Key Points
An academically developed IGHV leader-based NGS assay can routinely detect and quantify MRD to 10−5 and beyond in CLL.
MRD quantification below 10−4 using this assay improves prognostic stratification in CLL.
Abstract
The sensitivity of conventional techniques for reliable quantification of minimal/measurable residual disease (MRD) in chronic lymphocytic leukemia (CLL) is limited to MRD 10−4. Measuring MRD <10−4 could help to further distinguish between patients with CLL with durable remission and those at risk of early relapse. We herein present an academically developed immunoglobulin heavy-chain variable (IGHV) leader-based next-generation sequencing (NGS) assay for the quantification of MRD in CLL. We demonstrate, based on measurements in contrived MRD samples, that the linear range of detection and quantification of our assay reaches beyond MRD 10−5. If provided with sufficient DNA input, MRD can be detected down to MRD 10−6. There was high interassay concordance between measurements of the IGHV leader-based NGS assay and allele-specific oligonucleotide quantitative polymerase chain reaction (PCR) (r = 0.92 [95% confidence interval {CI}, 0.86-0.96]) and droplet digital PCR (r = 0.93 [95% CI, 0.88-0.96]) on contrived MRD samples. In a cohort of 67 patients from the CLL11 trial, using MRD 10−5 as a cutoff, undetectable MRD was associated with superior progression-free survival (PFS) and time to next treatment. More important, deeper MRD measurement allowed for additional stratification of patients with MRD <10−4 but ≥10−5. PFS of patients in this MRD range was significantly shorter, compared with patients with MRD <10−5 (hazard ratio [HR], 4.0 [95% CI, 1.6-10.3]; P = .004), but significantly longer, compared with patients with MRD ≥10−4 (HR, 0.44 [95% CI, 0.23-0.87]; P = .018). These results support the clinical utility of the IGHV leader-based NGS assay.
Introduction
Detectability of minimal/measurable residual disease (MRD) after treatment predicts shorter progression-free survival (PFS), time to next treatment (TTNT), and overall survival (OS) in chronic lymphocytic leukemia (CLL).1,2 Hence, the international workshop on CLL guideline recommends assessment of MRD in all clinical trials that aim at maximizing the depth of remission.3 Accurate measurement of MRD requires reliable differentiation between leukemic cells and their healthy counterparts. To this end, most recent clinical trials have used either multicolor flow cytometry (MFC) or allele-specific oligonucleotide quantitative polymerase chain reaction (ASO-qPCR).4-6
MFC is a standardized technique and produces rapid results, but requires samples to be freshly processed.4,7,8 Conversely, molecular techniques, such as ASO-qPCR, rely on the availability of DNA, which is suitable for long-term storage. The fundamental principle behind these techniques is that all leukemic cells are expected to carry the same, highly unique immunoglobulin heavy-chain (IGH) rearrangement. This patient-specific nucleotide sequence can be used as a DNA fingerprint to detect residual leukemic cells. Indeed, ASO-qPCR–based MRD assays use patient-specific primers to selectively amplify and detect their target rearrangement.5,6 Although ASO-qPCR has considerable sensitivity, it requires patient-tailored primer design. Moreover, in a minority of CLL cases, sensitive assay design is hampered because of somatic hypermutation (SHM) in primer-annealing sites.9
Both MFC and ASO-qPCR are routinely used in clinical trials to quantify MRD down to 1 CLL cell per 10 000 (10−4) healthy counterparts.4-7 MRD below this level is designated as undetectable MRD (<10−4) (uMRD4). Of note, although some patients with CLL who achieve uMRD4 after treatment with chemoimmunotherapy have durable disease remission, most patients who reach uMRD4 will eventually convert to MRD positivity and experience disease relapse.10-13 Moreover, landmark analysis from the MURANO trial has demonstrated that patients who reached uMRD4 in the chemoimmunotherapy arm had inferior PFS, compared with patients who reached uMRD4 in the rituximab-venetoclax arm.14 These observations suggest that not all uMRD4 is equal. Quantification of MRD <10−4 may further distinguish between patients with CLL who retain durable remission and those at risk of earlier relapse.
