Abstract
The t(16;21)(q24;q22) translocation is a rare but recurrent chromosomal abnormality associated with therapy-related myeloid malignancies and a variant of the t(8;21) translocation in which theAML1 gene on chromosome 21 is rearranged. Here we report the molecular definition of this chromosomal aberration in four patients. We cloned cDNAs from the leukemic cells of a patient carrying t(16;21) by the reverse transcription polymerase chain reaction using anAML1-specific primer. The structural analysis of the cDNAs showed that AML1 was fused to a novel gene named MTG16(Myeloid Translocation Gene on chromosome16) which shows high homology to MTG8(ETO/CDR) and MTGR1. Northern blot analysis usingMTG16 probes mainly detected 4.5 kb and 4.2 kb RNAs, along with several other minor RNAs in various human tissues. As in t(8;21), the t(16;21) breakpoints occurred between the exons 5 and 6 ofAML1, and between the exons 1 and 2 or the exons 3 and 4 ofMTG16. The two genes are fused in-frame, resulting in the characteristic chimeric transcripts of this translocation. Although the reciprocal chimeric product, MTG16-AML1, was also detected in one of the t(16;21) patients, its protein product was predicted to be truncated. Thus, the AML1-MTG16 gene fusion in t(16;21) leukemia results in the production of a protein that is very similar to the AML1-MTG8 chimeric protein.
SPECIFIC CHROMOSOMAL translocations are frequently found in hematopoietic malignant tumors and some types of solid tumors.1 Molecular analysis of the chromosomal translocations of leukemia has shown rearrangements of genes involved in the programmed regulation of proliferation and differentiation during hematopoietic development. In many myeloid leukemias, chromosomal alterations have been shown to result in the production of unusual chimeric proteins.2 3
A number of different and recurring aberrations involving chromosomal band 21q22 have been observed in human acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), and the blast crisis phase of chronic myelogenous leukemia (CML). Previously, we cloned the AML1 gene on chromosome 21q22 from patients with the t(8;21) translocation4 which occurs frequently (approximately 40%) in subtype M2 of AML.5 It was shown that the AML1gene is the most frequent target of chromosome translocations in human leukemia.6 In the t(8;21) translocation, the AML1gene was shown to be juxtaposed to the gene which encodes a zinc finger-containing protein, MTG8 (ETO/CDR), on chromosome 8q22, resulting in the expression of AML1-MTG8 chimeric proteins.7-10 In addition, the AML1 gene was found to be fused with the TEL gene, which encodes a member of the Ets family of transcription factors, to form a TEL-AML1 chimeric product by the t(12;21) translocation.11,12 The resultant chimeric transcripts are detected in pediatric B-cell progenitor acute lymphoblastic leukemia, the most common form of leukemia observed in children. Furthermore, AML1-containing fusion products are formed by the t(3;21) translocation which occurs in MDS and in the blast crisis phase of CML.13-17
Shimada et al18 have previously shown that a cosmid clone covering the region spanning exons 5 and 6 of the AML1 gene is split in fluorescence in situ hybridization (FISH) analysis of leukemic cells with t(16;21)(q24;q22) translocation. This is where the translocation breakpoints of t(8;21) AML are clustered. We therefore isolated the partner gene of AML1 by asymmetric polymerase chain reaction (PCR) using AML1-specific primers. The results show that the AML1 gene is juxtaposed to a novel gene,MTG16, on chromosome 16. Isolation and characterization of the wild-type MTG16 cDNA showed that MTG16 is another member of the MTG8 family of proteins, which display a high degree of sequence similarity. The AML1-MTG16 chimeric protein shares several structural features with AML1-MTG8, including the presence of the AML1 runt domain and the four evolutionary conserved motifs of MTG8.
MATERIALS AND METHODS
Patient samples.
Leukemia cells with t(16;21)(q24;q22) translocation were obtained from four patients suffering from malignant myeloid diseases. The clinical and cytogenetic data of the patients have been reported previously.18 19 One patient (no. 1) had a non-Hodgkin malignant lymphoma, but after receiving cytotoxic chemotherapy developed therapy-related AML in the absence of MDS. Two patients who had lung (no. 2) or oviductal (no. 3) cancer as their primary malignancies received cytotoxic chemotherapy. They were diagnosed as being in the transitional stage from therapy-related MDS to AML M2. One patient (no. 4) had de novo hypoplastic MDS.
Cloning of chimeric cDNA.
Total RNA was isolated from the peripheral lymphocytes of patient no. 1 by the acid guanidium thiocyanate/phenol/chloroform method.20 The poly(A)+ RNA was purified from total RNA using oligotex-dT30 (Daiichi kagaku-yakuhin, Tokyo, Japan). The cDNA was synthesized with random hexamer primers using the Marathon cDNA Synthesis Kit (Clontech, Palo Alto, CA) and was ligated with a cDNA adaptor according to the manufacturer's instruction. The fragments containing the chimeric cDNA were amplified by the asymmetric PCR method using an AML1 exon 5–specific primer, AMLex5f1 (CCACCTACCACAGAGCCATCAAAA) and adaptor-specific primers AP1 and/or AP2. PCR amplification was performed for a total of 35 cycles (94°C for 1 minute, 5 cycles of 94°C for 5 seconds, and 72°C for 4 minutes; 5 cycles of 94°C for 5 seconds, and 70°C for 4 minutes; and 25 cycles of 94°C for 5 seconds, and 68°C for 4 minutes) in a Gene Amp PCR system 9600 (Perkin-Elmer Japan, Chiba, Japan). Amplified fragments were size-fractionated by low melting temperature agarose gel electrophoresis and were cloned in a plasmid vector, pGEM-T Easy (Promega, Madison, WI). The AML1 exon 5- and exon 6-specific fragments were isolated using PCR primers AML1C (GAGGGAAAAGCTTCACTCTG) and AMLP (TTCGAGGTTCTCGGGGCCC), and ABF (GACATCGGCAGAAACTAGAT) and ABR (CCTGCATCTGACTCTGAGGC), respectively, and labeled with 32P using the Multiprime DNA labeling system (Amersham, Buckinghamshire, UK). PositiveAML1 exon 5 and negative AML1 exon 6 clones were selected by colony hybridization of the transformants. DNA sequencing was performed using the PRISM dye-terminator FS cycle sequencing kit and a ABI PRISM 377 DNA Sequencer (Perkin-Elmer Japan).
cDNA cloning.
Reverse transcription (RT)-PCR and primers.
From 1 μg of poly(A)+ RNA or total RNA of peripheral blood from patients and normal individuals, cDNAs were synthesized with random hexamer primers and reverse transcriptase using the Superscript Preamplification System (GlBCO-BRL, Rockville, MD). The reaction was diluted 20-fold and 0.5 μL was used for PCR. PCR amplification was performed for 35 cycles (94°C for 30 seconds, 58°C for 60 seconds, and 72°C for 60 seconds), followed by denaturation at 94°C for 3 minutes and extension at 72°C for 10 minutes. PCR products were separated by electrophoresis through a 1% Sea Plaque GTG agarose gel (FMC, Rockland, ME) in 1× TAE (Tris-acetate/EDTA electrophoresis buffer). Fragments were excised from the gel and sequenced directly. PCR primers for the AML1 and MTG16were designed according to the known cDNA sequences as follows: AMLex5f1 shown above, AMLex4f2 (GATGGCTGGCAATGATGAAAACTACTCG), AMLex6r2 (ACTCTGAGGCTGAGGGTTAAAGGCAGTG), MTG16r2 (GTTCTCGTTGACTTCCAGTAGCAG), and MF1 (GTGAAGACGCAGCCCCG).
Genomic cloning.
A P1 library of the total human genome (Du Pont, Wilmington, DE) was screened by the PCR method as described.23 Two P1 clones, P24H2 and P122F9, were isolated using PCR primers MTG16f8 (CGTCTCCATATGTGTAGGAAAGGAC) and MTG16r6 (CTATGTACACGGTCAGGGTCTTCC). The P1 clone P70A4 was isolated using PCR primers P122F9S-F1 (CTCTGCCTGGGATGATCC) and P122F9S-R (TCTGGCTGACCTGTCTTCG) obtained from the SP6-end sequence of P122F9. The location of exons was determined by restriction mapping and Southern blot analysis of P1 clones using various parts of MTG16 cDNA as probes. The exon-intron boundaries of the gene were determined by the direct sequencing of PCR-amplified fragments or subclones using primers taken from the cDNA sequence.
FISH.
P1 clones were labeled with biotin-16-dUTP and/or digoxigenin-11-dUTP by nick translation. Hybridization to metaphase cells was performed as described previously.24 The nuclear DNA was counter stained with 4,6-diamidino-2-phenylindole (DAPI).
Southern blot analysis.
Genomic DNAs were isolated from patient no. 1 and from a human normal lymphocyte cell line, C496. The DNAs were digested with EcoR1, separated by electrophoresis on an agarose gel, and transferred to NyTRAN 0,45 membrane (Schleicher & Schuell, Dassel, Germany). The PCR-amplified product (434 bp) obtained using primers 124r5R1 (AACAGTGCTGCCAGAACG) and MTG16r5 (CAGACCATAGACCATTTTAAGCAGCC), and the EagI-EcoRI restricted 2-kb fragment were used as probes. Hybridizations were performed at 42°C under stringent conditions. The final washing was in 0.1× standard saline citrate (SSC)/0.1% sodium dodecyl sulfate (SDS) at 65°C. Autoradiography was performed using a bioimage analyzer, Fujix BAS2000 (Fuji shashin film, Tokyo, Japan).
Northern blot analysis.
Membranes containing Poly(A)+ RNA from a wide variety of human tissues were purchased from Clontech. Hybridization and autoradiography were performed as for Southern blot analysis.
RESULTS
Cloning of fusion cDNA from a patient with t(16;21).
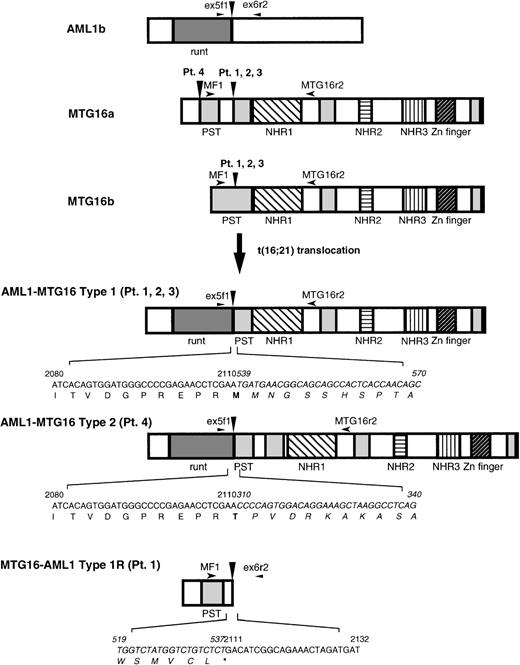
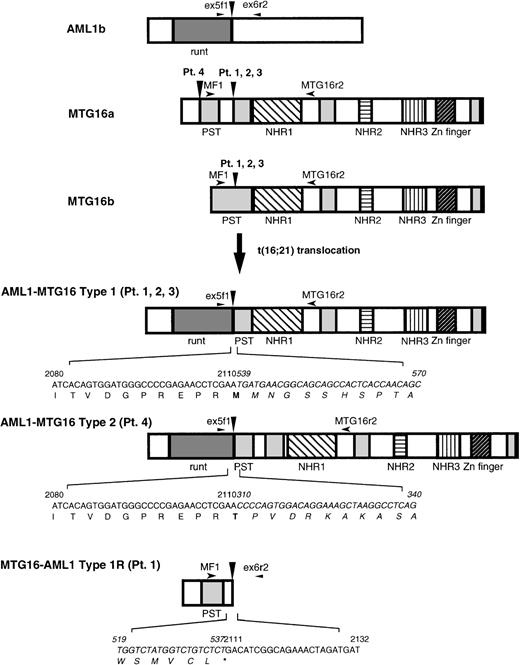
Because it was shown by FISH analysis18 that theAML1 gene is split in a region spanning exons 5 and 6 in t(16;21)(q24;q22) translocation, we performed asymmetric PCR using anAML1 exon 5-specific primer, AMLex5f1, to obtain the fusion cDNA. The fusion cDNA obtained from patient no. 1 shows that the 5′ portion of the AML1 gene is joined to a novel sequence in an in-frame manner. The 5′ portion of the fusion cDNA sequence corresponded to a nucleotide sequence present in AML1b mRNA (nucleotides 1983 to 2110; the nucleotide position 2110 corresponds to the end of exon 5).25 The sequence of the 3′ portion of the fusion cDNA was unknown, but showed significant homology toMTG8 (72% in nucleotide), a gene implicated in the t(8;21) translocation of acute myeloid leukemia.7 8 Hence we named this novel gene MTG16 (Myeloid TranslocationGene on chromosome 16).
Cloning and characterization of the wild-type MTG16 gene.
The wild-type MTG16 cDNA clones were isolated from the cDNA libraries of a human adult brain and a KG-1 myeloid cell line using the novel, partial sequence of MTG16 as a probe. Nucleotide sequence analysis of seven overlapping clones identified two types of composite cDNA sequences, named MTG16a and MTG16b, which differ in the sequences of their 5′ regions. MTG16a (identified as 4227 bp) and MTG16b (identified as 4024 bp) possess distinct 309-bp and 181-bp sequences, respectively, in their 5′-end portions. These sequences are then followed by a 153-bp sequence that is common to both cDNAs, by a 75-bp sequence specific toMTG16a, and by a common 3690-bp sequence present in the 3′ portion of both cDNAs (Fig 1; the identical 3′ sequence of 3690 bp in both types is not shown). We extensively searched MTG16b-type clones from a KG-1 cell line cDNA library, but could not find any clones containing the 75-bp sequence specific toMTG16a as shown in Fig 1. Each of the types was determined by sequencing at least two independent cDNA clones. Because these two types of MTG16 cDNAs were also detected in normal human peripheral blood using RT-PCR method (data not shown), both types probably correspond to alternatively spliced forms of MTG16. The predicted open reading frame of MTG16b cDNA has an in-frame stop codon before a potential methionine start codon at nucleotide position of 214 (Fig 1), and would code for a 567–amino acid protein.MTG16a cDNA, which has no in-frame stop codon and two potential, methionine start codons at nucleotide positions 149 and 222, would code for a 653–amino acid protein if translation started from the former methionine (Fig 1). We have no evidence in favor of the use of either methionine start codon, but the upstream sequence containing the first methionine codon has been confirmed repeatedly.
Nucleotide sequences of the 5′ portions of MTG16a and MTG16b cDNA. The 5′-end sequences that differ between MTG16a and MTG16b cDNA and the deduced amino acid sequences are shown. The shaded area of the sequence indicates the 153 nucleotide region common to both MTG16a andMTG16b cDNA and the underlined sequence indicates the 75 nucleotide region specific to MTG16a. ATG codons are boxed and their upstream stop codons are double-underlined. The entire nucleotide sequence data reported here has been deposited in the DDBJ/EMBL/Genebank database under the accession number AA010419 andAA010420.
Nucleotide sequences of the 5′ portions of MTG16a and MTG16b cDNA. The 5′-end sequences that differ between MTG16a and MTG16b cDNA and the deduced amino acid sequences are shown. The shaded area of the sequence indicates the 153 nucleotide region common to both MTG16a andMTG16b cDNA and the underlined sequence indicates the 75 nucleotide region specific to MTG16a. ATG codons are boxed and their upstream stop codons are double-underlined. The entire nucleotide sequence data reported here has been deposited in the DDBJ/EMBL/Genebank database under the accession number AA010419 andAA010420.
Both of the predicted protein products of MTG16a and MTG16b are highly homologous to MTG8b (67% and 75% identity, respectively)7,8 and MTGR1, another member of the MTG8 family (54% and 61% identity, respectively).26 From a comparison of the amino acid sequences of MTG8b, MTGR1, and Nervy,27 a putative Drosophila homologue of MTG8, we previously identified four evolutionary conserved regions termed NHR (Nervy homology region) 1, 2, and 3, and a zinc finger domain (or NHR4).26 Figure 2 shows that these NHRs and the C-terminal zinc finger domain are completely conserved in MTG16a and MTG16b. Three proline/serine/threonine-rich regions (designated as PST) found in MTG8 and MTGR1 are also conserved in MTG16 at the corresponding position, except that the first PST region is disrupted by an intervening 25–amino acid region in MTG16a.
Comparison of the amino acid sequences of MTG16a, MTG16b, MTG8b, and MTGR1. Identical residues shared by more than two proteins are shaded. Horizontal bars above the sequences indicate the positions of NHR1, NHR2, NHR3, and the zinc finger domain. Horizontal wavy lines also indicate the positions of PST region. The cysteine residues of the zinc finger motifs are marked by an asterisk below the sequence. The amino acid sequences of the two proteins are identical to one another after the 127th leucine of MTG16a and after the 41st leucine of MTG16b.
Comparison of the amino acid sequences of MTG16a, MTG16b, MTG8b, and MTGR1. Identical residues shared by more than two proteins are shaded. Horizontal bars above the sequences indicate the positions of NHR1, NHR2, NHR3, and the zinc finger domain. Horizontal wavy lines also indicate the positions of PST region. The cysteine residues of the zinc finger motifs are marked by an asterisk below the sequence. The amino acid sequences of the two proteins are identical to one another after the 127th leucine of MTG16a and after the 41st leucine of MTG16b.
MTG16 mRNA is expressed in many tissues with broad tissue specificity. The most common species were 4.5 kb, 4.2 kb, and 3.1 kb, with the exception of liver where only 2.95-kb and 2.5-kb transcripts were detected. The MTG16 expression level is relatively high in heart, pancreas, skeletal muscle, spleen, thymus, and peripheral blood leukocytes, and low in testis and ovary. No MTG16 RNA was detected in kidney (Fig 3). The 4.5-kb and 4.2-kb species of RNA seem to correspond to the MTG16a andMTG16b transcripts, respectively, because their lengths coincide well with the sizes of the corresponding cDNAs. The origin and significance of the 3.1-kb RNA, and the 2.95-kb and 2.5-kb RNAs specific to liver remain to be determined. These might represent alternative splicing forms of MTG16.
Northern blot analysis of the expression of MTG16in human tissues. Northern blot filters from human adult tissues (Clontech) were used for analysis. Probes: (A), whole MTG16a cDNA (nucleotides 1 to 4227); (B), G3PDH. Lanes 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, pancreas; 9, spleen; 10, thymus; 11, prostate; 12, testis; 13, ovary; 14, small intestine; 15, colon; 16, peripheral blood leukocyte.
Northern blot analysis of the expression of MTG16in human tissues. Northern blot filters from human adult tissues (Clontech) were used for analysis. Probes: (A), whole MTG16a cDNA (nucleotides 1 to 4227); (B), G3PDH. Lanes 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, pancreas; 9, spleen; 10, thymus; 11, prostate; 12, testis; 13, ovary; 14, small intestine; 15, colon; 16, peripheral blood leukocyte.
Genomic organization of MTG16 gene.
The genomic clones P24H2 and P122F9 were isolated from a human genomic P1 library using the PCR primers specific to the 3′ portion ofMTG16 cDNA. Restriction mapping and MTG16 exon mapping of P24H2 and P122F9 revealed that they overlapped in the 3′ region of the gene and that P122F9 contained most but not all of theMTG16 gene. Genomic walking from the SP6-end of P122F9 was performed and yielded the clone P70A4 containing the 5′-end sequence ofMTG16 cDNA. The MTG16 gene has 13 exons (1a, 1b, and 2-12) which have been aligned on the restriction map of P70A4 and P122F9 in Fig 4. Genomic sequencing analysis revealed that exon 1b was located at the terminus of the 18kbNotI-SalI fragment of P70A4. Detailed restriction analysis of the region containing exon 1a was not performed, but it was mapped to the 40 kb NotI fragment of P70A4 by Southern hybridization. The exon/intron boundary sequences of the MTG16gene were determined (Table 1). All of exon/intron boundaries determined possess consensus splice donor and acceptor sequences,28 including the GT-AG motif. The sizes of introns (intron 2 to 11) were also determined by genomic PCR. Based on this map, we estimate that the MTG16 gene is larger than 73 kb, although the location of exon 1a has not been determined precisely.
The structure of MTG16 gene. Three P1 clones covering the entire MTG16 gene are shown in the upper part of the figure. Exons of the MTG16 gene are represented by boxes on the restriction map. Horizontal bars indicate breakpoints in t(16;21) patients. Breakpoints in patients 2 and 3 indicate sites predicted from cDNA sequences. Sa, N, E, and Bg indicate the restriction site forSa/I, NotI, EcoRI, and BglII, respectively. Horizontal bars under the restriction map indicates the probes that were used in Southern blot analysis of the genomic DNAs. Schematic representation of alternative splicing forms of theMTG16 transcripts, MTG16a and MTG16b, are also shown. The positions of deduced start codons and stop codons are shown by ATG and TGA, respectively.
The structure of MTG16 gene. Three P1 clones covering the entire MTG16 gene are shown in the upper part of the figure. Exons of the MTG16 gene are represented by boxes on the restriction map. Horizontal bars indicate breakpoints in t(16;21) patients. Breakpoints in patients 2 and 3 indicate sites predicted from cDNA sequences. Sa, N, E, and Bg indicate the restriction site forSa/I, NotI, EcoRI, and BglII, respectively. Horizontal bars under the restriction map indicates the probes that were used in Southern blot analysis of the genomic DNAs. Schematic representation of alternative splicing forms of theMTG16 transcripts, MTG16a and MTG16b, are also shown. The positions of deduced start codons and stop codons are shown by ATG and TGA, respectively.
Expression of the AML1-MTG16 fusion gene in t(16;21) AML patients.
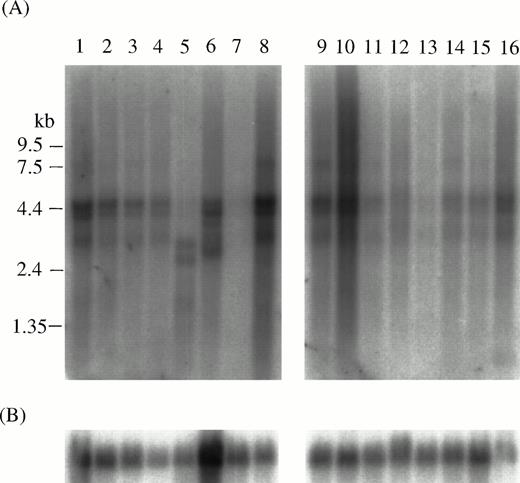
The expression of AML1, MTG16, AML1-MTG16, and MTG16-AML1 was examined by RT-PCR method using total RNA isolated from the peripheral blood of t(16;21) patients and a normal control. AML1 and MTG16were detected with the expected product size in both the t(16;21) patients and normal individuals (Fig 5C and D). On the other hand, AML1-MTG16 chimera was detected as a product of 545 bp in three t(16;21) AML patients (no. 1, 2, and 3) and as a product of 773 bp in patient no. 4 using AMLex5f1 and MTG16r2 primers (Fig 5A). Sequence analysis of the PCR-amplified chimericAML1-MTG16 fragments indicated that three of the four patients (no. 1, 2, and 3) had breaks between exon 3 and exon 4 (type 1), and that one patient (no. 4) had a break between exon 1 and exon 2 ofMTG16 (type 2) (Fig 6). The predicted products of AML1-MTG16 would be 704 amino acids (type 1) and 780 amino acids (type 2) in length.
RT-PCR analysis of the AML1-MTG16 and theMTG16-AML1 chimeric transcripts in four t(16;21) patients. Poly(A)+ RNA and total RNA samples from peripheral blood were reverse transcribed into cDNA. The primer pairs, AML1ex5f1 and MTG16r2, MF1 and AML1ex6r2, AML1ex4f2 and AML1ex6r2, and MF1 and MTG16r2 were used for amplification of AML1-MTG16 (A),MTG16-AML1 (B), AML1 (C), and MTG16 (D) transcripts, respectively (their locations are shown in Fig 6). Lane 1, poly(A)+ RNA from patient no. 1; lanes 2 through 5, total RNA from patients no. 1 through 4; lanes 6 and 7, total RNA from normal individuals; lane 8, template-free. Arrows indicate the detected transcripts with their sizes in bp. The two AML1-MTG16 chimeric products (545 bp in lanes 1 through 4 and 773 bp in lane 5 seen in [A]) and the one MTG16-AML1 chimeric product (357 bp in lanes 1 and 2 seen in [B]) were purified and sequenced.
RT-PCR analysis of the AML1-MTG16 and theMTG16-AML1 chimeric transcripts in four t(16;21) patients. Poly(A)+ RNA and total RNA samples from peripheral blood were reverse transcribed into cDNA. The primer pairs, AML1ex5f1 and MTG16r2, MF1 and AML1ex6r2, AML1ex4f2 and AML1ex6r2, and MF1 and MTG16r2 were used for amplification of AML1-MTG16 (A),MTG16-AML1 (B), AML1 (C), and MTG16 (D) transcripts, respectively (their locations are shown in Fig 6). Lane 1, poly(A)+ RNA from patient no. 1; lanes 2 through 5, total RNA from patients no. 1 through 4; lanes 6 and 7, total RNA from normal individuals; lane 8, template-free. Arrows indicate the detected transcripts with their sizes in bp. The two AML1-MTG16 chimeric products (545 bp in lanes 1 through 4 and 773 bp in lane 5 seen in [A]) and the one MTG16-AML1 chimeric product (357 bp in lanes 1 and 2 seen in [B]) were purified and sequenced.
Schematic diagram of the wild-type proteins of AML1 and MTG16, the chimeric proteins AML1-MTG16 type 1 and type 2, and the reciprocal chimeric protein MTG16-AML1 type 1R detected in patient no. 1. Vertical arrowheads indicate breakpoints of AML1 and MTG16. Horizontal arrowheads indicate the location of primers used for RT-PCR. Regions that correspond to the structural or functional domains of the proteins are shown: runt, runt domain; PST, proline/serine/threonine-rich region; NHR1-3, Drosophila Nervy homologous region 1-3; Zn finger, zinc finger domain. Nucleotide sequences and positions near the junction of chimeric proteins are also shown and those derived from MTG16 are indicated by italic.
Schematic diagram of the wild-type proteins of AML1 and MTG16, the chimeric proteins AML1-MTG16 type 1 and type 2, and the reciprocal chimeric protein MTG16-AML1 type 1R detected in patient no. 1. Vertical arrowheads indicate breakpoints of AML1 and MTG16. Horizontal arrowheads indicate the location of primers used for RT-PCR. Regions that correspond to the structural or functional domains of the proteins are shown: runt, runt domain; PST, proline/serine/threonine-rich region; NHR1-3, Drosophila Nervy homologous region 1-3; Zn finger, zinc finger domain. Nucleotide sequences and positions near the junction of chimeric proteins are also shown and those derived from MTG16 are indicated by italic.
The expression of the reciprocal MTG16-AML1 chimera was examined using MF1 and AMLex6r2 primers. A 357-bp PCR product of the expected size was observed specifically in the leukemic cells of patient no. 1, but it did not appear in the other patients (Fig 5B). Because patients no. 1, 2, and 3 expressed the same AML1-MTG16fusion product, we expected that the reciprocal MTG16-AML1product would be of the same size in these patients. However, the same 357-bp product was not repeatedly detected in the remaining two patients (no. 2 and 3) by RT-PCR. This fact might indicate that an additional aberration such as a deletion had occurred at the junction point on the der(21) chromosome. At least in the peripheral blood of patient no. 1, the MTG16-AML1 chimera derived from der(21) chromosome was expressed as well as AML1-MTG16 chimera derived from der(16) chromosome. However, sequence analysis predicted that the resulting MTG16-AML1 product would be truncated at the fusion point, because the UGA stop codon is located at the MTG16-AML1junction (Fig 6). The absence of MTG16-AML1 transcripts from most patients and the presence of a putative, truncatedMTG16-AML1 protein in patient no. 1 suggest thatAML1-MTG16 rather than MTG16-AML1 is involved in the pathogenesis of t(16;21) leukemia.
FISH analysis and detection of genomic rearrangements.
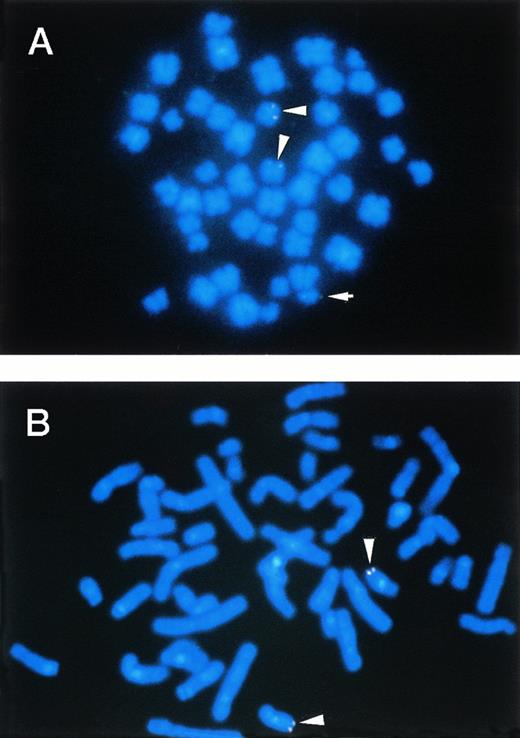
To confirm the rearrangements of the MTG16 gene in t(16;21) patients, FISH analysis and Southern blot analysis were performed using the genomic P1 clone, P122F9 and P24H2, or DNA fragments derived from P122F9. The location of P122F9 on chromosome 16q24 of a normal human individual was confirmed by metaphase FISH (Fig 7B). On the other hand, P122F9 signals were detected as triple signals in the t(16;21)(q24;q22) patient (no. 3) (Fig 7A). P24H2 containing the 3′ portion of MTG16 gene located distal to the breakpoints was mapped on the der(16) chromosome as well as on the normal chromosome 16 of patient no. 3 (data not shown), indicating that the direction of transcription of MTG16 is from telomere to centromere, like in the case of AML1.5
FISH. (A) and (B) illustrate the mapping of the P1 clone P122F9 in cells from the patient no. 3 and in cells from a normal individual, respectively. The P122F9 probe hybridized to both of the der(16) and the der(21) chromosomes, as well as to normal chromosome 16 in patient no. 3. The normal 16 and the der(16) chromosomes are indicated by an arrowhead, and the der(21) chromosome is indicated by an arrow.
FISH. (A) and (B) illustrate the mapping of the P1 clone P122F9 in cells from the patient no. 3 and in cells from a normal individual, respectively. The P122F9 probe hybridized to both of the der(16) and the der(21) chromosomes, as well as to normal chromosome 16 in patient no. 3. The normal 16 and the der(16) chromosomes are indicated by an arrowhead, and the der(21) chromosome is indicated by an arrow.
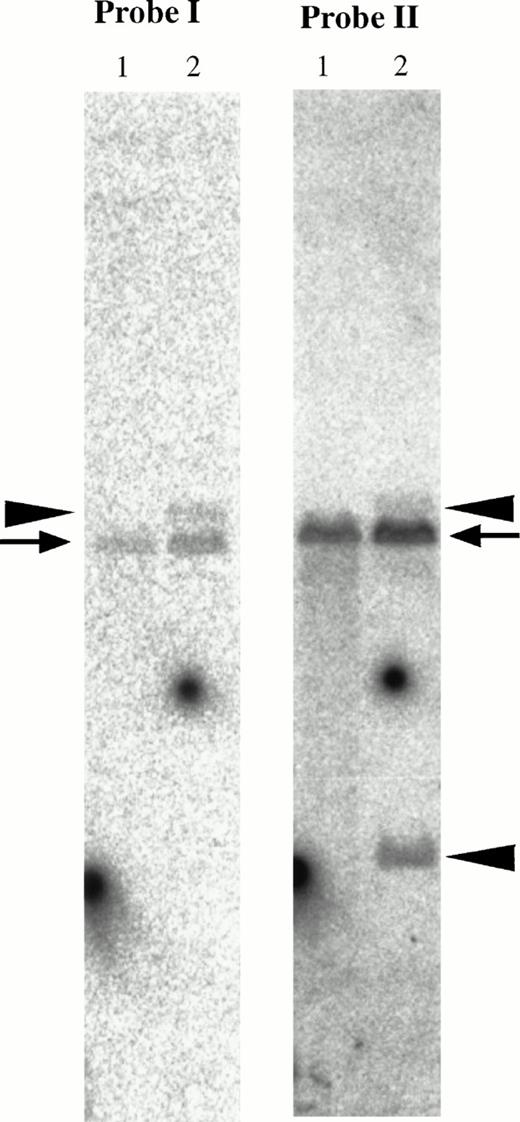
In genomic Southern hybridization of patient no. 1, the downstream fragment of MTG16 exon 3 (the 2-kb EagI-EcoRI fragment shown as probe II in Fig 4) detected rearranged bands of 15 kb and 3.7 kb in addition to a germline 12-kb EcoRI band. On the other hand, the upstream fragment of exon 3 (PCR-amplified 434-bp fragment shown as probe I in Fig 4) detected the 15-kb rearranged band and the normal band (Fig 8). As the translocation breakpoint in patient no. 1 was found to be located between exon 3 and exon 4 of MTG16 by sequence analysis of its fusion cDNA, these results suggest that the breakpoint is located within the 2-kb region, downstream of exon 3 of MTG16.
Southern blot analysis of the MTG16 gene. Genomic DNAs were digested with EcoRI and hybridized with probe I and II shown in Fig 4. Probe I; the PCR-amplified 434-bp genomic fragment using the primers 124r5R1 and MTG16r5. Probe II; theEagI-EcoRI 2-kb fragment. Rearranged bands (15-kb band derived from der(21) chromosome and 3.7-kb band derived from der(16) chromosome) and germline bands (12 kb) are shown by arrowheads and an arrow, respectively. Lane 1, a human normal leukocyte cell line C496 was used as a control; lane 2, patient no. 1.
Southern blot analysis of the MTG16 gene. Genomic DNAs were digested with EcoRI and hybridized with probe I and II shown in Fig 4. Probe I; the PCR-amplified 434-bp genomic fragment using the primers 124r5R1 and MTG16r5. Probe II; theEagI-EcoRI 2-kb fragment. Rearranged bands (15-kb band derived from der(21) chromosome and 3.7-kb band derived from der(16) chromosome) and germline bands (12 kb) are shown by arrowheads and an arrow, respectively. Lane 1, a human normal leukocyte cell line C496 was used as a control; lane 2, patient no. 1.
DISCUSSION
In this study, we analyzed the translocation breakpoint in t(16;21)(q24;q22) and identified a novel fusion gene consisting of a partial sequence from AML1 on chromosome 21 and MTG16on chromosome 16. The t(16;21)(q24;q22) translocation is a rare but recurrent chromosomal abnormality associated with therapy-related AML or MDS.18,19,29 Sequence analysis of MTG16 cDNA revealed two alternative splicing iso-forms, termed MTG16a andMTG16b, which contain different 5′-end exons. Sequence comparison of the predicted amino acid sequences of MTG16a and MTG16b with that of MTG8b shows 67% and 75% identity, respectively, and the sequence similarity extends over the entire coding region, with the exception of the N-terminal amino acid sequences. Interestingly, the MTG16 protein moiety of the fusion gene product faithfully comprises the four evolutionary conserved motifs, NHR1, NHR2, NHR3, and the zinc finger domain (NHR4) which were uncovered by sequences comparison between MTG8 and Nervy,26 as well as the corresponding MTG8 portion of AML1-MTG8. The presence of common structural features between MTG8 and MTG16 suggests that AML1-MTG16 may possess similar, if not the same biological activity as AML1-MTG8.
AML1 is a transcription factor and forms heterodimeric complex with CBFβ (also known as PEBP2β).30,31 The AML1 transcription factor complex regulates the transcription of target genes by binding to the DNA sequence TGT/cGGT.30-33Possible transcriptional targets include the T-cell antigen receptors,32,34,35 the colony-stimulating factor 1 receptor,36 myeloperoxidase, neutrophil elastase,37 granulocyte-macrophage colony-stimulating factor,38,39 and granzyme B.40 Targeted disruption has shown that both AML1 and CBFβ/PEBP2β are essential for all lineages of definitive hematopoiesis in mouse fetal liver.41-45 All of these studies indicated tight association between AML1 and hematopoiesis. On the contrary, little is known about the functional activity of MTG8, the fusion partner of the t(8;21) translocation, in leukemogenesis. The present finding that MTG16, a member of the MTG8 family of proteins, is targeted by a leukemia-causing translocation may indicate that the partner genes MTG8 and MTG16 are in some way important for the leukemogenic nature of the fusion gene products. AML1-MTG8 inhibits granulocytic differentiation in L-G mouse myeloid progenitor cells and AML1a, which lacks the transactivation domain, shows less inhibitory activity.26 Additional support for this prediction comes from another member of the MTG8 family, named MTGR1, which may be responsible for the repression of AML1-dependent transcription and, possibly, leukemogenesis.26 All of these observations strengthen the need for studies to elucidate how MTG8 family members contribute to the mechanism of myeloid leukemogenesis.
Three out of the four patients examined here developed therapy-related myeloid malignancies after chemotherapy treatment with agents that included etoposide or adriamycin. The breakpoint in the de novo MDS patient was mapped in the large intron (greater than 45 kb) between exons 1 and 2, whereas the breakpoints of therapy-related leukemia were mapped to another region. It will be interesting to examine if these breakpoints are associated with the presence of scaffold attachment regions and high-affinity topoisomerase II binding sites, which have been observed during the mapping of breakpoints within the MLLgene (also called ALL-1, HTRX1, orHRX).46-49 The t(16;21) translocation, which is predominantly found in therapy-related leukemia, might become another valuable system for the analysis of the molecular mechanism of recombination.
The t(8;21) chromosome translocation is one of the most frequent chromosome abnormalities in leukemia, whereas t(16;21) is quite rare. The molecular mechanism of recombination that leads to chromosome translocation remains unsolved. In the present study, we showed that the structural characteristics of AML1-MTG8 in t(8;21) are strongly conserved in AML1-MTG16 resulting from t(16;21). One of the parameters that might determine the frequency of occurrence of these translocations is the size of target introns. The breakpoints of both t(8;21) and t(16;21) occur within the same intron of AML1, which is located immediately downstream of a phylogenetically conserved DNA-binding domain (the runt box). Therefore, the frequency could be determined by the targeted intron size of the partner genes,MTG8 and MTG16. Most breakpoints on chromosome 8 fall within two introns, between two alternative 5′ MTG8 exons 1b and 1a and between exons 1a and 2, and the breakpoint clustered region is estimated about 20 kb in size.50 In t(16;21), chromosome breakages occur within two introns of MTG16, the sizes of which total more than 51 kb. These estimated sizes do not reflect the known frequency of both leukemias. Thus, the preferential occurrence of the t(8;21) translocation in leukemia would seem to suggest that the leukemogenic potential of AML1-MTG16 is not equivalent to that of AML1-MTG8 and leukemia with t(16;21) requires an another defect in addition to the formation of AML1-MTG16 fusion gene, or that there exist recombination hotspots or recombination-specific signals inMTG8 gene. Functional analysis of both pathogenic gene products as well as nucleotide sequence analysis of the recombinational junction sites should help to understand this difference in the frequency.
