Abstract
Interleukin-10 (IL-10) is a potent monocyte regulatory cytokine that inhibits gene expression of proinflammatory mediators. In this study, we investigated the mechanism by which IL-10 downregulates expression of intercellular adhesion molecule-1 (ICAM-1) on the cell surface of normal human monocytes activated with interferon-γ (IFN-γ). IL-10 inhibition of IFN-γ–induced ICAM-1 expression was apparent as early as 3 hours and was blocked by an anti–IL-10 antibody but not by an isotype-matched control antibody. Northern blot analysis showed that IL-10 reduced the accumulation of ICAM-1 mRNA in IFN-γ–stimulated monocytes. IL-10 inhibition of ICAM-1 steady-state mRNA was detected at 3 hours and remained at 24 hours. Nuclear run-on transcription assays showed that IL-10 inhibited the rate of IFN-γ–induced transcription of the ICAM-1 gene, and mRNA stability studies showed that IL-10 did not alter the half-life of IFN-γ–induced ICAM-1 message. Thus, IL-10 inhibits IFN-γ–induced ICAM-1 expression in monocytes primarily at the level of gene transcription. Activation of IFN-γ–responsive genes requires tyrosine phosphorylation of the transcriptional factor STAT-1α (signal transducer and activator of transcription-1α). However, IL-10 did not affect IFN-γ–induced tyrosine phosphorylation of STAT-1α or alter STAT-1α binding to the IFN-γ response element (IRE) in the ICAM-1 promoter. Instead, IL-10 prevented IFN-γ–induced binding activity at the NF-κB site of the tumor necrosis factor α (TNF-α)–responsive NF-κB/C-EBP composite element in the ICAM-1 promoter. These data indicate that IL-10 inhibits IFN-γ–induced transcription of the ICAM-1 gene by a regulatory mechanism that may involve NF-κB.
INTERLEUKIN-10 (IL-10) is a potent immunoregulatory cytokine attenuating a wide range of immune effector and inflammatory responses (reviewed in Moore et al1 and Mosmann2 ). IL-10 is produced by monocytes/macrophages, B cells, murine Th2 cells, human Th1 and Th2 cells, and some CD8+ cells.3-5 Activation of monocytes by lipopolysaccharide (LPS) and interferon-γ (IFN-γ) induces a broad range of immunologic activities, many of which are suppressed by IL-10. For example, IL-10 suppresses production of the proinflammatory cytokines IL-1, tumor necrosis factor α (TNF-α), IL-8, and IL-6, cell surface expression of major histocompatibility complex (MHC) class II, intercellular adhesion molecule-1 (ICAM-1), and B7 molecules, and release of reactive oxygen and nitrogen intermediates.6-12 However, the mechanisms responsible for IL-10 inhibition of monocyte/macrophage function are largely unknown. In LPS-activated monocytes, IL-10 inhibits cytokine secretion by transcriptional and posttranscriptional means.13 Recent data indicate that IL-10 prevents cytokine gene transcription by inhibiting the transcriptional factor NF-κB.14 IL-10 also inhibits LPS-induced p56 lyn activity and the Ras signaling pathway.15
The mechanism by which IL-10 inhibits IFN-γ–induced ICAM-1 in normal human monocytes has not been defined. ICAM-1 is a glycosylated, integral membrane protein that facilitates selective cellular adhesion. ICAM-1 is constitutively expressed at low levels on a wide variety of cells and can be induced in monocytes by LPS and IFN-γ.16,17 ICAM-1 expression is transcriptionally regulated18,19; the promoter contains binding sites for AP-1, NF-κB, and C/EBP.20-22 Recently, Look et al22 identified a consensus IFN-γ response element (IRE) located at nucleotides −116 to −106 (relative to the transcription initiation site) that is necessary for induction of ICAM-1 transcription by IFN-γ in epithelial cells.22 The ICAM-1 IRE is similar to IFN-γ response sequences of the FcγRI and IRF-1 genes.22-24
IFN-γ activates the Janus family tyrosine kinases (JAKs) JAK1 and JAK2,25-27 which in turn leads to tyrosine phosphorylation of the latent transcription factor STAT-1α (signal transducer and activator of transcription).28,29 Phosphorylated STAT-1α forms a homodimer that translocates to the nucleus and binds to the IRE in promoter regions of IFN-γ–responsive genes.30,31 Feldman et al32 reported that IgG complexes inhibit IFN-γ activation of FcγRI gene transcription by preventing phosphorylation and binding of STAT-1α to an IRE in the FcγRI promoter. We were therefore interested in determining whether IL-10 inhibited IFN-γ–induced ICAM-1 expression through a similar mechanism. We demonstrate herein that IL-10 inhibits IFN-γ–induced ICAM-1 expression at the level of gene transcription. In contrast to the mechanism of IgG inhibition of the FcγRI gene, IL-10 did not alter the phosphorylation or binding activity of STAT-1α. However, IL-10 does prevent NF-κB binding to a proximal DNA element in the ICAM-1 promoter, providing a possible mechanism by which IL-10 could inhibit IFN-γ–induced ICAM-1 gene transcription in human monocytes.
MATERIALS AND METHODS
Reagents.Human recombinant IL-10 (specific activity, 1 × 107 U/mg) was a gift from Dr Kevin Moore, DNAX Research Institute (Palo Alto, CA). Human IFN-γ (specific activity, 1 × 107 U/mg) was purchased from Sigma Chemical Co (St Louis, MO). A rat anti–human IL-10 was a gift from Dr Kevin Moore. Complete culture media contained RPMI 1640 (GIBCO Laboratories, Grand Island, NY), 10% heat-inactivated fetal calf serum ([FCS] Atlanta Biologicals, Norcross, GA), 2 mmol/L-glutamine, penicillin-streptomycin (100 μg/mL), and 10 mmol/L HEPES (Sigma Chemical Co) (complete media). Anti–STAT-1α (p91) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Oligonucleotides were synthesized by Integrated DNA Technologies Inc (Coralville, IA). Actinomycin D was purchased from Sigma.
Cells.Human mononuclear cells were prepared from human venous blood of healthy donors by density-gradient separation using Ficoll-Hypaque (Pharmacia, Piscataway, NJ). Monocytes were obtained by adherence to 150-mm tissue culture dishes (Corning Inc, Corning, NY) for 1.5 hours at 37°C in complete media. Nonadherent cells were removed by vigorous washing in cold RPMI 1640 in 3% FCS. Cell preparations yielded greater than 85% CD14+ monocytes as determined by flow cytometry.
FACS analysis.Peripheral blood mononuclear cells (2 × 106) were cultured in 24-well Falcon culture dishes (Becton Dickinson Co, Lincoln Park, NJ) for 1.5 hours, and then nonadherent cells were removed by vigorously washing with cold RPMI 1640 with 3% FCS. Monocytes were stimulated in the presence or absence of IFN-γ, IL-10, or both in combination in complete media. Adherent cells were harvested using a cell scraper and then washed with buffer (phosphate-buffered saline, 3% bovine serum albumin, and 0.01% NaN3 ). Cells were incubated with phycoerythrin (PE)- and FITC-conjugated monoclonal antibodies for 30 minutes on ice, washed twice in buffer, and fixed with 1% paraformaldehyde. Monocytes were identified as CD14+ using PE-conjugated anti-CD14 monoclonal antibody, and electronically gated CD14+ cells were analyzed for intensity of green fluorescence with FITC-conjugated anti–ICAM-1 and FITC-conjugated anti–HLA-DR monoclonal antibody. PE-conjugated anti-CD14 monoclonal antibody, FITC-conjugated anti-DR, anti–ICAM-1, and isotype control were purchased from Coulter (Hialeah, FL).