Achieving reliable quantification beyond MRD 10−4 requires novel approaches. A promising modality for the measurement of MRD is next-generation sequencing (NGS).15 Herein, consensus primers are used to amplify all IGH rearrangements from a pool of DNA, obviating the need for laborious, patient-specific primer design. Subsequently, massively parallel deep sequencing is performed to detect the presence of the leukemia-specific target. Previously, the NGS-based clonoSEQ immunoglobulin κ/immunoglobulin λ (IGH/IGK/IGL) assay proved capable of measuring beyond MRD 10−4 in CLL.8,16,17 However, this assay is exclusively commercially operated, and protocols and primer sequences are not publicly accessible.18 In addition, although it has been suggested that measurement beyond MRD 10−4 in peripheral blood more accurately predicts survival, this remains to be clearly demonstrated.16
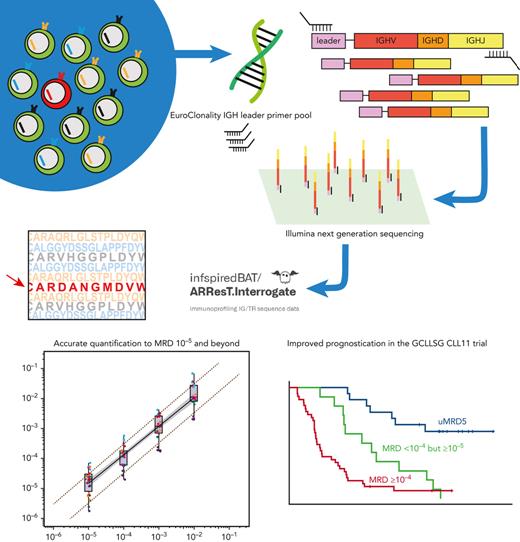
In this article, we present an academically developed IGHV leader-based NGS assay for the measurement of MRD in CLL, based on the pioneering work of the Euroclonality-NGS working group.19,20 We demonstrate that this assay can reliably detect and quantify MRD beyond the level of MRD 10−5 and up to MRD 10−6, provided that enough DNA is available. Moreover, we demonstrate that this assay shows good concordance with ASO-qPCR and droplet digital polymerase chain reaction (ddPCR). Finally, using patient samples from the CLL11 trial, we assessed the clinical utility of measuring MRD to 10−5 using the IGHV leader-based NGS assay.
Materials and methods
Patient materials
All primary material used in this study was collected in the CLL11 trial (NCT01010061).21 In this multicenter phase 3 trial, patients with treatment-naïve CLL and a significant burden of coexisting conditions and/or decreased creatinine clearance were randomized in a 1:2:2 manner to receive six 28-day cycles of chlorambucil alone, rituximab and chlorambucil, or obinutuzumab and chlorambucil. Detailed trial design and inclusion and exclusion criteria have been published previously.21 Per trial protocol, DNA, isolated from peripheral blood (PB) mononuclear cells (PBMCs) or bone marrow mononuclear cells preceding and following treatment was stored. All samples were pseudonymized. For this study, patients were selected on the basis of the availability of sufficient DNA. The CLL11 trial was approved by the institutional review board or independent ethics committee at each participating institution and was conducted in accordance with the Declaration of Helsinki and the International Conference on Harmonization Guidelines for Good Clinical Practice. All patients provided written informed consent.
IGH target definition and identification
The clonal IGH target was defined as the most abundant productive IGH rearrangement present in a pretreatment sample, as detected by Sanger sequencing, in accordance with recommendations of the European Research Initiative on CLL.22,23 Specifically, the target was defined as a clonotype: an identity based on the specific utilization of IGHV, IGHD, and IGHJ genes, combined with the complementarity determining region 3 amino acid sequence.
Serial dilutions
Contrived MRD samples, ranging from MRD 10−2 to <10−6, were created using pretreatment DNA from 9 patients from the CLL11 trial (supplemental Table 1; available on the Blood website). DNA was serially diluted (g/g) in pooled PBMC DNA from healthy donors (Hoffmann-La Roche, Basel, Switzerland). MRD depth was normalized to initial tumor load, quantified by pretreatment flow cytometry.
Primers
To amplify all IGH rearrangements present in the end-of-treatment (EOT) DNA pool, the IGHV leader primer set developed by the EuroClonality-NGS working group was used.24 This set consists of 24 forward primers, targeting all IGHV leader sequences in 5’, and a consensus reverse primer, targeting a conserved IGHJ region in 3’. Primers contained a forward and reverse adapter sequence for a 1-step polymerase chain reaction (PCR) library preparation workflow for Illumina-based sequencing (supplemental Table 2). For multiplexing purposes, the adapter sequences included TruSeq DNA CD i5 or i7 indexes (Illumina, San Diego, CA).
DNA input and PCR conditions
DNA concentrations were determined with the Qubit dsDNA BR assay on a Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA). Each 50 μL PCR contained 5 μL High Fidelity 10× Buffer, 3.5 mM MgSO4, 1 U High-Fidelity Taq polymerase (all from Thermo Fisher Scientific), 400 μM deoxynucleoside triphosphates, and 0.1 μM of each forward and reverse primer. Per PCR tube, IGH rearrangements were amplified from a maximum of 1 μg of DNA input: for larger amounts of input, replicate reactions with identical indexes were used. Following hot-start initial denaturation at 95°C for 3 minutes, PCR amplification was achieved by denaturation at 95°C for 45 seconds, primer annealing at 63°C for 45 seconds, and subsequent extension at 72°C for 60 seconds, repeated for 35 cycles. Final extension was performed at 72°C for 10 minutes.
Purification and library preparation
After PCR cycling, amplification success was checked by 1% agarose gel electrophoresis or by using the D1000 DNA ScreenTape assay on a 4200 Agilent TapeStation (Agilent, Santa Clara, CA). PCRs with detectable amplicons in the 500 to 600 bp range were purified using the AMPure XP kit (Beckman Coulter, Brea, CA) with a volume ratio varying between 1× and 1.8×. In some samples with lowly abundant PCR product, tandem purification was performed. Following purification, amplicon size and concentration were remeasured on a TapeStation. Subsequently, PCR products were diluted to 4 nM and pooled in equimolar manner into a single multiplexed library pool.
