Abstract
In each case of acute promyelocytic leukemia (APL) one of three PML-RARα mRNA types is produced, depending on the break/fusion site in the PML gene that is linked to a common RARα gene segment: a short (S)-form type, PML exon 3 RARα exon 3; a long (L)-form type, PML exon 6 RARα exon 3; or a variable (V)-form type, variably deleted PML exon 6 RARα exon 3. We evaluated whether PML-RARα mRNA type is associated with distinct pretreatment clinical characteristics and therapeutic outcome in previously untreated adult APL patients registered to protocol INT 0129 by the Eastern Cooperative Oncology Group, the Southwest Oncology Group, and the Cancer and Leukemia Group B. Of 279 clinically eligible cases, 230 were molecularly evaluable, and of these, 111 were randomized to receive remission induction therapy with all-trans retinoic acid (ATRA) and 119 with conventional chemotherapy. Nine cases not excluded by central pathology review were PML-RARα negative, and notably, none of five of these cases treated with ATRA achieved complete remission (CR). Among 221 PML-RARα–positive cases, there were 82 S-form cases (37%), 121 L-form cases (55%), and 18 V-form cases (8%). Before any antileukemic therapy, the S-form type, compared with the L-form type, was associated with higher values for the white blood cell (WBC) count (median 2,500/μL v 1,600/μL; P = .009), the percentage of blood blasts plus promyelocytes (median 29% v 8.5%; P = .03), and the absolute blood blasts plus promyelocytes (884/μL v 126/μL; P = .019). Also, an increased percentage of S-form versus L-form cases had the M3 variant phenotype, 24% v 12% (P = .036). There were no differences between S-form and L-form cases in either CR rate (79% v 69%; P = .14) or disease free survival distribution (multivariate analysis adjusting for the association of S-form type and higher WBC count; P = .40). We conclude that the S-form type is associated with previously-identified adverse risk WBC parameters but that the identification of the S-form or L-form type of PML-RARα mRNA, per se, does not predict clinical outcome or add to the value of an increased WBC count as a negative prognostic indicator in APL patients.
THE CHROMOSOME translocation (15; 17) (q22; q21) is uniquely associated with acute promyelocytic leukemia (APL) and results in the fusion of two genes: PML (15q22) and RARα (17q21) (reviewed in Grignani et al1 ). Three different types of PML-RARα mRNA fusion transcripts can be formed because of different breakpoints in the PML gene: in intron 3, intron 6, or exon 6. In each APL patient, the 3′-end truncated PML transcript is fused to a common 5′-truncated RARα component generated by breaksites in intron 2 of the RARα gene to form the corresponding three types of PML-RARα mRNA: a short (S)-form type (PML exon 3 RARα exon 3), a long (L)-form type (PML exon 6 RARα exon 3) or a variable (V)-form type (PML partial exon 6 RARα exon 3).2-8
An important clinical consideration is whether PML-RARα mRNA type is associated with differences in presenting patient characteristics and/or is predictive of therapeutic responses in individual APL cases. This seems crucial to establish in APL, because this form of acute myeloid leukemia is unique in its responsiveness to the natural product all-trans retinoic acid (ATRA), which functions primarily by inducing terminal differentiation of APL cells.9-11 These variations in PML-RARα type might differentially affect the ability of this oncoprotein to form critical oligomeric interactions with other macromolecules involved in the transmission of ATRA-mediated signals related to cell differentiation and, thereby, alter therapeutic responsiveness in different molecular types of APL.2-4,12 13
In the current study, we sought to determine if PML-RARα mRNA type is associated with differences in pretreatment clinical parameters or response to therapy in adult APL patients entered on the cooperative intergroup clinical trial, Protocol INT 0129. In this trial, previously untreated patients with morphologically defined APL were randomized to receive induction therapy with either ATRA or chemotherapy and, after consolidation chemotherapy, complete remission (CR) cases were rerandomized to observation or maintenance ATRA for 1 year.14,14a The 230 study cases provided by INT 0129 represented the largest cohort of uniformly treated and evaluated patients so far subjected to correlative clinical and molecular analysis. Three previously reported studies with 28 to 53 cases have suggested that the S-form type of PML-RARα may be associated with an adverse clinical outcome in previously untreated APL patients after somewhat variably administered ATRA and/or chemotherapy remission induction therapy.15-17 In a fourth report, no difference in prognosis between S-form and L-form type PML-RARα cases was found in either previously untreated patients (45 cases) or patients relapsing from and/or refractory to chemotherapy (51 cases).18 As a further refinement, we also sought to distinguish and separately analyze L-form cases from a minor fraction of V-form cases, which was not accomplished in previously reported series,15-18 because some V-form cases may have distinct biological features.8
MATERIALS AND METHODS
Patient materials and clinical studies.Patient materials used for this study were obtained from adult patients (age ≥ 15 years) entered on intergroup protocol INT 0129 by the Eastern Cooperative Oncology Group (ECOG), Southwest Oncology Group (SWOG), and Cancer and Leukemia Group B (CALGB). As detailed elsewhere,14 14a protocol INT 0129 involved initial randomization of previously untreated patients with the clinical diagnosis of APL to receive remission induction therapy with either ATRA or conventional chemotherapy with daunorubicin and cytarabine. After achieving CR and after consolidation chemotherapy, patients were rerandomized to receive either no further therapy or maintenance therapy with ATRA for 1 year.
Of 319 cases registered by ECOG, SWOG, and CALGB on protocol INT 0129, 279 cases were clinically eligible for this study, ie, they were not cancelled, had on-study clinical information, were not ineligible by clinical criteria, and were not excluded by pathological reclassification of leukemia type. PML-RARα mRNA typing was not accomplished in 49 eligible cases, because no sample was available (38 cases) or because of some technical shortcoming, most frequently poor RNA recovery (11 cases). Thus, 230 eligible cases had evaluable reverse-transcription polymerase chain reaction (RT-PCR) results for PML-RARα mRNA type and are included in this report.
All on-study data were obtained from the central data registry at the ECOG Statistical Center based on clinical data flow sheet reports submitted by each participating institution and cooperative group. Patient materials were obtained under a protocol INT 0129 consent form approved by the institutional review boards of all participating institutions. On-study hematological data included in this article represent the presenting hemogram, ie, before any form of therapy including hydroxyurea, which was required under protocol INT 0129 to reduce the white blood cell (WBC) count to less than 10,000 in patients randomized to the ATRA induction therapy arm of the protocol.14 14a Differential cell counts on peripheral blood and bone marrow samples were performed at each individual institution, and, because of variable classification of immature leukemic blood cells, combined blast and promyelocyte counts are presented in this communication. On the other hand, all bone marrow specimens were subjected to central pathology review by each cooperative group for subclassification as either the classical M3 or M3 variant (M3v) form of APL.
RT-PCR analysis. Heparinized bone marrow and peripheral blood samples were received by overnight express mail. A low-density WBC fraction (density ≤ 1.077 g/mL) was prepared by standard sodium metrizamide step gradient centrifugation.19 In many instances, the low-density WBCs were viably frozen in dimethylsulfoxide-containing medium and thawed for RNA extraction at a later date. Total RNA was prepared from the WBCs by modifications of either the guanidine-isothiocyanate extraction-cesium chloride gradient ultracentrifugation procedure20 or by the acid phenol extraction procedure.21
Tests performed by ECOG and SWOG used the same primers and reaction conditions, as previously described,8 whereas those performed by CALGB used a modification of an alternative RT-PCR procedure.6 Both procedures shared the common feature of initiating the reverse transcription (RT) phase of the reaction from random primers22 and performing the polymerase chain reaction (PCR) phase using upstream primers anchored in PML gene exon 3 and downstream primers anchored in RARα gene exons 3 or 4 (Fig 1), which allowed the classification of PML-RARα mRNA type as L-form, V-form, or S-form from the initial amplification procedure in the great majority of cases. The following primers were used: ECOG and SWOG laboratories, upstream primer 5′-ACCGATGGCTTCGACGAGTTC-3′; downstream primer, 5′-AGCCCTTGCAGCCCTCACAG-3′8; CALGB laboratory, upstream primer, 5′-AGCGCGACTACGAGGAGATG-3′; downstream, 5′-CCATAGTGGTAGCCTGAGGACT-3′ (modified from Miller et al6 ). The consistency of results obtained by these various RT-PCR procedures was verified by an interlaboratory exchange of 5 unknown samples per laboratory in which there was 100% concordance of PML-RARα mRNA classification results. All L-form and V-form cases were further distinguished by Southern blot transfer of the PCR products and hybridization to a probe that crosses the PML exon 6 RARα exon 3 junction (5′-TCAATGGCTGCCTCC-3′ ), yielding a positive signal in L-form and no signal in V-form cases.8 V-form cases were further verified by sequence analysis. Atypical PCR bands, which were sometimes observed, particularly in PML-RARα -negative cases, were excluded as potential unusual PML-RARα mRNA products by a variety of procedures, including use of alternative or nested PCR primer sets, hybridization to internal PML or RARα gene probes, and sequence analysis with exchange between two or more of the participating laboratories in these cases.
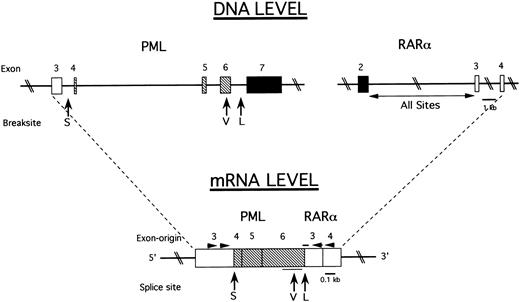
Intron-exon structure of relevant PML and RARα gene regions and the generation of three PML-RARα mRNA types in APL. (DNA Level) Solid lines indicate introns and rectangles indicate exons. (□), exons preserved in all measured PML-RARα mRNA transcripts; (▪), exons excluded from all PML-RARα mRNA transcripts; (▧), exons present in L-form and V-form but not S-form PML-RARα mRNA transcripts. The vertical arrows indicate the three breakpoint cluster regions of the PML gene as follows: S-form (S) in intron 3 (bcr3); V-form (V), variably, in the distal half of exon 6 (bcr2); and L-form (L) in intron 6 (bcr1). The RARα gene is consistently broken in intron 2, labeled “All Sites.” (mRNA Level) Joined rectangles show how exons are spliced together to form three types of PML-RARα mRNA at the splice junctions indicated by the vertical arrows. The thin horizontal line below the rectangles indicates the sites of variable breakage in exon 6 in V-form cases. The thick horizontal line above the rectangles indicates the site of the oligonucleotide hybridization probe that spans the PML exon 6 RARα exon 3 junction in L-form cases. The arrowheads indicate the approximate sites of PCR primers used to amplify all three types of PML-RARα mRNA.
Intron-exon structure of relevant PML and RARα gene regions and the generation of three PML-RARα mRNA types in APL. (DNA Level) Solid lines indicate introns and rectangles indicate exons. (□), exons preserved in all measured PML-RARα mRNA transcripts; (▪), exons excluded from all PML-RARα mRNA transcripts; (▧), exons present in L-form and V-form but not S-form PML-RARα mRNA transcripts. The vertical arrows indicate the three breakpoint cluster regions of the PML gene as follows: S-form (S) in intron 3 (bcr3); V-form (V), variably, in the distal half of exon 6 (bcr2); and L-form (L) in intron 6 (bcr1). The RARα gene is consistently broken in intron 2, labeled “All Sites.” (mRNA Level) Joined rectangles show how exons are spliced together to form three types of PML-RARα mRNA at the splice junctions indicated by the vertical arrows. The thin horizontal line below the rectangles indicates the sites of variable breakage in exon 6 in V-form cases. The thick horizontal line above the rectangles indicates the site of the oligonucleotide hybridization probe that spans the PML exon 6 RARα exon 3 junction in L-form cases. The arrowheads indicate the approximate sites of PCR primers used to amplify all three types of PML-RARα mRNA.
Statistical methods. Univariate differences between dichotomous variables were evaluated with Fisher's Exact Test.23 Differences in the distribution of disease-free survival (DFS; time from achievement of a CR to relapse, death, or last follow-up) were compared between groups with a logrank test.24 Multivariate analyses of DFS were performed with proportional hazards regression.25 Survival curves were estimated by the method of Kaplan and Meier.26 Differences between continuous variables, eg, WBC count, were evaluated with a Wilcoxon Rank Sum Test.27 This analysis has 90% power to detect a 20% difference in 2-year DFS between S- and L-form complete responders, and this has a high probability of detecting a clinically important difference.
RESULTS
PML-RARα mRNA analysis.Figure 1 shows previously established breakpoint cluster regions (bcrs) in the PML and RARα genes and the location of primer and probe sites used in this report to identify the S-, L-, and V-form types of PML-RARα mRNA by RT-PCR and to distinguish the L- and V-forms of PML-RARα mRNA from one another by molecular hybridization analysis (see Materials and Methods). PML-RARα mRNA typing was determined in 230 cases out of 279 clinically eligible adult cases (age ≥ 15) registered on protocol INT 0129 by ECOG, SWOG, and CALGB (82%). As assessed by on-study clinical parameters (discussed later), there were no significant differences between the 230 analyzable cases and the entire group of 279 eligible cases. Nine analyzable cases (4%) were PML-RARα negative. These cases were considered to have “APL-like” morphology and were not excluded by central pathology review. These few, somewhat phenotypically and karyotypically heterogeneous, cases are under separate study.
A total of 221 cases were PML-RARα positive with the following breakdown of PML-RARα mRNA types: S-form, 82 (37%); L-form, 121 (55%); and V-form, 18 (8%). The V-form cases were too few for confident statistical analysis of clinical characteristics and outcome as a group. Thus, detailed analysis in this report is confined to a comparison of APL cases with the S-form or L-form of PML-RARα.
On-study patient characteristics. There was no indication of any association of PML-RARα mRNA type with age, sex, race, initial performance status, or signs of clinically diagnosed coagulation/bleeding disorder (Table 1). In contrast, notable differences were observed for five presenting hematologic characteristics of S-form– compared with L-form–type cases (Table 2). There was a significantly increased proportion of S-form cases with a WBC count > 5,000 (44% v 21%; P = .0008). Less dramatically, increased proportions of S-form cases had blood blasts plus promyelocytes > 10% (67% v 47%; P = .04), an absolute number of blood blasts plus promyelocytes > 100/ μL (72% v 52%; P = .04), expression of the M3v rather than the classical M3 phenotype (24% v 12%; P = .036), and a platelet count ≤ 20,000 (39% v 26%; P = .06). There were no statistically significant differences in the percentage of leukemic blasts plus promyelocytes in presenting bone marrow aspirates (P = .95).
The finding that a greater proportion of S-form cases have higher levels of circulating WBCs and leukemic blasts plus promyelocytes than L-form cases (Table 2) was complemented by the finding of a similar relationship for the median values of these parameters. That is, the median values (interquartile ranges) for S-form cases versus L-form cases, respectively, were WBC count, 2,500/μL (1,200 to 13,000) versus 1,600/μL (1,000 to 3,900; P = .009); percentage of blood blasts plus promyelocytes, 29% (7% to 66%) versus 8.5% (0% to 49.5%; P = .03); and absolute number of blood blasts plus promyelocytes, 884/μL (63 to 5,502) versus 126/μL (0 to 1,865; P = .019).
Relationship between PML-RARα mRNA type and clinical outcome. The overall CR rate for the 221 PML-RARα–positive protocol cases was 73%. There were no significant differences in the CR rates for the S-form cases versus L-form cases considered either as an overall group, ie, regardless of therapy (79% v 69%; P = .14), or as a function of randomization to either the chemotherapy (77% v 66%) or ATRA (82% v 73%) induction arms of protocol INT 0129 (Table 3). Although the small number of V-form cases (N = 18) did not permit statistical analysis, there were no apparent major differences between the CR rates of these cases compared with S- and L-form cases. On the other hand, a difference did appear to exist for the small number of PML-RARα–negative cases (N = 9), because none of five cases randomized to ATRA achieved CR; in contrast, CR was achieved in three of four cases randomized to chemotherapy.
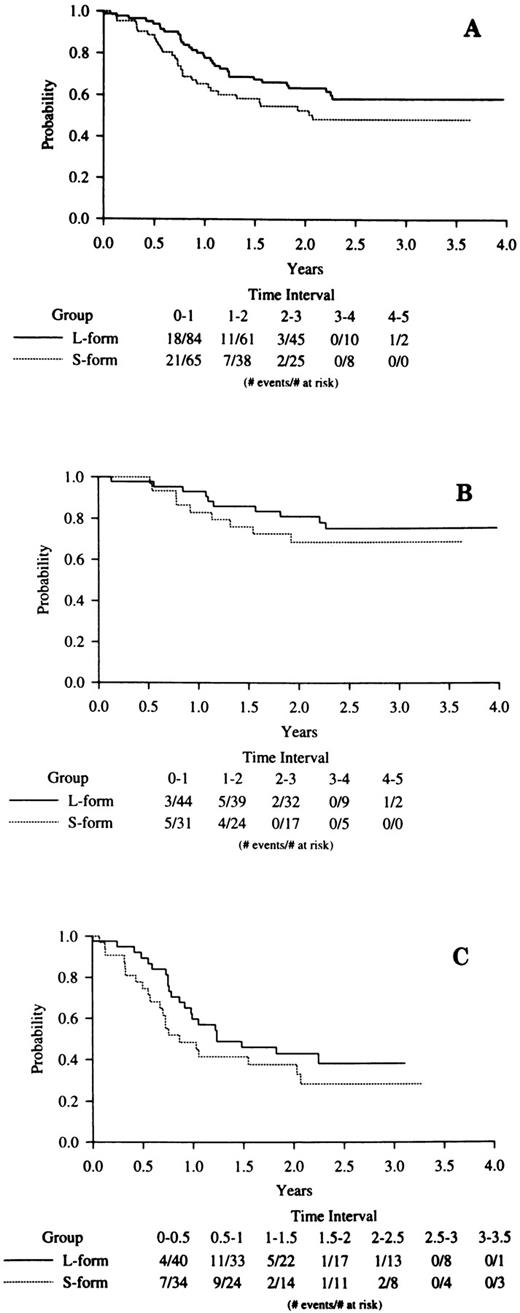
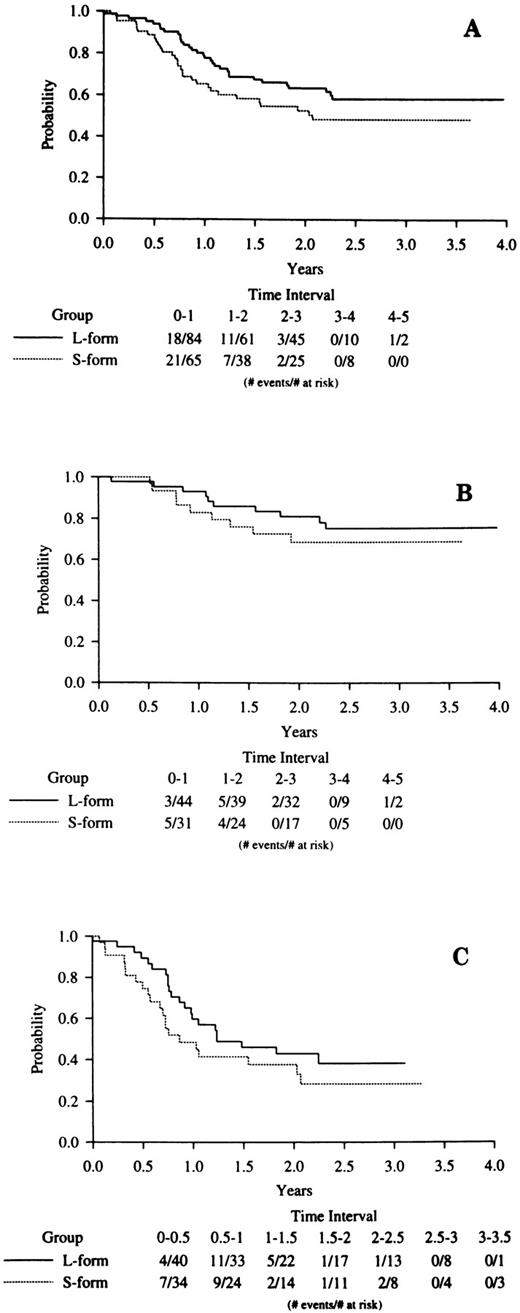
DFS was analyzed with a median follow-up of 30 months after the achievement of CR. A Kaplan-Meier plot of overall DFS (Fig 2A) by PML-RARα type suggests decreased remission duration among S-form cases, although this did not reach statistical significance (P = .12). However, as reported elsewhere,14 14a there was a significant association between increased WBC count and poor DFS in the overall clinical trial cases, and this was also seen among the 221 cases determined to be PML-RARα positive in this study (P = .0006). Additionally, as shown above, there is a significant association between increased WBC and the occurrence of the S-form fusion type. A multivariate proportional hazards model showed that WBC (P = .0014) but not PML-RARα mRNA type (P = .40) was associated with DFS. Figure 2B and C present DFS by PML-RARα fusion type within induction treatment. With small numbers, neither comparison is significant, especially when the analyses are adjusted for presentation WBC count.
Kaplan-Meier analyses of DFS in S-form versus L-form PML-RARα mRNA type cases. (A) All cases; (B) ATRA induction treatment cases; (C) chemotherapy induction treatment cases. The legend denominator indicates the number of protocol cases at risk in successive 6-month intervals up to 4 years of follow-up beginning from the time of the clinical diagnosis of CR. The legend numerator indicates the number of cases at risk that experienced adverse events leading to an off-protocol status (relapse or death).
Kaplan-Meier analyses of DFS in S-form versus L-form PML-RARα mRNA type cases. (A) All cases; (B) ATRA induction treatment cases; (C) chemotherapy induction treatment cases. The legend denominator indicates the number of protocol cases at risk in successive 6-month intervals up to 4 years of follow-up beginning from the time of the clinical diagnosis of CR. The legend numerator indicates the number of cases at risk that experienced adverse events leading to an off-protocol status (relapse or death).
DISCUSSION
The evaluation of clinical outcome of previously-untreated APL patients entered on the intergroup clinical trial, INT 0129, indicated that, among patients randomized to receive induction therapy with either ATRA or chemotherapy, respectively, there was no difference in CR rates (72% v 69%; P = .56) but that, after consolidation with chemotherapy, there was a notable difference in DFS over 3 years of follow-up (67% for ATRA patients versus 32% for chemotherapy patients; P < .0001, logrank test).14,14a These data, obtained under uniform treatment conditions, are consistent with those of a previous smaller randomized study28 and establish that there is no difference in CR rates after ATRA or chemotherapy induction but markedly improved DFS in patients who achieve CR on ATRA. Additionally, in the INT 0129 study, DFS was as favorable in patients achieving CR on chemotherapy who were rerandomized to receive ATRA maintenance therapy as in patients who achieved CR on ATRA induction therapy, whereas those rerandomized to observation, ie, CR patients who never received ATRA, had a poorer prognosis.14 14a These clinical observations support the notion that the long-term superiority of ATRA-based treatment compared with purely chemotherapy-based treatment is related to the unique activity of ATRA that induces differentiation of APL cells and acts synergistically with chemotherapeutic agents.
The differentiative activity of ATRA in APL cells involves the activation of PML-RARα by specific binding to the ligand binding domain of the RARα component of the fusion protein.2-4,12,13 However, the PML portion of PML-RARα has also been shown to be important in mediating the APL cell response to ATRA.12,13,29-31 In recombinant vector transfection-reporter assays, the two major types of PML-RARα due to variations in the PML component, ie, the S-form and L-form types, have been shown to have modestly different molecular activities.2-4 12 However, these assays only tested activity mediated through the RARα component of PML-RARα, because no effective bioassay has yet been reported for directly testing the PML portion of the molecule. Thus, they could underestimate biological differences related to variations in PML-RARα type. Based on these molecular considerations and the clinical observations, a reasonable hypothesis is that variations in clinical outcome after ATRA therapy might be associated with specific PML-RARα type. A corollary is that such an association related to an ATRA-targeted molecule would not be expected after chemotherapy alone.
Although the INT 0129 study was primarily designed to test clinical questions related to the efficacy of ATRA and chemotherapy in APL, the large cohort size for this infrequent form of acute myeloid leukemia permitted a more critical evaluation of the possible association of PML-RARα type with pretreatment characteristics and treatment outcome results than previous, smaller studies. From our analysis of 221 PML-RARα–positive cases, we found no association between PML-RARα type and potentially adverse on-study clinical characteristics, such as low performance status or evidence of increased coagulation/bleeding problems (Table 1). Nor was the overall CR rate any different between 82 S-form– and 121 L-form–type cases (79% v 69%; P = .14; Table 3). Further, there was no indication of a difference in CR rates of these PML-RARα types after induction with either ATRA (82% v 73%) or chemotherapy (77% v 66%). However, the analysis was more complex for certain pretreatment hematological parameters and DFS, which were partially inter-related.
We found that APL cases with the S-form type compared with the L-form type of PML-RARα mRNA had a relatively high presenting WBC count and increased immature WBCs (blasts plus promyelocytes) in the peripheral blood, documented by either their absolute numbers or as the percent of total WBCs (Table 2). Also, a greater proportion of S-form cases had APL cells with the M3v rather than the classical hypergranular M3 phenotype.32 An association between a relatively high WBC count and the S-form PML-RARα type was suggested in one previous report of 52 cases16 but was not found in two other reports with 97 total previously untreated APL cases.17,18 An association of the M3v APL cell phenotype with S-form PML-RARα type is also consistent with one previous report of 43 cases33 but was not observed in another series with 52 cases.34 Our ability to discern a relationship between the S-form vis-a-vis the L-form of PML-RARα and these WBC parameters may be related to several factors, including our relatively large study group size, our certification through a central registry that all hematologic data were obtained before any antileukemic therapy and our molecular discrimination of L-form from V-form cases.
All of the S-form–associated WBC-related characteristics that we identified in this study, most consistently an increased total WBC count, have previously been associated with reduced short- or long-term clinical outcome in APL patients treated with chemotherapy.35-38 An increased WBC count, present either before or developing soon after the initiation of ATRA induction therapy, was also linked to poor short-term outcome related to development of the “retinoic acid syndrome.”39,40 However, data have been presented against this association during ATRA induction therapy,17,41 and with the introduction of treatment strategies to control the WBC count or its potential adverse effects, the importance of this element appears to have been minimized,17,41 including on protocol INT 0129.14,14a Previous reports involving ATRA induction therapy in combination with concurrent and/or consolidation chemotherapy did not evaluate the possible association of a relatively high presenting WBC count or the M3v phenotype on longer-term clinical outcome.16-18,28,41 In the intergroup trial INT 0129, there was a highly significant association between a presenting WBC count > 2,000 and DFS for the overall group of patients (P = .0006), and this association was consistent across treatment arms.14 14a
These WBC count considerations had an important impact on our analysis of DFS related to PML-RARα type. The post-CR Kaplan-Meier DFS curve was lower for S-form than for L-form cases (Fig 2A; P = .12), suggesting a slight trend towards a poorer long-term prognosis for S-form cases. However, a proportional hazards model that adjusts for presentation WBC, a factor known to be associated with outcome in APL,35-38 showed no compelling evidence for a difference between the DFS of S-form compared with L-form complete responders (P = .40). Additionally, there was no evidence of an interaction: DFS was shorter with a high presenting WBC count in both S-form and L-form cases. This lack of difference in DFS between S-form and L-form cases after adjustment for the presenting WBC count extended to both the ATRA (P = .30) and chemotherapy (P = .91) induction arms of the protocol. With small case numbers, there was also no indication of an association between PML-RARα mRNA type and treatment outcome in INT 0129 cases initially treated with chemotherapy and then rerandomized to either ATRA maintenance or observation (data not shown). From these data, we conclude that there is no evidence from this large randomized study to support the hypothesis that differences in the specific ATRA-target PML-RARα affect treatment outcome. We cannot exclude the possibility that the high effectiveness of the ATRA-chemotherapy combinations used in the INT 0129 clinical trial could have obscured a difference in biological response related to PML-RARα type that might be observed under less effective ATRA treatment conditions.