Northern blot analysis.Total cellular RNA was isolated using RNAzol (Tel-test “B” Inc, Friendswood, TX) from adherent monocytes (1 × 107) stimulated with cytokines in complete media for the times and cytokine concentrations indicated in the figure. Total RNA samples (5 to 10 μg) were loaded onto a 1.2% agarose gel containing formaldehyde, and then blotted onto BA-S NC nitrocellulose membranes (Midwest Scientific, St Louis, MO). The membrane was baked for 2 hours at 80°C and probed in a 50% formamide hybridization solution overnight at 42°C. The probes, 1,400-basepair (bp) ICAM-1 (SalI-BglII digestion fragment) and 1,400-bp glyceraldehyde-3-phosphate dehydrogenase ([GAPDH] PstI-PstI digestion fragment), were labeled by random priming (Stratagene, La Jolla, CA) with 50 μCi [α-32P]dCTP (3,000 Ci/mmol; Amersham, Arlington Heights, IL). The membrane was washed and autoradiographed at −70°C for 1 to 3 days. Filters were stripped and rehybridized with 32P-labeled GAPDH under the same conditions. Levels of ICAM-1 mRNA are expressed as the ratio of ICAM-1 to GAPDH mRNA determined by laser densitometry (Personal Densitometer SI; Molecular Dynamics Inc, Sunnyvale, CA).
ICAM-1 mRNA stability analysis.Monocytes were stimulated for 16 hours with IFN-γ (100 U/mL) in the presence or absence of IL-10 (100 U/mL) in complete media. Actinomycin D (5 μg/mL) was added to the cells, and RNA was extracted at 0, 1, 2, and 3 hours. Northern blot analysis was performed as described earlier. The maximum levels of ICAM-1 mRNA determined by densitometry for IFN-γ and IFN-γ plus IL-10 were set at 100% and plotted against the percent reduction in the amount of mRNA.
Nuclear run-on analysis.Monocytes were unstimulated or stimulated with IFN-γ (100 U/mL), IL-10 (100 U/mL), or a combination of both for 3 hours at 37°C in complete media. Cells were washed, scraped from the culture dishes, centrifuged, resuspended in Nonidet P-40 (NP-40) lysis buffer (10 mmol/L Tris hydrochloride, pH 7.4, 10 mmol/L NaCl, 3 mmol/L MgCl2 , and 0.5% NP-40) for 5 minutes at 4°C, and pelleted to isolate the nuclei. Nuclei were resuspended in 200 μL glycerol buffer (0.1 mmol/L EDTA, 40% glycerol, 5 mmol/L MgCl2 , and 50 mmol/L Tris hydrochloride, pH 8.3) and flash-frozen in liquid nitrogen for storage at −80°C. Intact nuclei (200 μL) were thawed, and 200 μL transcription buffer (10 mmol/L hydrochloride, Tris, 5 mmol/L MgCl2 , 0.3 mol/L KCl, 1 mmol/L unlabeled ATP, CTP, and UTP ribonucleotides, and 5 mmol/L DTT), 250 mCi [α-32P]GTP (3,000 Ci/mmol; Amersham Corp), and 500 U RNasin (Promega, Madison, WI) were incubated at 30°C for 30 minutes. For RNA extraction, nuclei were mixed with 30 μL (1 mg/mL) RNase-free DNase I (Promega) and 600 μL HSB buffer (0.5 mol/L NaCl, 50 mmol/L MgCl2 , 2 mmol/L CaCl2 , and 10 mmol/L Tris hydrochloride, pH 7.4) and vigorously pipetted and incubated for 5 minutes at 30°C. For RNA purification, 200 μL sodium dodecyl sulfate (SDS)/Tris buffer (5% SDS, 0.5 mol/L Tris hydrochloride, pH 7.4, and 0.125 mol/L EDTA) and 5 μL (50 mg/mL) proteinase K (Promega) were incubated for 30 minutes at 42°C. RNA was extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1). Nitrocellulose was slotted with 5 μg linearized ICAM-1 cDNA and GAPDH cDNA, washed six times with SSC (1× SSC is 0.15 mol/L NaCl plus 0.015 mol/L sodium citrate), dried, and baked for 2 hours at 80°C. Hybridization was performed at 42°C for 24 to 36 hours with 107 cpm labeled RNA. Filters were washed with 2× SSC/0.1% SDS for 15 minutes at room temperature, followed by 0.2× SSC/0.5% SDS for 10 minutes at 42°C, and ending with 0.2× SSC/0.1% SDS for 10 minutes at 42°C. Membranes were autoradiographed at −70°C for 4 to 7 days. Levels of ICAM-1 RNA were normalized by expressing the ratio of ICAM-1 RNA to GAPDH RNA determined by laser densitometry.
Electrophoretic mobility shift analysis.Electrophoretic mobility shift assays (EMSAs) were performed as we have previously described.33 Nuclear extracts were prepared by treating the cell pellet with cold buffer (10 mmol/L HEPES, pH 7.9, 10 mmol/L KCl, 0.1 mmol/L EDTA, 0.1 mmol/L EGTA, 1 mmol/L DTT, and 0.5 mmol/L phenylmethylsulfonyl fluoride [PMSF ]) for 15 minutes to allow cells to swell, and then 25 μL 10% NP-40 was added and the nuclei were pelleted. The nuclear pellet was resuspended in buffer (20 mmol/L HEPES, pH 7.9, 0.4 mol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1 mmol/L DTT, and 1 mmol/L PMSF ) for 15 minutes at 4°C. Debris was pelleted, and supernatants were frozen at −70°C. Nuclear extracts (5 to 7 μg protein) prepared from cytokine-activated monocytes were incubated with 50,000 cpm (0.5 ng) 32P-end-labeled double-stranded synthetic deoxyoligonucleotide probes for 30 minutes at room temperature in a 20-μL reaction volume containing 12% glycerol, 12 mmol/L HEPES-NaOH (pH 7.9), 60 mmol/L KCl, 5 mmol/L MgCl2 , 4 mmol/L Tris hydrochloride (pH 7.9), 0.6 mmol/L EDTA (pH 7.9), 0.6 mmol/L DTT, and 1 μg poly(dI).(dC). Protein-DNA complexes were resolved in 5% native polyacrylamide gels preelectrophoresed for 30 minutes at room temperature in 0.25× TBE buffer (22.5 mm Tris-borate and 0.5 mm EDTA, pH 8.3). Gels were dried and exposed overnight to x-ray film (Eastman Kodak Co, Rochester, NY) with an intensifying screen at −70°C. For supershift, nuclear extracts (2 to 5 μg protein) were incubated with 2 μg anti–STAT-1α or anti-STAT3 antibodies for 30 minutes before incubation with labeled probe. The oligonucleotides used in these studies were as follows: ICAM-1 IRE, 5′-GAGGTTTCCGGGAAAGCAGC-3′; c-fos SIE, 5′-GTGCATTTCCCGTAAATCTTGTCTACA-3′; ICAM-1 Sp-1, 5′-ACCGCCGCCCGCTTGC-3; and ICAM-1 proximal NF-κB, 5′-GCTCCGGAATTTCCAAGC-3′.
