Abstract
Segmental jumping translocations are chromosomal abnormalities in treatment-related leukemias characterized by multiple copies of theABL and/or MLL oncogenes dispersed throughout the genome and extrachromosomally. Because gene amplification potential accompanies loss of wild-type p53, we examined the p53 gene in a case of treatment-related acute myeloid leukemia (t-AML) with MLLsegmental jumping translocation. The child was diagnosed with ganglioneuroma and embryonal rhabdomyosarcoma (ERMS) at 2 years of age. Therapy for ERMS included alkylating agents, DNA topoisomerase I and DNA topoisomerase II inhibitors, and local radiation. t-AML was diagnosed at 4 years of age. The complex karyotype of the t-AML showed structural and numerical abnormalities. Fluorescence in situ hybridization analysis showed multiple copies of the MLL gene, consistent with segmental jumping translocation. A genomic region including CD3, MLL, and a segment of band 11q24 was unrearranged and amplified by Southern blot analysis. There was no family history of a cancer predisposing syndrome, but single-strand conformation polymorphism (SSCP) analysis detected identical band shifts in the leukemia, ganglioneuroma, ERMS, and normal tissues, consistent with a germline p53 mutation, and there was loss of heterozygosity in the ERMS and the t-AML. Sequencing showed a CGA→TGA nonsense mutation at codon 306 in exon 8. The results of this analysis indicate that loss of wild-type p53 may be associated with genomic instability after DNA-damaging chemotherapy and radiation, manifest as a complex karyotype and gene amplification in some cases of t-AML.
USING FLUORESCENCE in situ hybridization (FISH) analysis, Tanaka et al1 recently identified jumping translocations of the chromosomal segments containing the ABLor MLL oncogenes as a new form of gene amplification in three treatment-related leukemias with complex karyotypes. The leukemias were of French-American-British (FAB) M1, M2, or M4 morphologic subtypes and followed various classes of DNA-damaging chemotherapy and radiation used for the management of adult solid tumors.1 The karyotypes contained chromosome 5 or 7 monosomy typical of alkylating agent-induced leukemias, as well as several other numerical and structural abnormalities.1 The segmental jumping translocations were characterized by multiple copies of the ABLor MLL oncogenes dispersed throughout the genome and extrachromosomally.1
The complex karyotypes and ABL and MLL gene amplification in treatment-related leukemias with segmental jumping translocations suggest genomic instability. The G1 cell cycle checkpoint function of wild-type p53 maintains genomic stability and ploidy. With DNA-damaging chemotherapy and γ-radiation, the altered cell cycle arrest associated with loss of wild-type p53 is associated with genomic instability.2-5
In the present study, we identified MLL gene amplification consistent with segmental jumping translocation in a pediatric case of treatment-related acute myeloid leukemia (t-AML) with a complex karyotype. The child previously received multiagent chemotherapy and local radiation for nasopharyngeal embryonal rhabdomyosarcoma (ERMS) diagnosed at 2 years of age. We examined the p53 gene for mutation because germline p53 mutations are associated with early onset ERMS6 and because loss of wild-type p53 is associated with gene amplification in in vitro model systems.2 The mechanism for gene amplification in leukemias with segmental jumping translocations previously was unknown. Detection of a germline p53 mutation with loss of heterozygosity (LOH) in this t-AML indicates that one mechanism is loss of wild-type p53.
MATERIALS AND METHODS
The Institutional Review Boards at the Children's Hospital of Buffalo and the Children's Hospital of Philadelphia approved this research and the parents gave informed consent. Karyotype, FISH, and Southern blot analyses were performed on the t-AML of patient RUPN 84 and on the cell line 2L1, which was derived from the marrow of patient RUPN 84 at diagnosis of t-AML.
Characterization of the cell line 2L1.
The cell line 2L1 was continuously passaged for 14 months in Iscove's modified Minimum Essential Medium (MEM; Life Technologies, Inc, Gaithersburg, MD) containing 20% fetal bovine serum, 2 mmol/L glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. The diagnostic marrow and the 2L1 cells expressed CD33, CD13, CD15, CD11b, and CD38; however, the 2L1 cells were also strongly positive (>90%) for CD2, CD10, and CD56. The karyotypes of the 2L1 cells and of the diagnostic marrow of patient RUPN 84 were the same (see Results).
Cytogenetic analysis.
FISH analysis.
Probes used for FISH analysis included a YAC clone containing MLL(Oncor, Gaithersburg, MD), a CD3ε probe for analysis of the region centromeric to the MLL gene (a kind gift from Dr Thomas Shows, Roswell Park Cancer Institute, Buffalo, NY), and the more telomeric cosmid, C11q7q24, from chromosome band 11q24.9The probes were labeled with digoxigenin, and metaphase spreads from the marrow of patient RUPN 84 at diagnosis of t-AML and from the cell line 2L1 were examined by FISH analysis using standard methods.10
Southern blot analysis of the MLL gene.
Genomic DNAs from the diagnostic marrow of patient RUPN 84, from the cell line 2L1, and from control peripheral blood mononuclear cells were examined by Southern blot analysis. BamHI-, HindIII-,Sst I-, EcoRI-, and Bgl II-digested DNAs were hybridized with a 0.7-kb cDNA probe from the MLLbcr.11BamHI- and HindIII-digested DNAs were also hybridized with a 2.2-kb genomic fragment of the SCLgene at chromosome band 1p33-1p34.12 To assess equivalence in loading, HindIII-digested DNAs were simultaneously hybridized with the MLL bcr cDNA probe11 and theSCL genomic probe.12
BamHI-, HindIII-, and EcoRI-digested DNAs were hybridized with polymerase chain reaction (PCR)-generated fragments from MLL exon 3, exon 25, and exon 34. The sense and antisense PCR primers used to amplify MLL exon 3 were 5′-GTC AGT GCT ATC TCC TCG CG-3′ and 5′-GCA GAA GTT CGA TTA CTA GGC-3′, respectively. The sense and antisense PCR primers used to amplify MLL exon 25 were 5′-CTT ACC ACA GGA CTA AAT CC-3′ and 5′-TTA TGA TGT TGG GGA CAG TTC G-3′, respectively. The sense and antisense PCR primers used to amplifyMLL exon 34 were 5′-CAG AGA CAG AGT TGA GGT CTC G-3′ and 5′-CAG AAG TGA ACT CTC GAG TGG-3′, respectively. BamHI-, HindIII-, and SstI-digested DNAs were hybridized with a 1.3-kb cDNA probe from the CD3ε gene, which is centromeric to the MLL gene at chromosome band 11q23 (a kind gift from Dr Thomas Shows). Southern blot analysis also was performed on BamHI- and HindIII-digested DNAs using a 1.5-kb nonreiterated genomic fragment, C11q1.5SS, from the cosmid C11q7q24, which maps to chromosome band 11q24.9
Signal intensity on the autoradiographs was quantitated using a Molecular Dynamics computing densitometer and ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The MLL, CD3ε, and band q24 chromosome 11 signals were normalized by comparison with the signal from hybridization with the SCL probe from chromosome band 1p33-1p34.
p53 single-strand conformation polymorphism (SSCP) analysis.
Genomic DNAs from the marrow of patient RUPN 84 at diagnosis of t-AML and from the cell line 2L1 were screened by PCR/SSCP analysis.13 The oligonucleotide primers have been reported.14 PCR fragments containing p53 exons 5 and 6 or exons 7 and 8 and incorporating [α32P]-dCTP were amplified using 100 ng genomic DNA as template. Aliquots of the products containing exons 5 and 6 or exons 7 and 8 were digested withAat I or Dra I, respectively, to reduce the fragment sizes and separate the exons. Aliquots of the digested products were diluted with loading buffer, denatured by heating at 90°C for 5 minutes and electrophoresed at 4°C in nondenaturing polyacrylamide at constant power as previously described.13
Characterization of p53 mutation suggested by SSCP.
Fresh aliquots of genomic DNA from the marrow of patient RUPN 84 at diagnosis of t-AML were amplified in 3 separate PCR reactions with the same SSCP primers encompassing exons 7 and 8. The 100 μL PCR reaction mixtures contained 1 μg genomic DNA, 2.5 U AmpliTaq DNA polymerase, 200 mmol/L of each dNTP, PCR reaction buffer at 1× final concentration (Perkin Elmer, Norwalk, CT), and 100 pmol of each primer. After initial denaturation at 94°C for 9 minutes, 35 cycles at 94°C for 1 minute, 55°C for 1 minute, and 72°C for 2 minutes were used, followed by a final elongation at 72°C for 7 minutes. Products of 3 separate PCR reactions were subcloned into pBluescript II SK+ vector (Stratagene, La Jolla, CA). Six separate subclones from the 3 independent PCR reactions were sequenced by automated methods.
Extraction of genomic DNAs from paraffin-embedded tissues for SSCP analysis of p53 exon 8.
Genomic DNAs were extracted from formalin-fixed paraffin-embedded tissue blocks of the ERMS and ganglioneuroma of patient RUPN 84. After thorough cleaning of the microtome and installation of a new disposable knife, 15- to 20-μm sections providing a 1 cm2 area were cut from a control blank block and from the blocks containing tissue. Microdissection was performed to isolate the tumor from the surrounding normal tissue. The sections were deparaffinized with a 1:1 mixture of xylene and ethanol. Deparaffinized sections were incubated at 55°C for 1 to 3 hours in 100 μL of a solution containing 6 μg proteinase K, 10 mmol/L Tris HCl, pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2, and 0.01% gelatin. The solutions then were incubated at 100°C for 10 minutes, followed by microcentrifugation at 14,000 rpm for 10 minutes. The supernatant containing the DNA was removed to a clean microcentrifuge tube and diluted with dH2O to a final volume of 200 μL.
SSCP analysis of p53 exon 8 in genomic DNAs from paraffin-embedded tissues.
A new PCR/SSCP sense primer beginning at the Dra I site in p53 intron 7 was designed to amplify a 245-bp product containing p53 exon 8 when used with the same intron 8 antisense primer as described above.14 The sense primer was homologous to positions 13937-13954 of the p53 genomic sequence. One hundred nanograms of genomic DNA from the marrow of patient RUPN 84 at diagnosis of t-AML and 2 μL of genomic DNAs prepared from the paraffin-embedded tissues were amplified in 10 μL reactions incorporating [α32P]-dCTP using exactly the same PCR conditions as described above for SSCP.13 Blank blocks processed in the same manner as the paraffin-embedded tissues and dH2O were negative controls. One microliter of each PCR reaction was diluted with 9 μL of loading buffer and denatured by heating at 90°C for 5 minutes. Two microliters of each heat-denatured sample (1/50 of the initial PCR reaction) was electrophoresed at 4°C in nondenaturing polyacrylamide at constant power for 5 hours.
RESULTS
Case history.
Patient RUPN 84 was diagnosed simultaneously with ganglioneuroma and nonmetastatic nasopharyngeal ERMS at 2 years of age. Treatment for ERMS included multiagent chemotherapy and local radiation. Cumulative chemotherapy doses for ERMS were vincristine (40 mg/m2), actinomycin D (17.5 mg/m2), ifosfamide (7,300 mg/m2), and cyclophosphamide (8,600 mg/m2). Additional therapy doses for locally recurrent ERMS were etoposide (1,200 mg/m2), doxorubicin (300 mg/m2), and carboplatinum (3,810 mg/m2). The child was diagnosed with t-AML at 4 years of age after 10 months off therapy for ERMS. There was no family history of a cancer-predisposing syndrome. The diagnostic marrow contained 60% blasts of the FAB M5 monoblastic subtype that expressed CD33, CD13, CD15, CD11b, and CD38. The complex karyotype of the t-AML was 45,XY, der(5)t(5;11)(5pter→5q12::11q23→11qter),der(11)(pter→q12::q24→q12::q13→qter), −17. Remission induction with daunomycin, cytosine arabinoside, thioguanine, and teniposide was unsuccessful. The patient underwent HLA-matched allogeneic marrow transplantation but died from his disease.
Evidence for MLL gene amplification in the t-AML of patient RUPN 84.
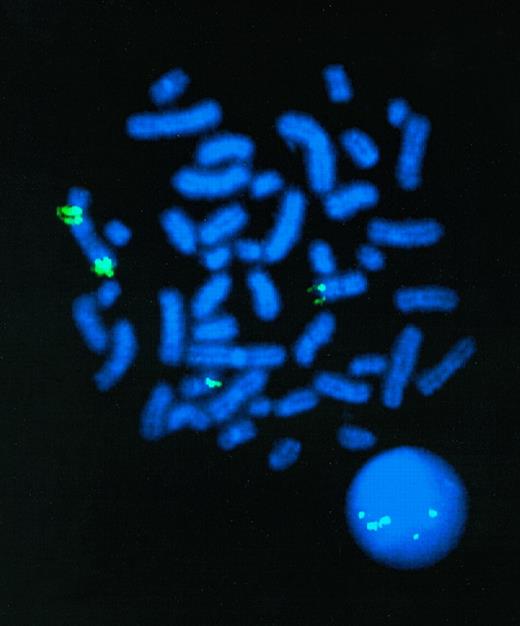
Consistent with segmental jumping translocation, 5 or 6 signals consistently were identified on hybridization of the MLL YAC probe with a total of 20 metaphase spreads prepared from the diagnostic marrow of patient RUPN 84. In the example shown in Fig 1, the MLL YAC probe detected signals on the der(5) chromosome, on the normal chromosome 11, and at distinct centromeric and telomeric regions on the der(11) chromosome. The telomeric and centromeric signals from the der(11) chromosome were consistently more intense than the MLL signal from the normal chromosome 11, suggesting that the centromeric and telomeric regions on the der(11) chromosome contained multiple copies of the MLLgene. In addition, at least 5 discrete signals were observed in the nearby interphase nucleus (Fig 1). Similar results were obtained by FISH analysis with CD3ε and 11q24 probes, indicating that the amplified genomic region extended centromeric and telomeric of theMLL gene. FISH analysis of the cell line 2L1 derived from the leukemia showed the same results (data not shown).
FISH analysis demonstrating multiple copies ofMLL in t-AML of patient RUPN 84. Digoxygenin-labeledMLL YAC probe (Oncor) was hybridized to metaphase spread. Centromeric and telomeric signals are detected on der(11) chromosome (9 o'clock). Signals also are detected on der(5) chromosome (6 o'clock) and on normal chromosome 11 (3 o'clock). There are 5 discrete signals on nearby interphase nucleus. Five or 6 signals consistently were detected on hybridization of MLL YAC probe with other metaphase spreads from the t-AML.
FISH analysis demonstrating multiple copies ofMLL in t-AML of patient RUPN 84. Digoxygenin-labeledMLL YAC probe (Oncor) was hybridized to metaphase spread. Centromeric and telomeric signals are detected on der(11) chromosome (9 o'clock). Signals also are detected on der(5) chromosome (6 o'clock) and on normal chromosome 11 (3 o'clock). There are 5 discrete signals on nearby interphase nucleus. Five or 6 signals consistently were detected on hybridization of MLL YAC probe with other metaphase spreads from the t-AML.
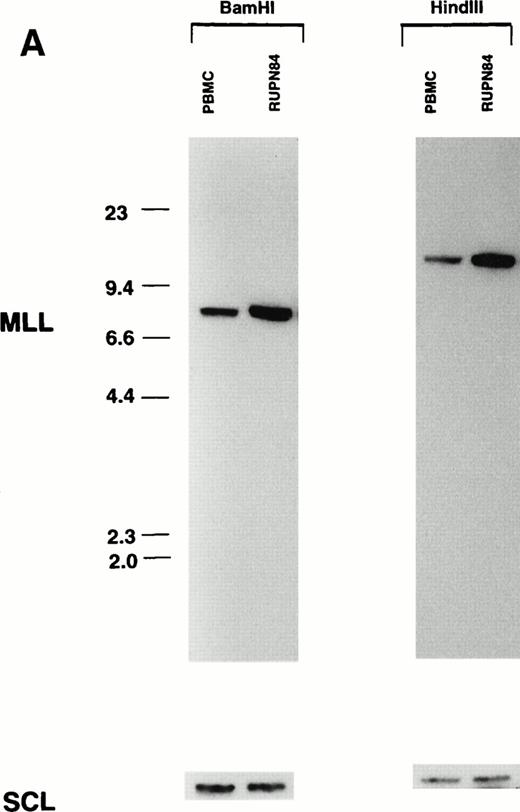
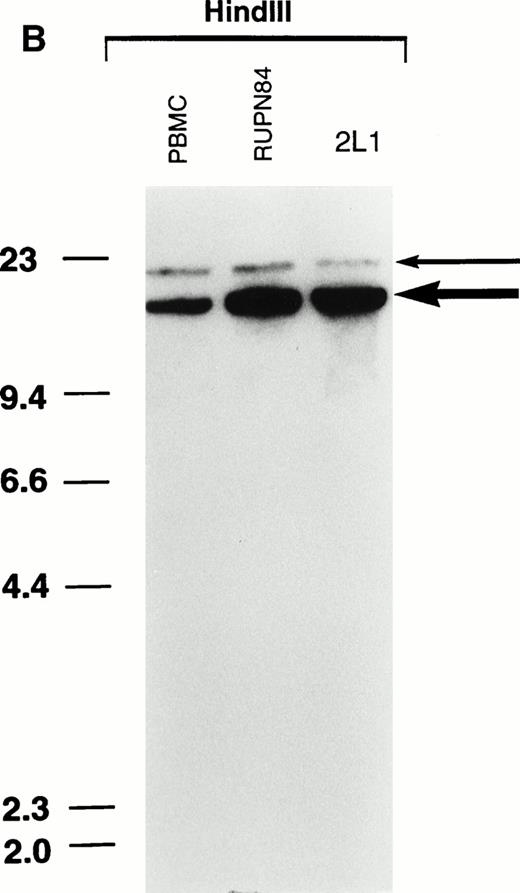
Consistent with the FISH analysis, Southern blot analysis suggested that there were multiple copies of the MLL gene in the t-AML of patient RUPN 84 and in the cell line 2L1 (Fig 2). The ratio of MLL signal intensities in the t-AML and in the cell line 2L1 were 4.3:1 compared with the peripheral blood mononuclear cell (PBMC) control when normalized for equal loading by hybridization with the SCLprobe. These results suggest approximately 8 to 9 copies of theMLL gene in the t-AML of patient RUPN 84 and in the cell line 2L1 derived from the leukemia (Fig 2). Although the patient received DNA topoisomerase II-targeted chemotherapy and had monoblastic leukemia, the MLL gene was not rearranged by Southern blot analysis. The results of Southern blot analysis with multiple restriction digests used in combination with MLL exon 3, exon 25, and exon 34 probes and a CD3ε probe showed similar unrearranged, amplified patterns, whereas hybridization with an 11q24 probe showed the unrearranged pattern but less amplification (2.0:1), again indicating that the amplified region extended both centromeric and telomeric of the MLL gene (data not shown).
Southern blot analysis demonstrating multiple copies of the MLL gene in t-AML of patient RUPN 84 and cell line 2L1.BamHI- and HindIII-digested DNAs were hybridized separately with MLL bcr cDNA and SCL genomic probes (A). Although the patient received DNA topoisomerase II-targeted chemotherapy and had monoblastic leukemia, the MLL gene was not rearranged. HindIII-digested DNAs were hybridized simultaneously with MLL bcr cDNA and SCL genomic probes to assess equivalence in loading (B). The bold arrow in (B) indicatesMLL signals; the thin arrow indicates SCL signals. The increased MLL signal intensity compared with PBMC control, 4.3:1 when normalized for loading by hybridization with SCLprobe, is consistent with approximately 8 to 9 copies of theMLL gene in the t-AML and cell line 2L1.
Southern blot analysis demonstrating multiple copies of the MLL gene in t-AML of patient RUPN 84 and cell line 2L1.BamHI- and HindIII-digested DNAs were hybridized separately with MLL bcr cDNA and SCL genomic probes (A). Although the patient received DNA topoisomerase II-targeted chemotherapy and had monoblastic leukemia, the MLL gene was not rearranged. HindIII-digested DNAs were hybridized simultaneously with MLL bcr cDNA and SCL genomic probes to assess equivalence in loading (B). The bold arrow in (B) indicatesMLL signals; the thin arrow indicates SCL signals. The increased MLL signal intensity compared with PBMC control, 4.3:1 when normalized for loading by hybridization with SCLprobe, is consistent with approximately 8 to 9 copies of theMLL gene in the t-AML and cell line 2L1.
Association of MLL gene amplification with p53 mutation.
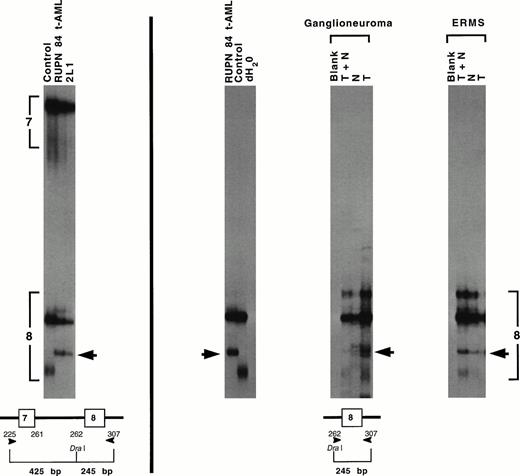
SSCP analysis of genomic DNAs from the marrow of patient RUPN 84 at diagnosis of t-AML and from the cell line 2L1 detected identical band shift patterns and LOH in the region of p53 exon 8 (Fig 3). Sequencing of 6 individual genomic subclones from 3 independent PCR reactions performed on the marrow DNA identified a CGA→TGA nonsense mutation at codon 306 that created a premature termination codon and would foreshorten the predicted protein. SSCP analysis of p53 exon 8 in DNA prepared from paraffin-embedded tissues showed the same band shift pattern in the normal tissues and in the sarcoma and ganglioneuroma, indicating that the p53 mutation was of germline origin. There was LOH in the sarcoma but not in the ganglioneuroma. As predicted with a truncated protein, p53 immunostaining was negative in the ganglioneuroma and in the sarcoma (data not shown).
Detection of germline p53 mutation in patient RUPN 84. SSCP analysis of genomic DNA from the t-AML and from cell line 2L1 showed identical band shift patterns with LOH in the region of p53 exon 8 (arrow, left). Schematic of strategy for PCR amplification andDra I restriction enzyme cleavage and resultant sizes of genomic DNA fragments containing p53 exon 7 and p53 exon 8 are shown below. SSCP analysis of p53 exon 8 in t-AML DNA and DNAs prepared from paraffin-embedded tissues showed the same band shift pattern in the ganglioneuroma, ERMS, and surrounding normal tissues, indicating that the p53 mutation was of germline origin (right). T, tumor tissue; N, normal tissue. There was LOH in the t-AML and the ERMS, but not in the ganglioneuroma. Sequencing of individual genomic subclones from t-AML DNA detected a CGA→TGA nonsense mutation at p53 codon 306 that created a premature termination codon and would foreshorten the predicted protein.
Detection of germline p53 mutation in patient RUPN 84. SSCP analysis of genomic DNA from the t-AML and from cell line 2L1 showed identical band shift patterns with LOH in the region of p53 exon 8 (arrow, left). Schematic of strategy for PCR amplification andDra I restriction enzyme cleavage and resultant sizes of genomic DNA fragments containing p53 exon 7 and p53 exon 8 are shown below. SSCP analysis of p53 exon 8 in t-AML DNA and DNAs prepared from paraffin-embedded tissues showed the same band shift pattern in the ganglioneuroma, ERMS, and surrounding normal tissues, indicating that the p53 mutation was of germline origin (right). T, tumor tissue; N, normal tissue. There was LOH in the t-AML and the ERMS, but not in the ganglioneuroma. Sequencing of individual genomic subclones from t-AML DNA detected a CGA→TGA nonsense mutation at p53 codon 306 that created a premature termination codon and would foreshorten the predicted protein.
DISCUSSION
We used FISH and Southern blot analysis to determine MLL gene copy number in a t-AML with a complex karyotype and monoblastic features and detected MLL gene amplification consistent with the jumping translocations of chromosomal segments containingMLL or ABL that were first discovered by Tanaka et al1 in 1997. Detailed molecular analyses demonstrated that the amplified MLL gene was not rearranged. The prior history of early onset ERMS and DNA damaging chemotherapy and radiation, and the central role of wild type p53 in maintaining genomic stability and ploidy,15 led to the investigation of the p53 gene and detection of the germline codon 306 mutation and LOH in the ERMS and the t-AML.
This case brings the total number of treatment-related leukemias with segmental jumping translocations that have been described to four. With additional detailed FISH and molecular analyses, the incidence of segmental jumping translocations in treatment-related leukemias should become apparent, because the abnormalities are not detected by karyotype alone. Consistent with the FISH analyses of Tanaka et al,1 we determined that there was intrachromosomal amplification of the specific segment containingMLL and that the segment containing MLL had moved to at least one other chromosome. Although the t-AML was monoblastic, Southern blot analysis showed that the amplified MLL gene, including regions 5′ and 3′ of the breakpoint cluster region, was not rearranged. The results of FISH and Southern blot analyses with CD3ε and 11q24 probes indicate that the amplified, unrearranged genomic region extended centromeric and telomeric of theMLL gene. Disruption of the breakpoint cluster region of theMLL gene by chromosomal translocation specifically is associated with the development of leukemia,16 but the role of MLL gene amplification in leukemogenesis currently is unknown.