Next-generation sequencing
The multiplexed library was denatured using a fresh 0.2 M NaOH dilution and diluted to a final loading concentration, varying between 12 and 15 pM. 5% to 20% PhiX control v3 (Illumina) was spiked in the diluted library pool to increase amplicon diversity and monitor sequence run quality. Paired-end sequencing (2 × 300 cycles) was performed on a MiSeq system using the MiSeq Reagent 600-cycle v3 kit (Illumina). Per run, the theoretical read coverage per sample was calculated to exceed the number of cell equivalents of input DNA. In practice, because in EOT samples only a small proportion of PBMCs is expected to carry an IGH amplicon, the read coverage exceeded the amount of input B cells manifold in each sample. To avoid cross-contamination and index miscalling of target reads, highly and lowly abundant samples from the same dilution series or patient were not analyzed in the same run.
Quality control
A central polytarget quality control (cPT-QC) was included in every run.20 The cPT-QC covers a broad immunoglobulin/T-cell receptor (IG/TR) repertoire and is included in a separate PCR to monitor primer performance and sequencing. In addition, in every PCR, a central in-tube quality control (cIT-QC) was spiked.20 This cIT-QC consists of DNA from 9 selected cell lines bearing 46 rearrangements, 7 of which are IGH rearrangements. A total of 280 IGH copies (2 μL) are spiked in every PCR replicate before amplification, allowing calculation of a conversion factor from reads to cells. This conversion factor can be used to partly correct for differential amplification bias and overamplification in B-cell depleted samples. Both the cPT-QC and the cIT-QC were developed by the EuroClonality-NGS working group.20
Data analysis in ARResT/Interrogate
NGS output was converted to FASTQ format and uploaded to ARResT/Interrogate, an immunoprofiling platform that annotates IG/TR sequences based on IMmunoGeneTics references.25,26 MRD depth was calculated by dividing the number of cells bearing the target clonotype by the total number of PBMCs/bone marrow mononuclear cells equivalent to the DNA input. In this study, 6.5 pg DNA/PBMC was used as conversion factor.27
Allele-specific oligonucleotide quantitative PCR
ASO-qPCR was performed in accordance with the European Study Group on MRD detection in acute lymphoblastic leukemia guidelines.6 Briefly, primers were developed matching the IGHV, IGHD, or IGK region of the IGH target, and were used in combination with a reverse consensus heavy-chain joining gene primer and a FAM-TAMRA probe (both from Hoffmann-La Roche) on a LightCycler 480 Real Time PCR system. DNA from pretreatment samples was serially diluted, corrected for initial tumor load, in a pool of healthy donor PBMC DNA to generate standard curves. Subsequently, cycle-threshold values from the serially diluted samples were compared with the standard curve to calculate MRD depth. ASO-qPCR–based MRD measurements, previously performed on EOT samples from the CLL11 trial, were used to cross-validate the IGHV leader-based NGS assay.9
Droplet digital PCR
ddPCR analysis was performed using a QX200 system (BioRad, Hercules, CA). Primers and probes used in the ddPCR were identical to those used in the ASO-qPCR experiments. ddPCR amplification was performed in a reaction volume of 20 μL, containing 340 ng of DNA and 1× ddPCR supermix for probes (BioRad). A single reaction was used for measurements to MRD 10−4, whereas 10 parallel reactions were used for measurements to MRD 10−5. Each 20 μL sample was partitioned into 20 000 water-oil emulsion droplets by a droplet generator (BioRad), following PCR amplification on a T100 thermocycler (BioRad) in a 96-well plate. Thermal conditions were as follows: initial denaturation at 94°C for 30 seconds, followed by 39 cycles of denaturation at 94°C for 30 seconds, then annealing and extension at 62°C for 60 seconds, followed by final inactivation at 98°C for 10 minutes. After amplification, the PCR plate was loaded on a QX200 droplet reader (BioRad). Data analysis was performed using Quantasoft software (version 1.7.4). The limit of blank was established on the basis of duplicate nontemplate control wells for each primer-probe combination. MRD depth was calculated by dividing the number of target copies/μL by the total amount of PBMCs/μL, inferred from the DNA concentration. All measurements were performed in duplicate.