Immunoprecipitation of STAT-1α.Adherent monocytes were stimulated in the presence or absence of cytokines for 5 minutes at 37°C in complete media. Cells were lysed with lysis buffer (50 mmol/L Tris hydrochloride, 1 mmol/L EDTA, 0.15 mol/L NaCl, 1% NP-40, 200 μmol/L Na3VO4 , 1 mmol/L PMSF, 1 mg/mL aprotinin, and 1 μg/mL leupeptin) for 15 minutes at 4°C. Particulate debris was removed by centrifugation at 12,000g for 15 minutes at 4°C. Protein content of the lysates was measured using the Bio Rad Protein Assay (Bio Rad, Hercules, CA). The lysates were precleared with Pansorbin (Calbiochem, La Jolla, CA) for 2 hours at 4°C with shaking. Anti–STAT-1α antibody (2 μg) (Santa Cruz Biotechnology) was added to precleared lysates and incubated for 16 hours at 4°C. Washed protein A/G sepharose beads (Calbiochem) were added to the lysates and incubated at room temperature for 4 hours. The beads were washed with lysis buffer and then boiled with equal amounts of 2× Laemmli sample buffer for 5 minutes. Proteins were resolved on 7.5% SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred to Hybond-ECL nitrocellulose membrane (Amersham). The membrane was blocked in 5% nonfat dry milk in TBST (20 mmol/L Tris-base, 137 mmol/L NaCl, and 0.1% TWEEN) for 90 minutes, followed by three 5-minute washes in TBST. The membrane was incubated with antiphosphotyrosine antibody (4G10; Upstate Biotechnology, Lake Placid, NY) diluted 1:1,000 in 5% nonfat dry milk in TBST for 1 hour, followed by three washes in TBST. The membrane was incubated with goat anti–mouse IgG horseradish peroxidase (HRP) for 1 hour, followed by 10 washes in TBST, and then incubated with equal volumes of enhanced chemiluminescence detection reagents 1 and 2 (Amersham) for 1 minute. The excess reagent was poured off and the membrane exposed to film. To identify STAT-1α, protein membranes were stripped and reprobed as described earlier with anti–STAT-1α diluted (1:1,000) in 5% nonfat dry milk in TBST, and developed with goat antimouse IgG HRP as already described.
RESULTS
IL-10 inhibits ICAM-1 cell surface expression.To determine the mechanism by which IL-10 inhibits IFN-γ–induced ICAM-1 expression, we first characterized cell surface expression of ICAM-1 on human monocytes stimulated with IFN-γ or IFN-γ plus IL-10. Human monocytes were incubated for 16 hours in the presence or absence of recombinant IFN-γ (100 U/mL), IL-10 (100 U/mL), or both in combination and then analyzed by flow cytometry for ICAM-1 and MHC class II DR cell surface expression. CD14+ monocytes were electronically gated, and the fluorescence intensity of ICAM-1 and HLA-DR was determined. In five separate experiments, overnight incubation of monocytes spontaneously induced a 4.7-fold increase in ICAM-1 and a 2.6-fold increase in DR expression over that of freshly isolated monocytes (Table 1).
Incubation of the cells overnight with IFN-γ further enhanced this spontaneous expression, producing an additional twofold increase in ICAM-1 and 3.6-fold increase in DR. Overnight incubation of monocytes with IL-10 inhibited both the spontaneous increase in ICAM-1 and DR and the IFN-γ–induced increase. In the absence of IFN-γ, inhibition of ICAM-1 by IL-10 ranged from 24% to 48%, and DR inhibition was less than that detected on freshly isolated monocytes (Table 1). For IFN-γ–stimulated monocytes, IL-10 inhibited 16% to 74% of the increase in ICAM-1 expression and 50% to 100% of the increase in DR expression. The inhibitory activity of IL-10 was specific, since anti–human IL-10 antibodies but not an isotype-matched control antibody blocked the IL-10 effect (Table 1). IL-10 inhibition was concentration-dependent, and the IC50 was approximately 10 U/mL (data not shown). For subsequent experiments, we chose to use 100 U/mL, which produced greater than 80% inhibition.
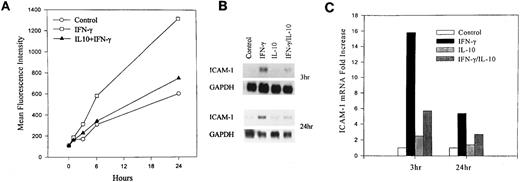
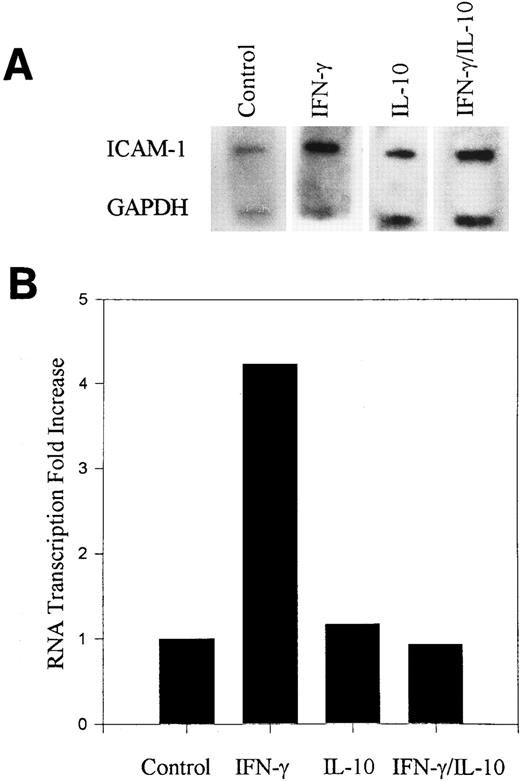
Kinetics of IL-10 inhibition of ICAM-1 expression.To further characterize the effects of IL-10 in monocytes, we examined the kinetics of IL-10 inhibition of ICAM-1 expression at the protein and mRNA levels. Human monocytes were incubated for 0, 1, 3, 6, and 24 hours in the presence or absence of recombinant IFN-γ (100 U/mL), IL-10 (100 U/mL), or both in combination and then analyzed by flow cytometry for ICAM-1 cell surface expression. IFN-γ induction of ICAM-1 was detected as early as 3 hours and continued to increase over the 24-hour time course (Fig 1A). IL-10 inhibited IFN-γ induction of ICAM-1 at all time points examined. To determine if IL-10 inhibited ICAM-1 mRNA expression, we examined steady-state levels of ICAM-1 mRNA by Northern blot analysis. Monocytes were stimulated in the presence or absence of IFN-γ (100 U/mL) and/or IL-10 (100 U/mL) for 3 or 24 hours. IFN-γ increased ICAM-1 mRNA, and IL-10 inhibited this induced expression at both time points (Fig 1B). The correlation between cell surface expression and steady-state levels of mRNA suggests that IL-10 inhibits ICAM-1 expression by reducing the accumulation of ICAM-1 mRNA.
IL-10 inhibits IFN-γ–induced ICAM-1 cell surface expression and mRNA accumulation. Monocytes were untreated (control) or treated with IFN-γ (100 U/mL), IL-10 (100 U/mL) or the combination for the indicated times. (A) Cell surface expression was measured by flow cytometry. Monocytes were identified by staining with PE-labeled anti-CD14 monoclonal antibody (mAb), and ICAM-1 was identified on CD14+ cells by staining with FITC-labeled anti–ICAM-1 mAb. (B) RNA was isolated from cytokine-stimulated monocytes, and total RNA was analyzed by Northern blot with ICAM-1 and GAPDH probes. (C) Normalized absorption values were obtained by densitometry scanning of ICAM-1 and GAPDH mRNA bands. From the ratio of ICAM-1 to GAPDH, the fold increase over untreated control cells was calculated. Data are representative of 3 experiments.
IL-10 inhibits IFN-γ–induced ICAM-1 cell surface expression and mRNA accumulation. Monocytes were untreated (control) or treated with IFN-γ (100 U/mL), IL-10 (100 U/mL) or the combination for the indicated times. (A) Cell surface expression was measured by flow cytometry. Monocytes were identified by staining with PE-labeled anti-CD14 monoclonal antibody (mAb), and ICAM-1 was identified on CD14+ cells by staining with FITC-labeled anti–ICAM-1 mAb. (B) RNA was isolated from cytokine-stimulated monocytes, and total RNA was analyzed by Northern blot with ICAM-1 and GAPDH probes. (C) Normalized absorption values were obtained by densitometry scanning of ICAM-1 and GAPDH mRNA bands. From the ratio of ICAM-1 to GAPDH, the fold increase over untreated control cells was calculated. Data are representative of 3 experiments.
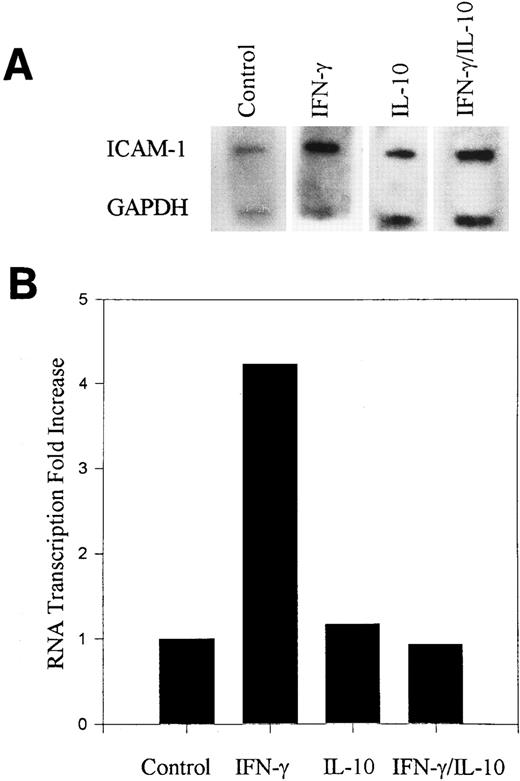
IL-10 inhibits ICAM-1 gene transcription.IL-10 inhibition of ICAM-1 mRNA may result from either a reduction in transcription of the ICAM-1 gene and/or an increase in degradation of ICAM-1 mRNA. We performed nuclear run-on assays to determine if IL-10 inhibited ICAM-1 gene transcription. Nuclei isolated from monocytes activated with IFN-γ in the presence or absence of IL-10 for 3 hours were monitored for ICAM-1 transcription. IFN-γ increased the rate of ICAM-1 gene transcription in monocytes, and IL-10 prevented this increased transcription (Fig 2). These data demonstrate that IL-10 regulation of ICAM-1 in monocytes is mediated at least in part by transcription.
IL-10 inhibits IFN-γ–induced ICAM-1 gene transcription. (A) Nuclei were isolated from monocytes stimulated with IFN-γ (100 U/mL) in the presence or absence of IL-10 (100 U/mL) for 3 hours. Transcription occurred in the presence of labeled ribonucleotides. Isolated RNA was hybridized to ICAM-1 cDNA and GAPDH cDNA that was slot blotted onto nitrocellulose. (B) Normalized absorption values were obtained by densitometry scanning of the ICAM-1. From the ratio of ICAM-1 to GAPDH, the fold increase over untreated control cells was calculated. Data are representative of 2 experiments.
IL-10 inhibits IFN-γ–induced ICAM-1 gene transcription. (A) Nuclei were isolated from monocytes stimulated with IFN-γ (100 U/mL) in the presence or absence of IL-10 (100 U/mL) for 3 hours. Transcription occurred in the presence of labeled ribonucleotides. Isolated RNA was hybridized to ICAM-1 cDNA and GAPDH cDNA that was slot blotted onto nitrocellulose. (B) Normalized absorption values were obtained by densitometry scanning of the ICAM-1. From the ratio of ICAM-1 to GAPDH, the fold increase over untreated control cells was calculated. Data are representative of 2 experiments.
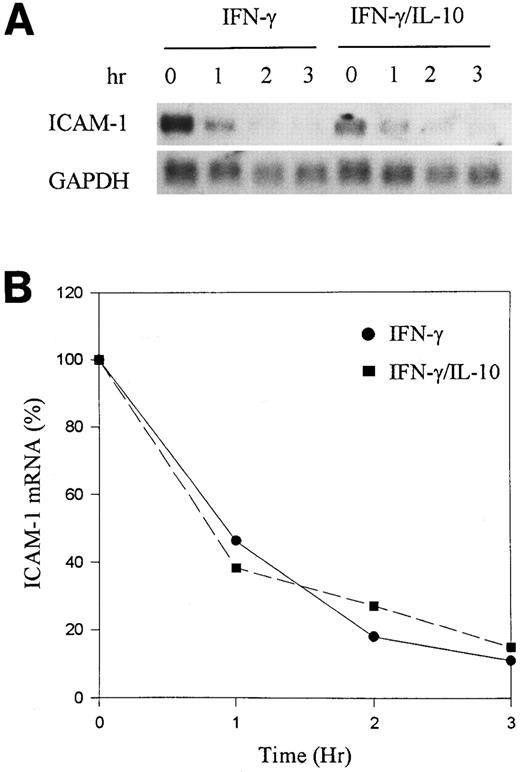
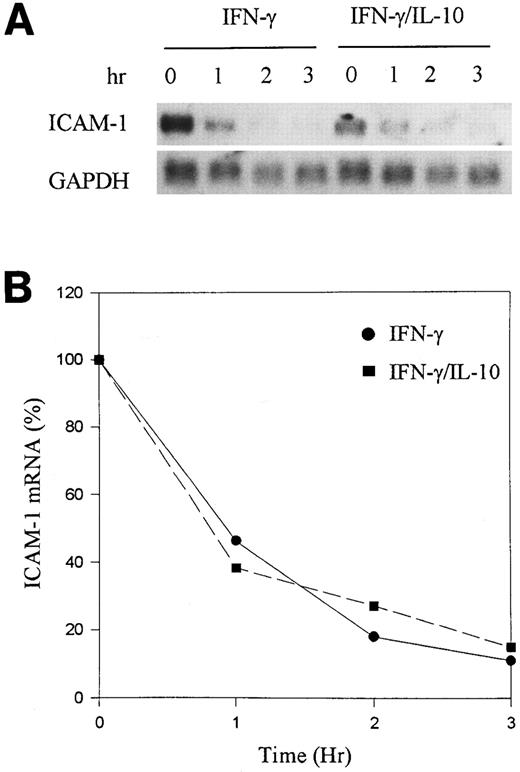
It has been reported that IL-10, in addition to inhibiting cytokine gene transcription, in some instances enhanced mRNA degradation.13 To determine if IL-10 alters the half-life (t1/2 ) of ICAM-1 mRNA, we used Actinomycin D to block transcription in activated monocytes and determined the rate of mRNA degradation by Northern blot analysis. Monocytes were stimulated with IFN-γ alone or IFN-γ plus IL-10 for 16 hours. Actinomycin D (5 μg/mL) was then added to prevent further mRNA synthesis. RNA was isolated at different time points, and ICAM-1 mRNA was analyzed by Northern blot. There was a steady reduction in ICAM-1 mRNA over the 3-hour period, with a t1/2 of about 1 hour (Fig 3). However, IL-10 did not enhance the rate of ICAM-1 mRNA degradation, suggesting that IL-10 does not alter ICAM-1 mRNA stability. Taken together, these data demonstrate that IL-10 inhibits IFN-γ–induced ICAM-1 expression primarily at the level of gene transcription.
IL-10 does not enhance mRNA degradation. (A) Monocytes were stimulated for 24 hours with IFN-γ (100 U/mL) in the presence or absence of IL-10 (100 U/mL). Actinomycin D (5 μg/mL) was added to the cells, and RNA was extracted at 0, 1, 2, and 3 hours. Northern blot analysis was performed. (B) Induced levels of ICAM-1 mRNA at time zero were determined by densitometry for IFN-γ and IFN-γ plus IL-10. The values were set at 100% and plotted against the percent reduction in the amount of mRNA. The RNA half-life measured the time at which mRNA declined by 50%. Data are representative of 3 experiments.
IL-10 does not enhance mRNA degradation. (A) Monocytes were stimulated for 24 hours with IFN-γ (100 U/mL) in the presence or absence of IL-10 (100 U/mL). Actinomycin D (5 μg/mL) was added to the cells, and RNA was extracted at 0, 1, 2, and 3 hours. Northern blot analysis was performed. (B) Induced levels of ICAM-1 mRNA at time zero were determined by densitometry for IFN-γ and IFN-γ plus IL-10. The values were set at 100% and plotted against the percent reduction in the amount of mRNA. The RNA half-life measured the time at which mRNA declined by 50%. Data are representative of 3 experiments.
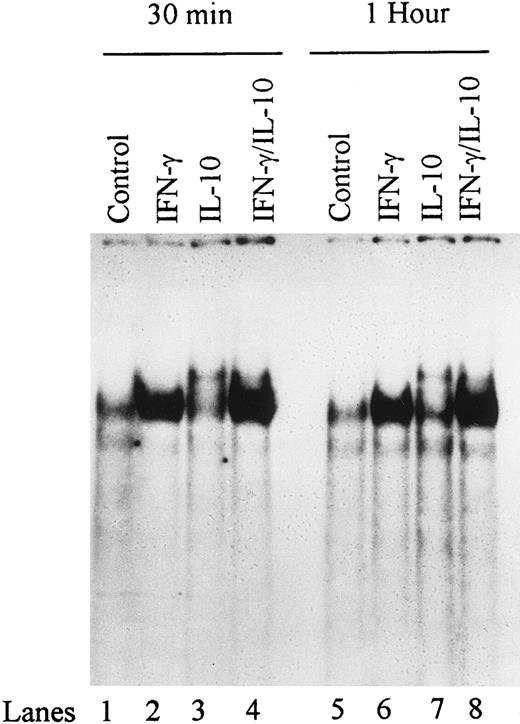
Effect of IL-10 on STAT-1α binding activity.IFN-γ has been shown to activate the transcription factor STAT-1α.28,29 Look et al22 described an IRE in the ICAM-1 promoter between nucleotide positions −116 and −106 that is required for ICAM-1 promoter induction by IFN-γ. To determine whether the IRE motif could bind IFN-γ–induced or IL-10–induced DNA binding proteins in monocytes, we synthesized a 20-bp oligonucleotide encompassing the IRE, prepared nuclear extracts from IFN-γ–treated monocytes in the presence or absence of IL-10 for 0.5 and 1 hour, and assessed DNA binding activity by EMSA. IFN-γ induced a complex that was evident as early as 30 minutes (Fig 4). In contrast, IL-10 induced two weak complexes: one comigrated with the IFN-γ–induced complex, and the second migrated more slowly. When IFN-γ and IL-10 were combined to stimulate monocytes, we detected a dominant complex corresponding to the IFN-γ–induced complex and a much weaker complex corresponding to the slower-migrating IL-10 complex. These data show that IL-10 does not inhibit the IFN-γ–induced complex.
Kinetics of IFN-γ and IL-10 induction of DNA binding activity on ICAM-1 IRE. Nuclear extracts were obtained from monocytes unstimulated (control) or stimulated with IFN-γ (100 U/mL), IL-10 (100 U/mL), or the combination of IFN-γ and IL-10 and analyzed for DNA binding activity in gel mobility shifts using labeled ICAM-1 IRE. Arrows indicate binding complexes. Data are representative of 3 experiments.
Kinetics of IFN-γ and IL-10 induction of DNA binding activity on ICAM-1 IRE. Nuclear extracts were obtained from monocytes unstimulated (control) or stimulated with IFN-γ (100 U/mL), IL-10 (100 U/mL), or the combination of IFN-γ and IL-10 and analyzed for DNA binding activity in gel mobility shifts using labeled ICAM-1 IRE. Arrows indicate binding complexes. Data are representative of 3 experiments.
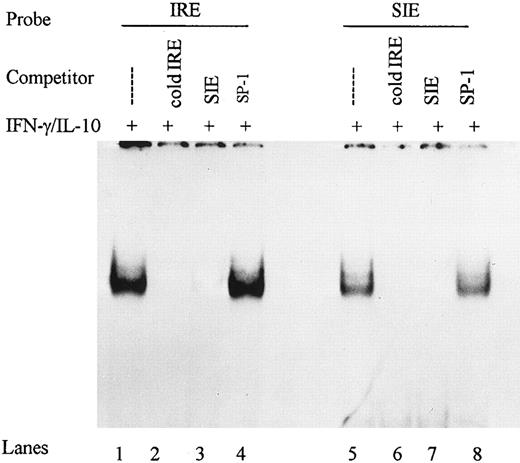
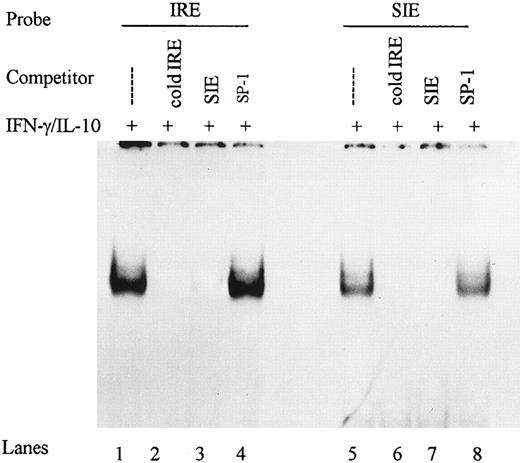
Competitive-inhibition studies demonstrated the specificity of the IFN-γ–activated IRE binding complex. We used the c-sis inducible element (SIE) from the c-fos promoter shown previously to bind STAT-1α,34 to assess whether the IRE is also recognized by STAT-1α. Nuclear extracts from monocytes stimulated with IFN-γ plus IL-10 were incubated with labeled IRE and a 100-fold molar excess of IRE or SIE. Both the IRE and the SIE competed for proteins binding to the IRE, whereas no competition was observed for an unrelated ICAM-1 Sp1 binding sequence (Fig 5). These data demonstrate that IFN-γ and IL-10 induced specific gel shift complexes on the ICAM-1 IRE that may contain STAT-1α.
DNA binding complexes are specific for IRE and SIE. Nuclear extracts from monocytes stimulated with IFN-γ (100 U/mL) and IL-10 (100 U/mL) were incubated with labeled IRE and competed with 100-fold molar excess of IRE, SIE, or SP-1 for DNA binding activity in gel mobility shifts (lanes 1 to 4). Reciprocally, nuclear extracts were incubated with labeled SIE and competed with 100-fold molar excess IRE, SIE, and SP-1 (lanes 5 to 8). Data are representative of 2 experiments.
DNA binding complexes are specific for IRE and SIE. Nuclear extracts from monocytes stimulated with IFN-γ (100 U/mL) and IL-10 (100 U/mL) were incubated with labeled IRE and competed with 100-fold molar excess of IRE, SIE, or SP-1 for DNA binding activity in gel mobility shifts (lanes 1 to 4). Reciprocally, nuclear extracts were incubated with labeled SIE and competed with 100-fold molar excess IRE, SIE, and SP-1 (lanes 5 to 8). Data are representative of 2 experiments.
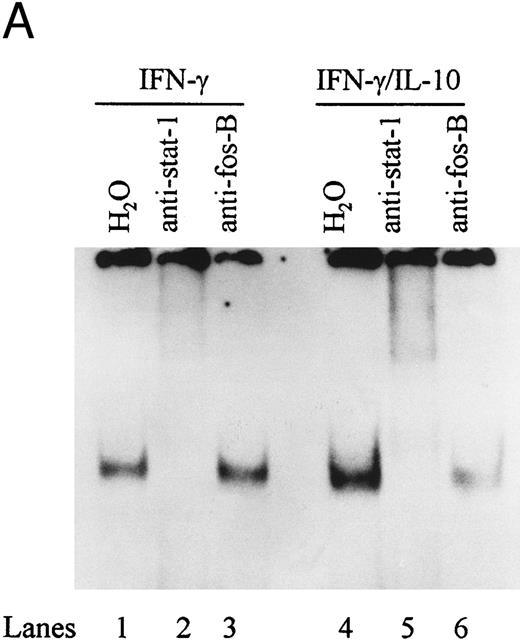
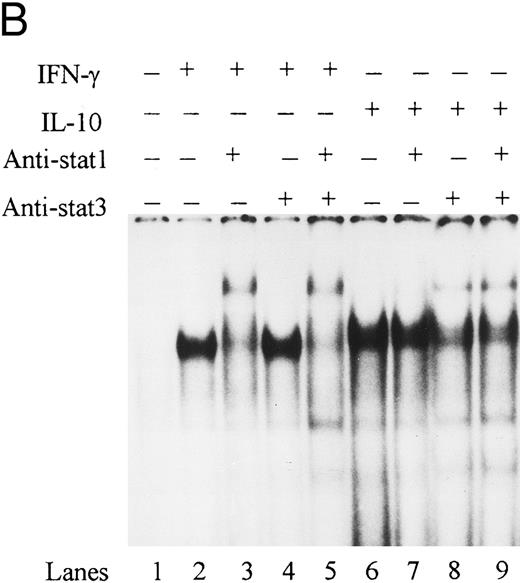
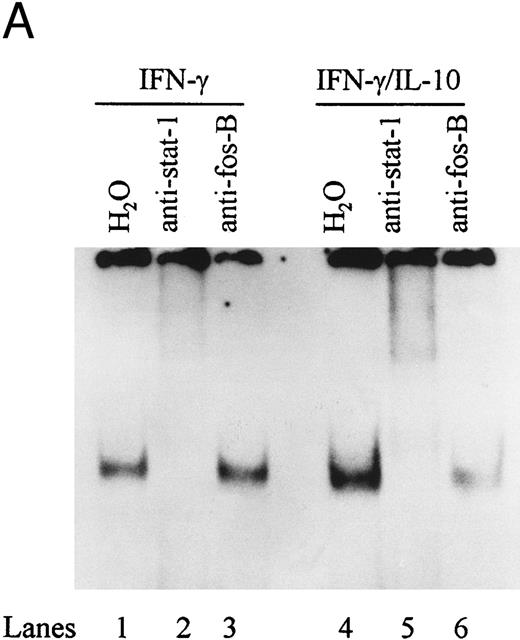
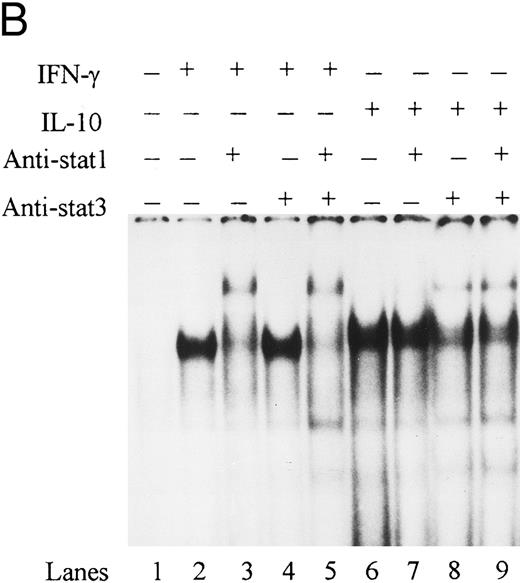
To directly demonstrate the presence of STAT-1α, we incubated STAT-1α antibody with nuclear extracts and assessed the effects on binding activity by EMSA. Anti–STAT-1α antibody supershifted the IFN-γ–induced IRE binding complex, whereas the anti–fos B antibody had no effect on the binding complex (Fig 6A). The combination of IFN-γ and IL-10 also induced an IRE binding complex that was supershifted by the anti–STAT-1α antibody. These data demonstrate that IL-10 does not alter STAT-1α binding to the ICAM-1 IRE. To determine if STAT-3 was present in the ICAM-1 IRE binding complexes, antibody to STAT-3 was incubated with nuclear extracts and assessed by EMSA. Anti–STAT-3 antibody did not supershift the IFN-γ–induced binding complex (lane 4), consistent with previous reports that IFN-γ specifically activates STAT-1α (Fig 6B). However, STAT-3 and STAT-1α were identified in the IRE binding complexes in IL-10–activated monocytes (lanes 7 and 8).
IL-10 does not block IFN-γ–induced STAT-1α binding to the ICAM-1 IRE. (A) Nuclear extracts from monocytes stimulated with IFN-γ (100 U/mL) in the presence (lanes 4 to 6) or absence of IL-10 (100 U/mL) (lanes 1 to 3) were incubated with anti–STAT-1α or anti–fos B antibody. (B) Nuclear extracts from monocytes stimulated with IFN-γ (100 U/mL) (lanes 1 to 4) and IL-10 (100 U/mL) (lanes 5 to 8) were incubated with anti–STAT-1α, anti–STAT-3, or both antibodies combined. Antibody binding complexes were assessed by gel mobility supershift with labeled ICAM-1 IRE. Data are representative of 3 experiments.
IL-10 does not block IFN-γ–induced STAT-1α binding to the ICAM-1 IRE. (A) Nuclear extracts from monocytes stimulated with IFN-γ (100 U/mL) in the presence (lanes 4 to 6) or absence of IL-10 (100 U/mL) (lanes 1 to 3) were incubated with anti–STAT-1α or anti–fos B antibody. (B) Nuclear extracts from monocytes stimulated with IFN-γ (100 U/mL) (lanes 1 to 4) and IL-10 (100 U/mL) (lanes 5 to 8) were incubated with anti–STAT-1α, anti–STAT-3, or both antibodies combined. Antibody binding complexes were assessed by gel mobility supershift with labeled ICAM-1 IRE. Data are representative of 3 experiments.