The karyotype of the t-AML that we examined and the karyotypes of the other three leukemias with segmental jumping translocations were complex1 and suggest genomic instability. In this regard, the t-AML was similar to an alkylating agent-induced leukemia in which we previously detected a germline 2-bp deletion at p53 codon 209 that was inherited from the father.17 In the latter case, FISH analysis was not performed and it is not known whether there was a segmental jumping translocation, but the karyotype was 45, XY, hsr(2)(q22), −5, der(7)del(7)(q11.23)hsr(7)(q11.23), der(12)t(12;19)(p11.2;q12), der(17)t(5;17)(p12;p11.2),−19,+mar1.17
As was true for the child in the present study, the patient was diagnosed at an early age (1 year and 10 months) with primary ERMS, and there was not a family history of the Li-Fraumeni syndrome.17 In a study of patients with RMS without family histories of the Li-Fraumeni syndrome, Diller et al6detected germline p53 mutations in 3 of 13 children diagnosed before 3 years of age, but found no germline p53 mutations in 20 older children. These observations suggest that germline p53 mutations may predispose a fraction of young children undergoing therapy for RMS to t-AML, because the defective G1 cell cycle checkpoint that accompanies loss of wild-type p53 brings about genomic instability with DNA damaging chemotherapy and radiation.15
Wild-type p53 blocks cell cycle progression in late G1 in the presence of DNA damage caused by certain anticancer drugs and radiation and, depending on the level of the damage, either mediates apoptosis or permits DNA repair and cell cycle re-entry.3-5,18-23p53-dependent apoptosis is responsible, in part, for the cytotoxic activity of anticancer drugs and γ-radiation, while cells deficient in wild-type p53 are resistant to the induction of apoptosis by these agents.22-24
p53 mutant cells lose the ability to inhibit cell growth after DNA-damaging chemotherapy and γ-radiation.4,5 Thus, p53 was a candidate gene to examine in a t-AML with gene amplification because wild-type p53 maintains genomic stability and ploidy, whereas altered cell cycle arrest, gene amplification potential and aneuploidy occur with loss of wild-type p53.2,3 Furthermore, heterozygosity for mutant p53 does not result in gene amplification in experimental systems.2 3 In the leukemia in this study, p53 SSCP analysis showed both mutation and LOH and the karyotype showed chromosome 17 monosomy, which explains the LOH.
The child in the present study and 3 patients with t-AML withABL and MLL segmental jumping translocations reported on by Tanaka et al1 received heterogeneous chemotherapy and, in some cases, radiation. Our own observations suggest that young children with RMS and germline p53 mutations may be at increased risk for t-AML resulting from genomic instability on exposure to genotoxic agents.17 There is insufficient information to recommend treatment changes based on the current knowledge, but systematic study of p53 mutations and prior therapy in a larger cohort of patients with this form of t-AML may inform the rational design of individualized primary cancer treatment for at-risk individuals.
The results of this analysis establish that the pathogenesis of the gene amplification in treatment-related leukemias with segmental jumping translocations involves loss of wild-type p53. Just as gene amplification potential accompanies loss of wild-type p53 in Li-Fraumeni fibroblasts in vitro,2 germline p53 mutations with LOH may be associated with MLL gene amplification in t-AML. In vitro studies also indicate that there are alternative pathways that allow gene amplification when p53 is wild-type.2 The frequency of mutant p53 in treatment-related leukemias with segmental jumping translocations remains to be determined. Hartwell15 suggested that genomic instability is a genetic trait. The demonstration of a germline p53 mutation in association with the complex karyotype and MLL gene amplification in the t-AML in the present study proves that this is indeed the case. Future studies will explore the roles of the specific genetic changes resulting from the instability in the genesis of leukemia.
C.A.F. is supported by American Cancer Society Grant No. DHP143, National Institutes of Health Grant No. 1R29CA66140-03, a Leukemia Society of America Scholar Award (1996-2001), the National Childhood Cancer Foundation, a National Leukemia Research Association Grant in Memory of Maria Bernabe Garcia, and The Children's Hospital of Philadelphia High Risk High Impact Grant. P.D.A. is supported by National Institutes of Health Grants No. CA73773 and CA15606 and the Leukemia Society of America Scholar Award (1997-2002).
Address reprint requests to Carolyn A. Felix, MD, Division of Oncology, Leonard and Madlyn Abramson Pediatric Research Center, Room 902B, Children's Hospital of Philadelphia, 324 S 34th St, Philadelphia, PA 19104-4318.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.