Statistical analysis
The limit of detection (LoD) was defined as the threshold of malignant cell-equivalent DNA input at which the probability of detection was equal to 95%, estimated by a logistic regression model that modeled the proportion of MRD-positive samples as a function of DNA input. Similarly, the limit of quantitation (LoQ) was defined as the threshold of malignant cell-equivalent DNA input at which the percentage coefficient of variance (%CV) was equal to 70%. The LoQ was estimated by modeling the %CV as a function of DNA input, using a Sadler precision profile (Y = [β0 + β1X1]J).28-30 The limit of blank (LoB) was defined as the MRD frequency at which a given CLL-specific clonal target will be identified in up to 5% of healthy repertoires. To estimate the LoB, the presence and abundance of 71 clonal targets from patients in the CLL11 trial were assessed in 30 healthy repertoires from peripheral blood, sequenced at 500-ng input, taken from a previously published data set,31 as well as in 2 sets of 3 sequencing replicates of a DNA pool of PBMCs of 50 healthy donors (Roche), sequenced at 5- and 27-μg input, respectively. Variability was assessed by averaging the %CV over replicates of serial dilutions within the same run (intra-assay variability) or in different runs (interassay variability) at different amounts of DNA input. Assay linearity was evaluated by a linear regression model, estimating the log10-transformed observed MRD measurements as a function of the log10-transformed expected MRD values. If the model fit was not significantly improved by the inclusion of a second-degree polynomial, as assessed by a likelihood-ratio test, linearity was established. Pearson correlation coefficients were calculated using log10-transformed MRD values. Survival analyses were performed for PFS, TTNT, and OS, according to MRD status at EOT by applying landmark analyses from EOT. Kaplan-Meier estimation was used to generate survival curves and time-to-event parameters with comparisons between MRD groups via 2-sided nonstratified log-rank tests. Hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated by Cox proportional hazards regression. Significance was defined as a P < .05 without adjustments for multiple testing. All statistical analysis was performed in R version 4.0.3 and SPSS version 27.32
Results
The IGHV leader-based NGS assay can detect and quantify MRD down to 4 malignant cells
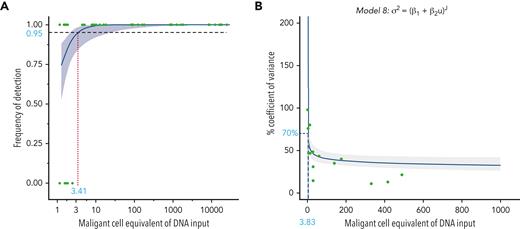
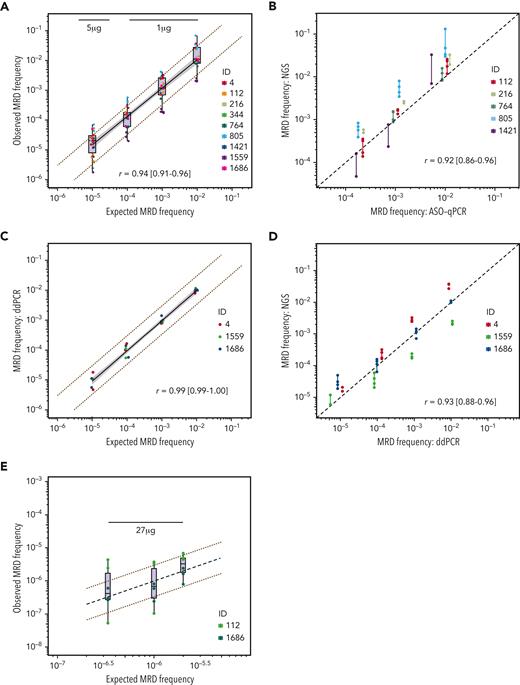
To determine the sensitivity of the IGHV leader-based NGS assay, we measured MRD on contrived samples using 500 to 1000 ng (MRD 10−2 to 10−5) or 5 μg (MRD 1 × 10−5 to 1.25 × 10−6) DNA input. In 176 of 183 (96%) measurements, the CLL-specific rearrangement was detected (Figure 1A). More specifically, the CLL-specific rearrangement was detected in all samples with MRD ≥10−4 (n = 108), and more important, in all samples with MRD ≥10−5 using at least 5 μg DNA input (n = 41). The LoD was estimated at 3.4 malignant cells per assay (95% CI, 1.9-16.0) (Figure 1A). To determine the precision of the leader-based NGS assay, we used the same set of serial dilutions. For 30 samples, ≥3 measurements were available, spanning 117 replicates and 6 different IGH rearrangements. The LoQ was estimated at 3.8 malignant cells per assay (95% CI, 1.7-8.2) (Figure 1B). The LoB was found to be 0, as no false-positive reads, corresponding to any of the 71 CLL-specific clonal targets, were identified in either the healthy PB repertoires or the PBMC pool replicates. Intra-assay variability ranged from 17.4% ± 13.6% at >1000 malignant cells input to 35.7% ± 19.9% at ≤10 malignant cells input (supplemental Table 3). Interassay variability ranged from 15.6% ± 6.5% at >1000 malignant cells input to 38.9% ± 28.0% at ≤10 malignant cells input (supplemental Table 3). Stratifying MRD measurements by IGHV gene usage, there was no evidence for strong amplification bias (supplemental Figure 1).
Sensitivity and precision of the IGHV leader-based NGS assay. (A) The probability of detection of the clonal IGH target in contrived MRD samples as a function of malignant cell-equivalent input. The y axis signifies detection (0, no; 1, yes), and the x axis signifies the MRD depth of the sample on the malignant cell-equivalent scale. A logistic regression model is fit to the data to estimate the limit of detection, defined as a probability of detection of ≥95%. (B) The percentage coefficient of variance (%CV) as a function of malignant cell-equivalent input. Data points represent the %CV from ≥3 replicate measurements on contrived MRD samples. The limit of quantitation (defined at 70%CV) is estimated using a Sadler precision profile, detailed above the graph.
Sensitivity and precision of the IGHV leader-based NGS assay. (A) The probability of detection of the clonal IGH target in contrived MRD samples as a function of malignant cell-equivalent input. The y axis signifies detection (0, no; 1, yes), and the x axis signifies the MRD depth of the sample on the malignant cell-equivalent scale. A logistic regression model is fit to the data to estimate the limit of detection, defined as a probability of detection of ≥95%. (B) The percentage coefficient of variance (%CV) as a function of malignant cell-equivalent input. Data points represent the %CV from ≥3 replicate measurements on contrived MRD samples. The limit of quantitation (defined at 70%CV) is estimated using a Sadler precision profile, detailed above the graph.
The IGHV leader-based NGS assay is linear in the range 10−2 to 10−5
Using the contrived samples described above, with DNA input exceeding the LoD, linearity was established in the MRD 10−2 to 10−5 range (r = 0.94 [95% CI, 0.91-0.96]) (Figure 2A). The introduction of a quadratic term did not significantly improve the fit of the estimated linear regression model, reinforcing the linear relationship between expected and observed measurements down to MRD 10−5.
Linearity and validation of the IGHV leader-based NGS assay. Comparisons between expected and observed MRD measurements, or cross-technique MRD comparisons. Each dot represents 1 measurement. All Pearson correlation coefficients were computed using the log10-transformed values. (A) Comparison between expected and observed MRD measurements obtained by measuring MRD on contrived samples using the IGHV leader-based NGS assay. The notch represents the median, the upper and lower hinges signify the first and third quartiles, respectively, and the whiskers extend to the largest value within 1.5 times the interquartile range. The value above the boxplots denotes the amount of input DNA used for this MRD depth. The black line indicates a linear regression between expected and observed values; the shaded area indicates the 95% confidence interval for the regression mean. The red dotted lines indicate a threefold difference between expected and observed data points. Data points have horizontal jitter to improve their visibility. (B) Comparison between measurements on contrived samples obtained by ASO-qPCR and IGHV leader-based NGS. The black dotted line represents a perfect linear relationship. The line between data points signifies the range of the replicates per rearrangement. (C) Comparison between expected and observed MRD measurements obtained by measuring MRD on contrived samples using ddPCR. The black line indicates a linear regression between expected and observed values; the shaded area indicates the 95% confidence interval for the regression mean. The red dotted lines indicate a threefold difference between expected and observed data points. (D) Comparison between measurements on contrived samples obtained by ASO-qPCR and IGHV leader-based NGS. The black dotted line represents a perfect linear relationship. The line between data points signifies the range of the replicates per rearrangement. (E) Comparison between measurements on contrived samples obtained by ASO-qPCR and IGHV leader-based NGS. The black dotted line represents a perfect linear relationship. The red dotted lines indicate a threefold difference between expected and observed data points. The value above the measurements denotes the amount of input DNA used for this MRD depth. ID, identifier.
Linearity and validation of the IGHV leader-based NGS assay. Comparisons between expected and observed MRD measurements, or cross-technique MRD comparisons. Each dot represents 1 measurement. All Pearson correlation coefficients were computed using the log10-transformed values. (A) Comparison between expected and observed MRD measurements obtained by measuring MRD on contrived samples using the IGHV leader-based NGS assay. The notch represents the median, the upper and lower hinges signify the first and third quartiles, respectively, and the whiskers extend to the largest value within 1.5 times the interquartile range. The value above the boxplots denotes the amount of input DNA used for this MRD depth. The black line indicates a linear regression between expected and observed values; the shaded area indicates the 95% confidence interval for the regression mean. The red dotted lines indicate a threefold difference between expected and observed data points. Data points have horizontal jitter to improve their visibility. (B) Comparison between measurements on contrived samples obtained by ASO-qPCR and IGHV leader-based NGS. The black dotted line represents a perfect linear relationship. The line between data points signifies the range of the replicates per rearrangement. (C) Comparison between expected and observed MRD measurements obtained by measuring MRD on contrived samples using ddPCR. The black line indicates a linear regression between expected and observed values; the shaded area indicates the 95% confidence interval for the regression mean. The red dotted lines indicate a threefold difference between expected and observed data points. (D) Comparison between measurements on contrived samples obtained by ASO-qPCR and IGHV leader-based NGS. The black dotted line represents a perfect linear relationship. The line between data points signifies the range of the replicates per rearrangement. (E) Comparison between measurements on contrived samples obtained by ASO-qPCR and IGHV leader-based NGS. The black dotted line represents a perfect linear relationship. The red dotted lines indicate a threefold difference between expected and observed data points. The value above the measurements denotes the amount of input DNA used for this MRD depth. ID, identifier.
