1/α-2–globin mRNAs was observed in B6SUtA cells. The expression of embryonic/fetal type globins in B6SUtA cells suggests that the origin of this cell line is from a primitive lineage of erythroid precursors. It is also possible that during the establishment or propagation of this cell line they may have been exposed to an inductive environment, ie, exposure to factors such as Epo and/or stem cell factor (SCF). Once the globin gene is activated, expression of the message persists after withdrawal of the stimulus. This phenomenon is exemplified by Ba/F3 cells, where the levels of βmajor-globin mRNA persist in the absence of Epo in EpoR+ Ba/F3 cells once the β-globin gene is activated by the cytokine.2 27
Epo predominantly induced adult types (βmaj/min and α-1/α-2) of globin mRNAs in EpoR+ Ba/F3 clones, whereas more striking effects on globin gene expression were obtained in parental Ba/F3 cells exposed to SB and TSA. Thus, SB and TSA allowed the expression of all species of globin mRNAs. Furthermore, as opposed to the augmentation of the preexisting program (ie, expression of ε- and α-1/α-2–globin genes) by Epo in EpoR+ B6SUtA clones, SB and TSA activated a latent phenotype (ie, expression of βS/T- and βh1-globin genes) in parental B6SUtA cells. The difference in the profile of globin mRNAs displayed in Epo-stimulated EpoR+ cells and in SB- and TSA-treated parental cells suggests that the mechanism by which Epo stimulates expression of globin genes is different from that used by SB and TSA.
The types and amounts of globin mRNAs induced in B6SUtA cells by SB and TSA were comparable (Fig 5), whereas the spectrum of globin mRNAs in Ba/F3 cells induced by SB and TSA were slightly different (Fig 4). Thus, SB was more prominent in inducing globin genes in the α-cluster (α-1/α-2 and ζ) than TSA, whereas TSA induced β-globin mRNAs (ε, βh1, and βmaj/min) more efficiently than SB. Because the RNA samples were prepared from cells exposed to TSA and SB at the concentrations that led to comparable degrees of growth inhibition and benzidine positivity (Table 2), the difference in the spectrum of globin mRNAs observed in Ba/F3 cells exposed to SB and TSA may be caused by competition between different globin loci for a factor(s) necessary for gene activation. Overall, the effects of SB and TSA were strikingly similar in these two types of IL-3–dependent cell lines, in that both agents allowed simultaneous activation of embryonic and adult globin genes, despite a difference in their effective concentrations in vitro by approximately five orders of magnitude (1 mmol/L SB v 10 nmol/L TSA). The mimicry of action shared by SB and TSA implies that (1) SB activates globin genes through an inhibition of histone deacetylase, and that (2) histone deacetylase is involved in the repression of both adult and embryonic globin genes in these IL-3–dependent cells.
The significance of the acetylation of core histones on the transcriptional activity of chromatin has begun to be recognized recently, with components of the transcriptional complex, which control the activation and repression of genes, being identified as histone acetylase and histone deacetylase, respectively.32-34Because a global increase in acetylation of core histones produced by inhibitors of histone deacetylase does not result in the widespread activation of genes,35 and information regarding which specific genes are under the control of histone deacetylase is limited, the present study, which identifies embryonic and adult globin genes in IL-3–dependent cells as genes under the control of histone deacetylase, is significant. Identification of additional components that tether histone deacetylase to the promoter region of globin genes is required. In this context, it is noteworthy that the retinoblastoma protein has been shown to recruit histone deacetylase to repress transcription of a subset of genes which are involved in progression from G1 to the S phase of the cell cycle and are controlled by the E2F family of transcription factors.36,37 Examination of whether the retinoblastoma protein is also involved in repression or derepression of globin genes and identification of an erythroid-specific transcription factor(s) which participates in these processes form the next focus of investigations. In connection with acetylation of chromatin and gene activation, it is interesting to note that signal transducers and activators of transcription, which bring about altered gene expression in response to cytokines, have been proposed to cooperate with p300/CBP,38,39 which possesses intrinsic histone acetylase activity.34
Heavily based on the types of hemoglobins produced in these cells by DMSO, it is generally accepted that Friend MEL cells possess an adult-type environment for globin gene expression, with this generalization forming the basis of many cell fusion experiments. Consistent with this notion, globin mRNAs in MEL 745-PC4 cells induced by DMSO were confined to adult types. In contrast to induction of adult types of globin mRNAs by DMSO, other groups have shown the induction by SB of small amounts of ε-globin in these cells.40-42 In our hands, embryonic species of globin mRNAs were detectable in SB-treated MEL 745-PC cells; however, the fraction of ε-globin was extremely low, estimated to be less than 1% of the βmajor/minor-globins. In contrast to Friend MEL 745 cell lines, the Friend MEL GM979 cell line, renamed after the T-3-Cl-2 clone originally isolated by Ikawa et al,43 contained ε-, βh1-, and βmajor/minor-globins in the uninduced state.16,25,41 Treatment of these cells with SB resulted in induction of large amounts of ε-globins.42 44 Thus, Friend MEL cell lines have varying environments for globin gene expression depending on the isolate.