ACKNOWLEDGMENT
We are grateful to Dr T. Matsumoto and Dr K. Matsushita of Imamura Hospital, and Dr T. Shimizu of Isehara Kyodo Hospital for providing patient samples. We thank Kazusa DNA Research Institute Foundation for support to a cDNA Research Program.
Supported in part by the Program of Fundamental Studies in Health Sciences of the Organization for Drug ADR Relief, R&D Promotion and Product Review of Japan; by a Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture; by a grant from the Special Coordination Funds for the Promotion of Science and Technology from Science and Technology Agency; by a Grant-in-Aid for the Comprehensive 10-year Strategy for Cancer Control; and by the Grant-in-Aid for Cancer Research from the Ministry of Health and Welfare of Japan.
Address reprint requests to Fumie Hosoda, PhD, Radiobiology Division, National Cancer Center Research Institute, 5-1-1, Tsukiji, Chuo-ku, Tokyo 104, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.





![Fig. 5. RT-PCR analysis of the AML1-MTG16 and theMTG16-AML1 chimeric transcripts in four t(16;21) patients. Poly(A)+ RNA and total RNA samples from peripheral blood were reverse transcribed into cDNA. The primer pairs, AML1ex5f1 and MTG16r2, MF1 and AML1ex6r2, AML1ex4f2 and AML1ex6r2, and MF1 and MTG16r2 were used for amplification of AML1-MTG16 (A),MTG16-AML1 (B), AML1 (C), and MTG16 (D) transcripts, respectively (their locations are shown in Fig 6). Lane 1, poly(A)+ RNA from patient no. 1; lanes 2 through 5, total RNA from patients no. 1 through 4; lanes 6 and 7, total RNA from normal individuals; lane 8, template-free. Arrows indicate the detected transcripts with their sizes in bp. The two AML1-MTG16 chimeric products (545 bp in lanes 1 through 4 and 773 bp in lane 5 seen in [A]) and the one MTG16-AML1 chimeric product (357 bp in lanes 1 and 2 seen in [B]) were purified and sequenced.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/11/10.1182_blood.v91.11.4028/4/m_blod41145005w.jpeg?Expires=1766019230&Signature=rb3WMR96I7jdkRqTULk0hkw-L7Dl8YRsZDa59vshthGjpxCtjK-wzBiV9xj3HM7lB4NfF5wQD6uAhxxOJetH2sbphfOuvF5-bdaYmtlAzH1qOsOtuyYGJ3MQENTlSLV7T7D8H8oXRaFnoLZL8246JnKIX-Qv8gzf9oKTzOlNNquQvCtwdA3OUKgEZO7e8QJWHJ8e2VbHFy-7zJQgt22p13issnQ-cAN7tXd3xWY6Adv~spJckGkr-fcrz~lqZKZL3KPkBYqgJ3eWkUjCkfrWnXnK8doSh5VqyO4W91DDvvG72oYCCZ~DmctgmEvEH3DzLNE9PBgbRcZzEjqI2latOQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)






![Fig. 5. RT-PCR analysis of the AML1-MTG16 and theMTG16-AML1 chimeric transcripts in four t(16;21) patients. Poly(A)+ RNA and total RNA samples from peripheral blood were reverse transcribed into cDNA. The primer pairs, AML1ex5f1 and MTG16r2, MF1 and AML1ex6r2, AML1ex4f2 and AML1ex6r2, and MF1 and MTG16r2 were used for amplification of AML1-MTG16 (A),MTG16-AML1 (B), AML1 (C), and MTG16 (D) transcripts, respectively (their locations are shown in Fig 6). Lane 1, poly(A)+ RNA from patient no. 1; lanes 2 through 5, total RNA from patients no. 1 through 4; lanes 6 and 7, total RNA from normal individuals; lane 8, template-free. Arrows indicate the detected transcripts with their sizes in bp. The two AML1-MTG16 chimeric products (545 bp in lanes 1 through 4 and 773 bp in lane 5 seen in [A]) and the one MTG16-AML1 chimeric product (357 bp in lanes 1 and 2 seen in [B]) were purified and sequenced.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/11/10.1182_blood.v91.11.4028/4/m_blod41145005w.jpeg?Expires=1766252264&Signature=qY3xwvHycgDgpNTX2PXUyb01CQ~aRk4fcVvy0MSgnZ1LivD2AcaaOUS~UuBVdjMtNgbkVijv69FKWWeEFbOGOwAM7bLEFg92JqjQgOHXuwjgRm3oqoVloDqSSyBuoaFL4UW8CCuiF4V5uNNoFqUOH~EJ6~ZGQrGgOYCcU47Suykyu4Q8CqiwqYjfchKdaOcnvnOy3znhca-zng3U4W50vJZWhvqnQfhH5M0OIR-1b7LUg0tCTySYPTLcknOEiyW5dKKHJpGAPxz9GdrZLI7lcjp9F60MIYAPKxtogBrWeP2gbWNxRDVBGasrEM4nveskMWaMgtwJT5JA-ALgaYFwyg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)