ASO-qPCR and ddPCR measurements cross-validate the IGHV leader-based NGS assay
To cross-validate the IGHV leader-based NGS assay against an established MRD assay in direct pairwise comparisons, we used ASO-qPCR to quantify MRD in the appropriate range (MRD ≥10−4) on contrived MRD samples representing 5 IGH rearrangements (identifiers [IDs] 112, 216, 764, 805, and 1421) (Figure 2B). The interassay agreement was high (r = 0.92 [95% CI, 0.86-0.96]). To cross-validate the IGHV leader-based NGS assay down to MRD 10−5, we measured MRD using ddPCR on contrived MRD samples representing 3 IGH rearrangements (IDs 4, 1559, and 1686). The ddPCR assay detected the CLL-specific rearrangement in all 18 replicates in the range 10−2 to 10−4 and in 5 of 6 replicates at MRD 10−5, with high concordance between expected and observed MRD values (r = 0.99 [95% CI, 0.99-1.00]) (Figure 2C). The interassay agreement between the IGHV leader-based NGS assay and ddPCR was high (r = 0.93 [95% CI, 0.88-0.96]) (Figure 2D).
The IGHV leader-based NGS assay can detect MRD beyond 10−6
To determine whether the IGHV leader-based NGS assay can detect MRD to 10−6, we created contrived MRD samples down to MRD 5 × 10−6, 1 × 10−6, and 3 × 10−7, representing 2 IGH rearrangements (IDs 112 and 1686). Next, we measured MRD on these samples in triplicate, using 27 μg DNA input per replicate, thereby exceeding the LoD of 3.4 malignant cells for measurements to MRD 10−6. In all 18 replicates, MRD was detected, even in those samples that were calculated to be below the LoD (MRD 3 × 10−7) (Figure 2E).
The IGHV leader-based NGS detects MRD beyond 10−5 in clinical samples
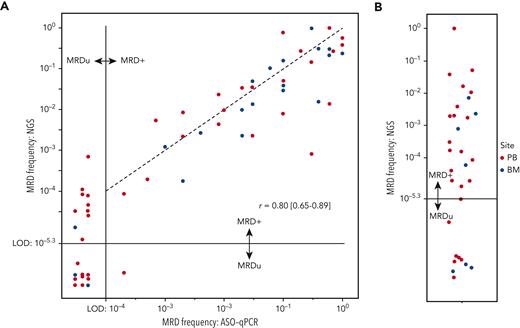
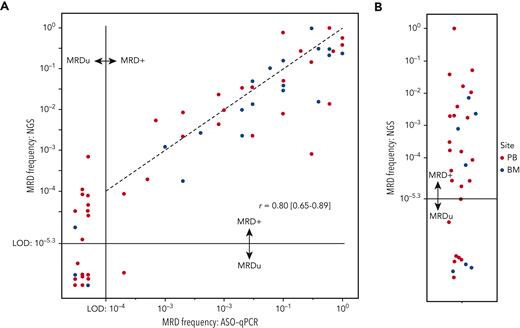
To validate the IGHV leader-based NGS assay on clinical samples, we quantified MRD using 43 PB and 20 bone marrow EOT samples from the CLL11 trial and compared these with measurements previously obtained through ASO-qPCR9 (Figure 3A). For each sample, 5 μg of input DNA was used, allowing for detection of MRD to 5 × 10−6. In 41 of 63 (65%) samples, MRD was detectable and quantifiable by both NGS and ASO-qPCR (r = 0.80 [95% CI, 0.65-0.89]) (Figure 3A, right upper quadrant). In 22 of 63 samples (35%), MRD depth was uMRD4 by ASO-qPCR. In 10 of 22 (45%) samples, however, MRD was detectable by NGS (Figure 3A, left upper quadrant). In the remaining 12 of 22 (55%) samples, MRD was undetectable by both ASO-qPCR and NGS, presumably signifying MRD <10−5, for which >5 μg DNA input would have been required. In a single sample, MRD was detectable by ASO-qPCR >10−4, but was undetectable by NGS.
Measuring beyond MRD 10−4in the CLL11 trial. (A) Comparison between MRD measurements obtained on samples from the CLL11 trial by ASO-qPCR or IGHV leader-based NGS. Each dot represents 1 measurement, and the color indicates the sampled anatomical compartment. The horizontal and vertical black lines indicate the limits of detection of the NGS assay and ASO-qPCR, respectively. Dots to the left of the vertical black lines signify MRD undetectability by ASO-qPCR, and dots below the horizontal dotted line signify MRD undetectability by leader-based NGS. The Pearson correlated coefficient was calculated using the log10-transformed values of samples in which MRD was quantifiable by both techniques. The black dotted line represents a perfect linear relationship. (B) Dot plot indicating MRD values obtained through IGHV leader-based NGS on samples in which ASO-qPCR was not feasible. Each dot represents 1 measurement, and the color indicates the sampled anatomical compartment. The horizontal black line indicates the limit of detection of the NGS assay. Dots below the horizontal dotted line signify MRD undetectability by leader-based NGS. BM, bone marrow; LoD, limit of detection; MRD+, detectable MRD; MRDu, undetectable MRD.
Measuring beyond MRD 10−4in the CLL11 trial. (A) Comparison between MRD measurements obtained on samples from the CLL11 trial by ASO-qPCR or IGHV leader-based NGS. Each dot represents 1 measurement, and the color indicates the sampled anatomical compartment. The horizontal and vertical black lines indicate the limits of detection of the NGS assay and ASO-qPCR, respectively. Dots to the left of the vertical black lines signify MRD undetectability by ASO-qPCR, and dots below the horizontal dotted line signify MRD undetectability by leader-based NGS. The Pearson correlated coefficient was calculated using the log10-transformed values of samples in which MRD was quantifiable by both techniques. The black dotted line represents a perfect linear relationship. (B) Dot plot indicating MRD values obtained through IGHV leader-based NGS on samples in which ASO-qPCR was not feasible. Each dot represents 1 measurement, and the color indicates the sampled anatomical compartment. The horizontal black line indicates the limit of detection of the NGS assay. Dots below the horizontal dotted line signify MRD undetectability by leader-based NGS. BM, bone marrow; LoD, limit of detection; MRD+, detectable MRD; MRDu, undetectable MRD.
In a subgroup of patients with CLL, intense SHM precludes the design of a functional ASO-qPCR assay. In a selection of 31 of such patients from the CLL11 trial, MRD was detectable in 22 of 31 samples (71%) using the IGHV leader-based NGS assay (Figure 3B). In the remaining 9 of 31 samples (29%), MRD was undetectable.
MRD measurement to 10−5 improves prognostic stratification
Using the MRD measurements obtained through the IGHV leader-based NGS assay on PB of patients in the CLL11 trial, we performed survival analysis from the EOT landmark to evaluate whether MRD measurement beyond 10−4 improves prognostic stratification. For this analysis, PB EOT samples from 67 patients were included (baseline characteristics are summarized in supplemental Table 4). Median follow-up from study enrollment was 70.4 months (range, 19.9-85.0 months), with 45 of 67 (67%) of patients remaining alive at time of the analysis.
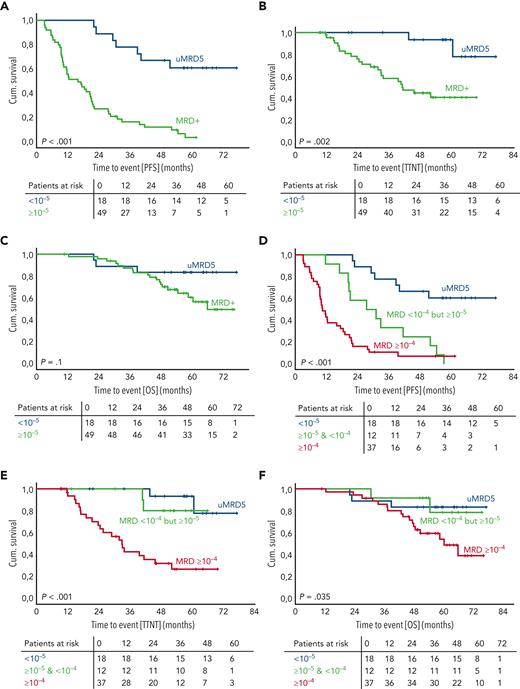
Patients with uMRD by IGHV leader-based NGS (MRD <10−5) (n = 18) had superior PFS, compared with patients with MRD ≥10−5 (n = 49) (median PFS: not reached [NR] vs 15.9 months; 4-year PFS rate: 66.7% vs 11.3%; HR, 0.15 [95% CI, 0.07-0.35]; P < .001) (Figure 4A). Similarly, TTNT was longer in the uMRD group, compared with the MRD ≥10−5 group (median TTNT: NR vs 41.2 months; 4-year TTNT rate: 93.3% vs 44.7%; HR, 0.14 [95% CI, 0.03-0.58]; P = .007) (Figure 4B). Regarding OS, no significant difference could be identified between the 2 groups (4-year OS rate: 83.3% vs 72.3%; HR, 0.38 [95% CI, 0.11-1.27]; P = .116) (Figure 4C).
MRD measurement beyond 10−4improves prognostic stratification. (A-F) Kaplan-Meier graphs and risk tables depicting the proportion of patients with PFS (A and D), treatment-free survival (B and E), or OS (C and F) over time. Patients are stratified by detectability of MRD by the IGHV leader-based NGS assay (A-C) or by MRD depth (D-F). P values were obtained using 2-sided, nonstratified log-rank tests. A cross indicates right censoring. Cum., cumulative; MRD+, detectable MRD; uMRD5, undetectable MRD <10−5.
MRD measurement beyond 10−4improves prognostic stratification. (A-F) Kaplan-Meier graphs and risk tables depicting the proportion of patients with PFS (A and D), treatment-free survival (B and E), or OS (C and F) over time. Patients are stratified by detectability of MRD by the IGHV leader-based NGS assay (A-C) or by MRD depth (D-F). P values were obtained using 2-sided, nonstratified log-rank tests. A cross indicates right censoring. Cum., cumulative; MRD+, detectable MRD; uMRD5, undetectable MRD <10−5.