Permissiveness for activation of embryonic as well as of adult globin genes on treatment with SB in the two IL-3–dependent cell lines used in this study has important implications in the assignment of the origin of these cells in the developmental stage. Coexpression of hemoglobins of different developmental stages has been shown in numerous systems. Thus, (1) small amounts of fetal hemoglobin are consistently present in the blood of normal human adults12; (2) burst-forming unit–erythroid colonies developed from the human yolk sac, as well as from fetal liver erythroid precursors, coexpress embryonic, fetal, and adult globins45; (3) embryonic and adult hemoglobins are produced in erythroid colonies from mouse embryos at an early gestational stage46; and (4) blast colonies of embryonic stem cells formed in response to vascular endothelial growth factor and SCF express βh1- and βmajor-globin mRNAs.47 These findings are interpreted as indications that (1) globin expression in erythroid cells is not strictly stage specific, (2) erythroid cells in a transitional state at the time of the hemoglobin switch have the potential to coexpress hemoglobins of different developmental origins, and (3) primitive and definitive erythropoiesis are derived from a common precursor. Human erythroid cell lines, such as K562 and HEL, established from adult leukemias with an erythroleukemic phenotype, characteristically express fetal and some embryonic globin chains.12 Deduced from the program of globin gene expression and the methylation pattern of globin genes in these cell lines, Enver et al48 have speculated that expression of fetal and/or embryonic globins in leukemia cell lines is not an aberration of neoplastic transformation, but is indicative of fetal or embryonic potential in normal adult hematopoietic progenitors. The fact that both Ba/F3 and B6SUtA cells are derived from adult mouse bone marrow and yet are permissive for the expression of both adult and embryonic globin genes adds direct evidence in support of this view, and suggests that (1) multipotent stem cells in adult bone marrow undergo a program of hemoglobin switching during erythroid differentiation analogous to that of developmental switching during ontogeny, and (2) these IL-3–dependent cells possess the characteristics of erythroid precursors at the transitional developmental stage. In contrast to human erythroid leukemia cell lines, which invariably express fetal/embryonic globin mRNAs to various extents in the uninduced state,12,48 the Ba/F3 mouse bone marrow cells described in this report are devoid of background globin expression, and fetal/adult globin mRNAs are induced only after exposure to inhibitors of histone deacetylase. Thus, the Ba/F3 cell line is a relatively clean model system to study the mechanism of activation of fetal/adult globin genes. Previous investigations have shown that methylation of genes is an important determinant of the control of activity of globin genes.48,49 Methylation of genes and hypoacetylation of chromatin may be coupled to contribute to the mechanism of repression of globin genes, because a protein that selectively binds to methylated DNA sequences has recently been shown to exist as a complex with histone deacetylase.50 51
In previously known cell systems where developmentally incorrect forms of globin(s) are expressed, the ratio of the incorrect form of globin relative to total globin is generally low.12,45,46 In contrast, SB and TSA simultaneously and efficaciously activated embryonic as well as adult-type β-globin genes in IL-3–dependent cells. Extensive investigations on tissue specific and developmental stage-specific regulation of globin genes have shown that sequences local to the individual globin genes are sufficient to direct developmental stage-specific and tissue-specific expression, whereas high-level expression of the globin genes depends on the distal regulatory elements LCR.52,53 Studies have also indicated that the LCR can only activate one gene at a time and that the LCR interacts directly with the gene it is enhancing through a looping mechanism. Thus, it is believed that the developmental switching of globin gene expression is regulated, at least in part, through a competition between the genes for interaction with the LCR.52 The occurrence of the phenomenon observed in this study, ie, efficacious activation of all of the genes (ε, βh1, βmajor/βminor) in the β-cluster in SB-treated B6SUtA cells, thus seems contradictory with this model. However, using erythroid cells of human embryonic liver origin, which coexpress γ- and β-globins, Wijgerde et al54 have shown that the LCR can interact with these two genes by a flip-flop mechanism. Based on in vivo dimethyl sulfate foot printing, Reddy and Shen55 have constructed three possible models for DMSO-induced globin gene expression, with or without the involvement of the LCR. Overall, the precise role of the LCR in the activation of globin genes by DMSO and SB is unclear.
In summary, our previous2-4 and current studies have shown the versatility of murine IL-3–dependent bone marrow–derived Ba/F3 and B6SUtA cells as models for comparative studies of growth and differentiation controlled by the physiological cytokine Epo and by chemical inducers in several ways. First, both cell lines yield two subsets of clones on transfection of the EpoR, one with sensitivity to the mitogenic action of Epo and the other with sensitivity to both the mitogenic and differentiation-inducing effects of Epo.2 4Therefore, these clones are capable of serving as model systems for the dissection of Epo-dependent growth and differentiation signals. Secondly, because these cell lines respond to both Epo and inhibitors of histone deacetylase by undergoing differentiation, they are excellent models to use to delineate identities and differences in differentiation signals utilized by these agents. Most importantly, the identification of histone deacetylase as an integral part of repression and derepression of globin genes in IL-3–dependent bone marrow–derived cells provides the initial step toward identification of factors involved in these processes which could serve as targets of therapeutics to correct hemoglobin disorders.
ACKNOWLEDGMENT
We are grateful to Drs Harvey Lodish and Joel Greenberger for gifts of the Ba/F3 and B6SUtA cell lines, respectively. We also thank Drs Murat Arcasoy and Bernard Forget for gifts of PBSMαT96 and pSP64Mζ and for helpful discussions. We are indebted to Drs Ajay Bhargava and Thalia Papayannopoulou for provision of pBluescript-εy2and pSP64-ζ, respectively. We are also grateful to Dr Aya Leder for providing the unpublished DNA sequence of the murine α-2–globin gene. We wish to thank the R. W. Johnson Pharmaceutical Research Institute for supplying human recombinant Epo.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Alan C. Sartorelli, PhD, Department of Pharmacology, Yale University School of Medicine, 333 Cedar St, New Haven, CT 06520.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal