In this issue of Blood, Thomas et al1 present a whole-genome sequencing analysis of 230 pediatric and adult Burkitt lymphomas (BLs). The researchers wished to discover genomic subgroups within this rare disease by feeding data on somatic mutations, copy number alterations, and structural variants into a consensus clustering algorithm. Previously, this approach established 5 subtypes of diffuse large B-cell lymphoma (DLBCL) with compelling biological and clinical differences.2 In BLs, the result is less clear-cut, and our imperfect diagnostic criteria may be largely to blame. Although some clusters emerged from the BL cases, the researchers obtained a more robust classification after adding and comparing 295 DLBCL genomes to the BL genomes.
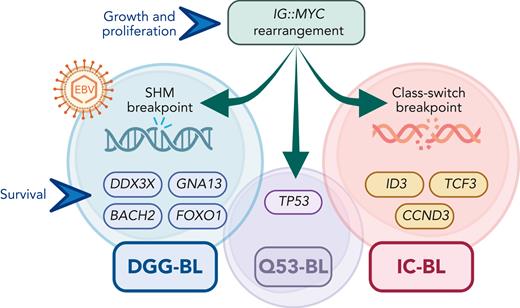
Consistent with prior experience from gene expression profiling (GEP) studies, quite a few histologically confirmed BL tumors turned out to group with DLBCL and vice versa. The 3 main identified subtypes (termed DGG-BL [defined by mutations in DDX3X, GNA13, GNAI2, BACH2, and FOXO1], IC-BL [defined by mutations in ID3, TCF3, and CCND3], and Q53-BL [defined by mutations in TP53]; see figure) illustrate various genomic pathways that allow BL cells to overcome vulnerability to cell death under the common MYC-driven cell growth and proliferation program. We can also glimpse hints of prognostic relevance for specific DNA alterations.
Main discerning features of the BL genomic subgroups: rearrangements between MYC and IG genes are universal, but the SHM breakpoints are more common in DGG-BL, which is strongly associated with EBV infections; DGG-BL and IC-BL acquire separate sets of additional mutations that act in concert with the MYC-induced cell growth and proliferation program; TP53 mutations occur frequently in all subtypes, but a small subset (termed Q53-BL) lacks any additional driver mutations. IG, immunoglobulin; SHM, somatic hypermutation; EBV, Epstein-Barr virus.
Main discerning features of the BL genomic subgroups: rearrangements between MYC and IG genes are universal, but the SHM breakpoints are more common in DGG-BL, which is strongly associated with EBV infections; DGG-BL and IC-BL acquire separate sets of additional mutations that act in concert with the MYC-induced cell growth and proliferation program; TP53 mutations occur frequently in all subtypes, but a small subset (termed Q53-BL) lacks any additional driver mutations. IG, immunoglobulin; SHM, somatic hypermutation; EBV, Epstein-Barr virus.
BL seems deceptively homogeneous: it has the nearly universal IG::MYC translocation, a monotonous morphology, and a simple karyotype, and it is consistently responsive to intensive, unsophisticated blasts of chemotherapy.3 Historical classifications of BL used patient age or epidemiologic attributes, distinguishing the endemic, immunosuppression-associated, and sporadic variants. Apart from the striking association between the endemic variant and an EBV infection, these approaches provided little insight to influence treatments or patient outcomes. Prognosis and therapeutic decisions rely on gross indicators of disease burden and have not taken advantage of any molecular factors.4 Uncovering driver alterations that could help replace cytotoxic agents with more targeted therapeutic approaches is a high research priority in BL. In this context, the utility of genomic subclassification in BL will depend on whether it can help us understand the clinical and epidemiologic associations, clarify the diagnostic conundrums, and inform prognostication or treatment.
Previous studies have treated BL as a fixed entity separable from DLBCL, with frequent mutations in genes involved in the germinal center dark zone program (TCF3, ID3, CCND3, and FOXO1). The EBV status proved more relevant for genomic profiling than age or epidemiologic context.5,6 As identified in this study, 72% of DGG-BL tumors were EBV positive. Consequently, they showed a relatively high mutation load related to overexpression of activation-induced cytidine deaminase and aberrant SHM. SHM-related IG::MYC breakpoints were also more common in EBV-positive cases, even though class-switch recombination had been considered the predominant mechanism for such breakpoints in BL.7 The most perplexing finding is that tumors classified as IC-BL unexpectedly overexpressed IRF4 and TNFRSF13B as well as gene sets associated with post–germinal center phenotypes. The clinicopathologic significance of this discovery awaits exploration. The differential therapeutic vulnerability of the DGG-BL and IC-BL subtypes to PI3K signaling inhibition or immunomodulation is an intriguing possibility.
The genomic subgroups of BL are perhaps best thought of as segregated pathogenic mechanisms that give rise to a common phenotype, rather than different diseases. The double-hit lymphoma story illustrates that high-grade B-cell lymphoma (HGBL) biology requires 2 concurrent genomic hits: one to facilitate uncontrolled proliferation (eg, IG::MYC) and another to provide protection from cell death (eg, BCL2::IGH). We can now discern that in BL, this second survival-promoting hit can be acquired through several pathways: the TCF3/ID3 axis supporting constitutive PI3K signaling (IC-BL), DDX3X-mediated attenuation of proteotoxic stress (DGG-BL), or even simple inactivation of p53 (Q53-BL). These mechanisms are not mutually exclusive, as evidenced by substantial overlap in the relevant gene mutations among the subgroups. TP53 mutations are common in all 3 BL subtypes and are overrepresented in relapsed disease, potentially contributing to treatment refractoriness regardless of the mutational profile. Nevertheless, the quiet Q53-BL subgroup intriguingly lacks any driver mutations beyond those in MYC and TP53, and it does not show the genomic complexity typically associated with p53 aberrations (in contrast to the aneuploid A53 subtype of DLBCL).
Approximately 9% of DLBCL tumors (as well as most BLs whose diagnosis were rejected upon central review) were classified within the BL genomic clusters. This finding replicates prior GEP experience and contributes to our understanding of the entire spectrum of mature HGBL. HGBLs include BL, double-hit lymphomas, HGBL with 11q aberration, and the rare HGBL, not otherwise specified, as well as perhaps 10% to 15% of tumors currently diagnosed as DLBCL. Despite the confusing range of histologic patterns, these aggressive lymphomas seem to use consistent sets of mutated transcription factors to hijack mechanisms that operate within the germinal center dark zone. The emerging GEP and genomic classifiers need translation into trials that would separate the HGBL-like tumors from DLBCL to identify more efficacious treatments and mitigate their refractoriness to the standard-intensity chemotherapy.
The study by Thomas et al does not paint an easily interpretable picture of prognostic relevance for the discovered BL subgroups. The overall survival of the pediatric and adult patients included in the study exceeded 80%, much higher than observed in clinical practice in either North America or Europe.4 In aggregate, outcomes did not differ between DGG-BL and IC-BL, and borderline (and contradictory) associations from the pediatric and adult cohorts are hard to interpret with as few as 5 to 8 patients in the subgroups and the lack of stratification in baseline variables or treatment. Nevertheless, the notable prognostic disadvantage for adult BL carrying TP53 mutations provides a hint that sequencing data might add value to clinical prognosis. The constellation of MYC rearrangement with TP53 mutation, regardless of histology, may define one of the worst categories of HGBL.8 Future research should include more cases of disseminated, extranodal BL, which is often diagnosed using blood, bone marrow, or cerebrospinal fluid. Such tumors carry significantly worse prognosis in both experimental and observational cohorts4,9,10 but may be underrepresented in sequencing studies due to unavailability of archival paraffin-embedded nodal tissue. Analysis of prospectively collected samples from patients who are uniformly treated on clinical trials will be the next critical step to examine the prognostic impact of specific genomic alterations in BL and the practical relevance of the subtypes.
Conflict-of-interest disclosure: A.J.O. consulted for Schrödinger, Genmab, and TG Therapeutics and received research funding from the Leukemia and Lymphoma Society, Adaptive Biotechnologies, CellDex, Genentech, Genmab, Precision BioSciences, Spectrum Pharmaceuticals, and TG Therapeutics.