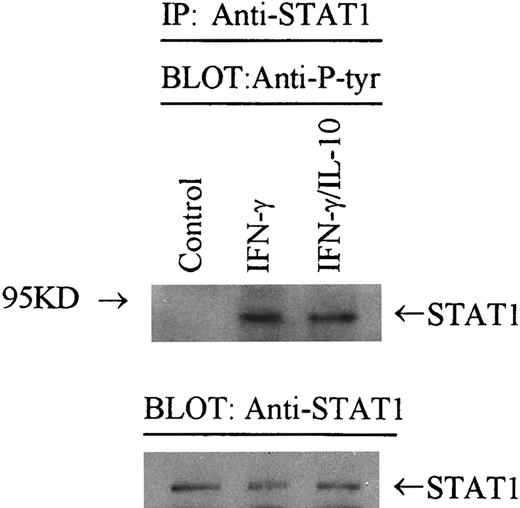
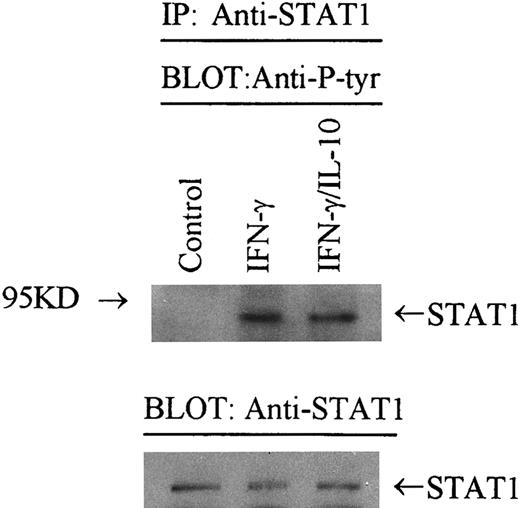
Effect of IL-10 on STAT-1α phosphorylation.STAT-1α is rapidly activated by tyrosine phosphorylation, which allows STAT-1α dimers to bind IRE sequences.28,29 31 Because we were unable to identify a difference in STAT-1α binding in the presence of IL-10, it appears that IL-10 does not inhibit tyrosine phosphorylation of STAT-1α. To directly demonstrate this, we immunoprecipitated STAT-1α from IFN-γ–activated monocytes in the presence or absence of IL-10. Adherent monocytes were treated with cytokines for 5 minutes, solubilized, and immunoprecipitated with anti–STAT-1α antibody. The immunocomplexes were washed, resolved on 8% SDS-PAGE, transferred to nitrocellulose, and probed with antiphosphotyrosine antibody 4G10. In monocytes treated with IFN-γ and IL-10 (Fig 7, lane 4), there was approximately the same level of STAT-1α phosphorylation versus IFN-γ alone (lane 2). Normalization of phosphorylated STAT-1α to the level of STAT-1α protein by laser densitometry showed that an equivalent amount of phosphorylated STAT-1α was detected from IFN-γ and IFN-γ plus IL-10 precipitates. These data in combination with the DNA binding experiments indicate that IL-10 does not inhibit the activity of IFN-γ by altering the binding activity of tyrosine phosphorylated STAT-1α protein.
IL-10 does not inhibit tyrosine phosphorylation of STAT-1α. Monocytes were stimulated with IFN-γ (100 U/mL) in the presence or absence of IL-10 (100 U/mL) for 5 minutes. Cells were lysed, and STAT-1α was immunoprecipitated with Sepharose-bound anti–STAT-1 antibody. Immunoprecipitates were resolved on SDS-PAGE, blotted to nitrocellulose, probed with an antiphosphotyrosine antibody (4G10), and developed by enhanced chemiluminescence. In the lower panel, the blot was stripped and reprobed with anti–STAT-1 antibody and developed. Data are representative of 2 separate experiments.
IL-10 does not inhibit tyrosine phosphorylation of STAT-1α. Monocytes were stimulated with IFN-γ (100 U/mL) in the presence or absence of IL-10 (100 U/mL) for 5 minutes. Cells were lysed, and STAT-1α was immunoprecipitated with Sepharose-bound anti–STAT-1 antibody. Immunoprecipitates were resolved on SDS-PAGE, blotted to nitrocellulose, probed with an antiphosphotyrosine antibody (4G10), and developed by enhanced chemiluminescence. In the lower panel, the blot was stripped and reprobed with anti–STAT-1 antibody and developed. Data are representative of 2 separate experiments.
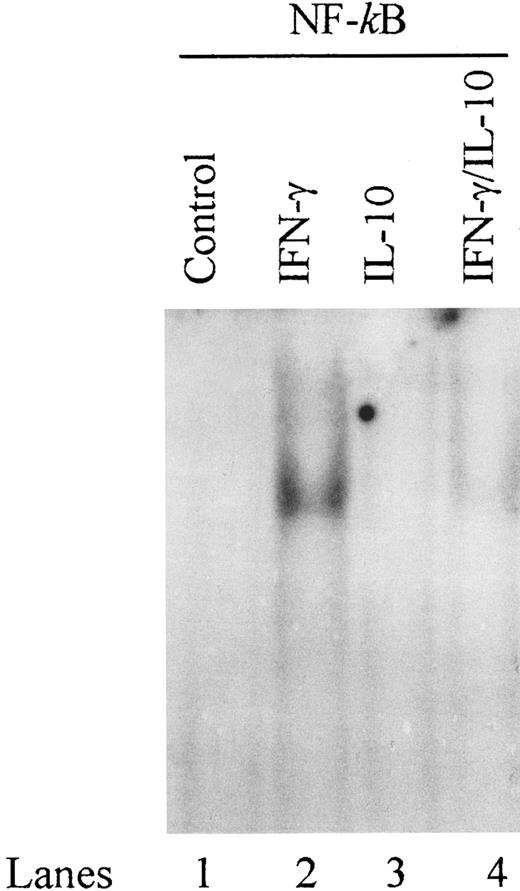
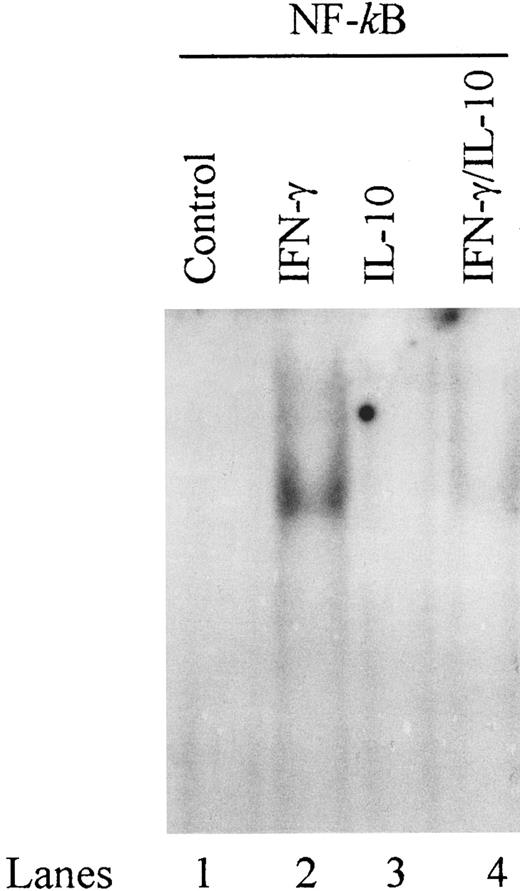
IL-10 inhibits binding of NF-κB to a proximal site in the ICAM-1 promoter.ICAM-1 promoter also contains binding sites for AP-1, AP-2, AP-3, NF-κB, and Sp1.20 21 In EMSAs, we were unable to detect binding of nuclear proteins from IFN-γ–stimulated monocytes to oligonucleotides corresponding to binding sites for AP-1, AP-2, AP-3, and Sp1 (data not shown). The ICAM-1 promoter contains two NF-κB sites located distal (nucleotides −540 to −528) and proximal (nucleotides −228 to −217) to the transcription start site.
These NF-κB binding sites have not been previously identified to be involved in IFN-γ–induced transcription of the ICAM-1 gene. However, because recent data indicate that IL-10 can inhibit NF-κB binding activity in LPS-stimulated monocytes,14 we assessed NF-κB binding activity in IFN-γ–activated monocytes. We synthesized oligonucleotides that correspond to the distal and proximal NF-κB elements in the ICAM-1 promoter and assessed their binding activity by EMSA. IFN-γ increased DNA binding activity of the proximal NF-κB site, and IL-10 markedly reduced this binding activity (Fig 8). In contrast, IFN-γ did not induce DNA binding activity of the distal NF-κB site (data not shown). These results demonstrate that IL-10 can inhibit IFN-γ–induced NF-κB binding activity of the ICAM-1 promoter in monocytes.
IL-10 inhibits binding of NF-κB to a proximal site in the ICAM-1 promoter. Monocytes were stimulated with IFN-γ (100 U/mL), IL-10 (100 U/mL), or IFN-γ plus IL-10 in combination for 5 minutes. Nuclear extracts were analyzed for gel mobility shifts with the labeled ICAM-1 proximal NF-κB oligonucleotide. Data are representative of 3 separate experiments.
IL-10 inhibits binding of NF-κB to a proximal site in the ICAM-1 promoter. Monocytes were stimulated with IFN-γ (100 U/mL), IL-10 (100 U/mL), or IFN-γ plus IL-10 in combination for 5 minutes. Nuclear extracts were analyzed for gel mobility shifts with the labeled ICAM-1 proximal NF-κB oligonucleotide. Data are representative of 3 separate experiments.
DISCUSSION
In this study, we investigated the mechanism by which IL-10 inhibits ICAM-1 expression in human monocytes activated with IFN-γ. We show that regulation of ICAM-1 by IL-10 occurs at the level of gene transcription and not at the level of mRNA stability. These observations suggest that IL-10 targets transcription factors interacting with the ICAM-1 promoter elements. Feldman et al32 reported that IgG complexes inhibit FcγRI gene transcription in monocytes by inhibiting phosphorylation and binding activity of STAT-1α. However, we were unable to detect any effect of IL-10 on IFN-γ induction of STAT-1α phosphorylation or binding to the ICAM-1 IRE. These findings suggest that IL-10 inhibition of ICAM-1 may involve a novel mechanism distinct from that used to downregulate the FcγRI gene.
It is interesting that ICAM-1 expression in microglial cells, macrophage-like cells in the central nervous system, is induced by IFN-γ stimulation and inhibited by IL-10.35 However, unlike the effect in human monocytes, IL-10 does not inhibit ICAM-1 mRNA accumulation. In microglial cells, IL-10 affects ICAM-1 expression at the translational and/or posttranslational level, whereas in human monocytes it affects transcription.35 There are several possible differences between these two cell populations. Microglial cells are a differentiated macrophage-like cell, and transcriptional activation of genes in monocytes is developmentally regulated.36 Also, since microglial cells are rodent-derived, the differences may be due to species differences.
There are a number of possible mechanisms that may be responsible for inhibition of IFN-γ–induced ICAM-1 transcription in monocytes. Several observations indicate that not only tyrosine phosphorylation but also serine phosphorylation is required for transcriptional activity of STAT proteins. Eilers et al36 showed that a reduction in STAT-1α serine phosphorylation correlates with decreased STAT-1α binding activity. Wen et al37 demonstrated that cells expressing a mutant form of STAT-1 in which Ser-727 was changed to alanine could be phosphorylated on tyrosine and bound specific IRE sites; however, serine phosphorylation was required for full IFN-γ activity. Thus, IL-10 may alter serine phosphorylation of STAT-1α proteins, which in turn would influence transcription of the ICAM-1 gene.
Another possible way IL-10 could influence IFN-γ–induced ICAM-1 transcription is through the induction of STAT-3 binding to the ICAM-1 IRE. The IRE in the ICAM-1 gene is similar to other IFN-γ–responsive elements in FcγRI, c-fos, and IRF-1 genes.23,24,34 The IFN-γ–responsive element (GRR) of the FcγRI gene has been used to identify DNA binding proteins that are induced by IFN-γ and IL-10.22 It has been reported that IFN-γ induces STAT-1α,28,29 and IL-10 induces predominately STAT-3.38 STAT-1α and STAT-3 can form both homodimers and heterodimers.38 It is possible that since IFN-γ induces STAT-1α and IL-10 induces STAT-3, IL-10 inhibits ICAM-1 transcription by altering the composition of dimers formed on the IRE. In IFN-γ–activated monocytes, STAT-1α would form homodimers capable of activating the ICAM-1 promoter, whereas in IL-10–activated monocytes, there would be mostly STAT-3 homodimers, which cannot activate the ICAM-1 promoter. Consequently, the combination of IFN-γ/IL-10 could produce heterodimers of STAT-1α and STAT-3 that may function as an inhibitory complex, thereby reducing ICAM-1 transcription.
One of the main outcomes of the inhibitory activity of IL-10 in monocytes is the reduction in cytokine synthesis. Although IL-10 controls cytokine production at the level of transcription, little is known about the mechanism. Experiments from Geng et al15 showed that LPS induces the early tyrosine kinase p56 lyn in monocytes and that IL-10 inhibits p56 lyn activity. Since tyrosine kinase activity is necessary for LPS activation of IL-1, IL-6, and TNF-α, this may be the mechanism by which IL-10 inhibits cytokine production. Wang et al14 also reported that LPS- and TNF-α–induced NF-κB activity is inhibited by IL-10. They postulated that since NF-κB activity is required for transcription of many cytokine genes, this may be a common mechanism by which IL-10 inhibits transcription. Consistent with a role for NF-κB in IL-10 inhibition, we showed that IL-10 inhibited binding of IFN-γ–induced nuclear factors to the proximal ICAM-1 NF-κB binding site. These data suggest that NF-κB may be necessary for maximum IFN-γ–induced ICAM-1 transcription in monocytes. It is of interest that the proximal NF-κB element is a functional element in ICAM-1 transcription regulating TNF-α responses.39,40 Recently, it was shown in epithelial and endothelial cells that mutation of the proximal NF-κB site reduced IFN-γ–induced ICAM-1 transcription.39 In addition, IFN-γ activation of the IP-10 gene is regulated by cooperative interactions between STAT-1α and NF-κB.41 Future experiments using transfection of the ICAM-1 promoter elements will determine if IFN-γ–induced ICAM-1 transcription is regulated by NF-κB and whether IL-10 inhibition specifically localizes to the proximal NF-κB binding site. Taken together, these observations suggest that IL-10 inhibition of IFN-γ–induced ICAM-1 transcription is complex and may involve the interaction of multiple transcription factor binding sites.
Supported by the Arthritis Foundation, Illinois Chapter.
Address reprint requests to Alison Finnegan, PhD, Section of Rheumatology, Rush Presbyterian-St Luke's Medical Center, 1653 W Congress Pkwy, Chicago, IL 60612.