The somewhat complex interaction of the S-form PML-RARα mRNA type and WBC count related to clinical prognosis that we have identified may have contributed to the inconsistent conclusions of previous correlative studies involving smaller numbers of less-uniformly treated patients.15-18 Our study also differed from previous studies by the use of special diagnostic procedures to identify and remove V-form cases from the comparative analysis of S-form versus L-form cases, because V-form cases are genetically distinct4,5,8 and because APL cells from a subset of V-form cases may have altered biological properties.8 There were too few V-form cases in the current study (N = 18) to statistically analyze for associated clinical parameters; however, analysis of these cases on an individual basis with sequence analysis of the altered region of PML exon 6 preliminarily supports the possibility that there could be an association between the extent of PML exon 6 deletion and poor clinical prognosis.42
An important clinical conclusion from our study is that the determination of PML-RARα mRNA type does not provide any basis for considering treatment modification, as has been suggested.17 Despite this conclusion and the consideration that detection of the S-form versus the L-form of PML-RARα mRNA type per se does not provide prognostic information, we recommend continued study of the PML-RARα mRNA type in new APL patients. This procedure expeditiously identifies PML-RARα–negative cases that phenotypically mimic true APL cases but fail to respond to ATRA,41 as we confirmed in five cases in the INT 0129 correlative study. Additionally, this determination provides a baseline for serial monitoring of residual leukemic disease by RT-PCR procedures after achieving CR, which may prove to be of significant value in predicting relapse and directing early therapeutic intervention (summarized in Diverio et al43 ). This is also needed for further basic research investigations, particularly to determine if as yet undefined therapy-dependent alterations may occur in the PML-RARα fusion gene that could contribute to the development of clinical resistance to ATRA.
ACKNOWLEDGMENT
The authors thank the physicians, nursing staffs, and data managers of ECOG-, SWOG-, and CALGB-affiliated institutions for their cooperation in obtaining and sending the clinical specimens and information used for our investigations. We also thank Drs John M. Bennett (ECOG), David Head (SWOG), and Frederick R. Davey (CALGB) for pathological analysis of bone marrow specimens used for the current studies.
Supported by grants from the National Institutes of Health (CA56771, CA21115, CA14958, CA23318, CA12296, CA32102, CA59518, and CA37027), The State of New Mexico Dedicated Health Fund and the Coleman Leukemia Research Fund. This study is based on an intergroup clinical trial (INT 0129) involving participation of the Eastern Cooperative Oncology Group (Robert L.Comis, MD, Group Chair), the Southwest Oncology Group (Charles A. Coltman, Jr, MD, Group Chair) and the Cancer and Leukemia Group B (Richard L. Schilsky, MD, Group Chair).
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
Address reprint requests to Robert E. Gallagher, MD, Department of Oncology, Montefiore Medical Center, 111 East 210th Street, Bronx, NY 10467.