Measurement to MRD 10−5 allowed for stratification of a patient group (MRD <10−4 but ≥10−5) (n = 12), with significantly longer PFS compared with patients with MRD ≥10−4 (n = 37), but significantly shorter PFS compared with patients with MRD <10−5 (n = 18) (median PFS: ≥10−4, 10.4 months; <10−4 but ≥10−5, 27.5 months; <10−5, NR; 4-year PFS rate: ≥10−4, 7.2%; <10−4 but ≥10−5, 25.0%; <10−5, 66.7%; univariate HRs are reported in Table 1) (Figure 4D). In addition, the TTNT of patients with MRD <10−4 but ≥10−5 was significantly longer, compared with patients with MRD ≥10−4 (median PFS: NR vs 32.9 months; 4-year TTNT rate: 80.0% vs 31.9%; HR, 0.16 [95% CI, 0.04-0.69]; P = .014). There was no significant difference in TTNT between patients with MRD <10−4 but ≥10−5 and patients with MRD <10−5 (Figure 4E; supplemental Table 5). Regarding OS, no significant differences were found between the 3 groups (Figure 4F; supplemental Table 6).
Discussion
In this article, we present an academically developed IGHV leader-based NGS assay for the quantification of MRD in CLL beyond 10−5. Conditional on the amount of input DNA, this assay can detect MRD to the level of 10−6, and is linear at least to MRD 10−5. More important, using samples from the CLL11 trial, we assessed the clinical utility of our assay by allowing for stratification of a patient group with MRD <10−4 but ≥10−5. Although these patients have a significantly longer PFS, compared with those with MRD ≥10−4, their PFS is significantly shorter, compared with those with MRD <10−5.
More important, our assay includes forward primers that target the IGHV leader sequence in 5’. As the leader sequence is generally unaffected by SHM, the risk of nucleotide substitutions in primer-annealing sites is low. Another major advantage of an IGHV leader-based NGS pipeline is that it allows for complete characterization of the IGHV sequence. As such, this assay, once implemented, can be used in one general workflow for both IGHV mutational status determination, as well as quantification of MRD. Of note, a revised iteration of the IGHV leader primer pool, which is intended to further harmonize amplification across all IGHV genes, has been under development by the EuroClonality consortium. Future endeavors will include evaluating the performance of this primer set, which is expected to increase precision and validity, in the context of MRD detection.
In a pilot experiment, we demonstrated the ability of the IGHV leader-based NGS assay to measure MRD to 10−6 and beyond, using a DNA input of 27 μg. Of note, the theoretical lower limit of the LoD of a perfect MRD assay can be estimated using the binomial distribution. Specifically, to detect residual tumor cells with high probability (≥95%), the coverage should always exceed the desired sensitivity by threefold, a dictum known as “the rule of three.”33,34 The LoD of the IGHV leader-based assay, estimated at 3.4×, closely approximates this theoretical lower limit. Thus, inevitably, the feasibility of deep MRD quantification is conditional on the availability of a large amount of input DNA and carries significant financial costs. Moreover, the added benefit of measurement to MRD 10−6 as prognostic marker has not yet been conclusively demonstrated. Therefore, in the current study, we have chosen to limit our measurements in the CLL11 trial to MRD ≥10−5.
The NGS-based ClonoSEQ platform has received regulatory approval for the detection of MRD in acute lymphoblastic leukemia, multiple myeloma, and, recently, CLL.35 Previously, this assay has been used to quantify MRD down to 10−6 following treatment with fludarabine, cyclophosphamide and rituximab, or rituximab and venetoclax.16,17 In this article, the performance of our leader-based NGS platform is similar compared with the ClonoSEQ platform, at least in the range MRD ≥10−5.18 However, although the ClonoSEQ platform is exclusively commercially operated, we commit to open publication of our protocols and primer sequences. This will allow for implementation in multiple centers, including interlaboratory proficiency testing to ensure stringent quality control.
Our current study has some limitations that need to be addressed. First, we have not been able to show the utility of the IGHV leader-based NGS assay in the prognostic stratification of OS, due to a relative paucity of OS events in a first-line setting. The availability of highly effective second-line therapeutic agents may further explain the significant association in our cohort between MRD detectability and TTNT, but not OS. Evaluation of the utility of the IGHV leader-based NGS platform in a larger, relapsed and refractory cohort is warranted to validate our findings and evaluate stratification for OS. Second, the maximum input of 1 μg per PCR tube may hamper assay applicability in routine practice, especially when performing measurements down to MRD 10−6, which would require many parallel reactions. Interestingly, in the setting of MRD detection in acute lymphoblastic leukemia, the EuroClonality-NGS working group has optimized the PCR up to 2 μg of DNA input36 (M. Svatoň, Charles University Prague, written communication, 1 December 2022). Similar optimization may further enhance the implementability of the IGHV leader-based NGS assay.
To cross-validate the leader-based NGS platform to MRD 10−5, we performed ddPCR on contrived MRD samples. Although the use of ddPCR for the cell-based quantification of MRD in CLL has previously been suggested, we believe we are the first to actually demonstrate its feasibility.37 ddPCR measurements in the range MRD 10−2 to MRD 10−5 were highly linear (r = 0.99), warranting further investigation of the applicability of ddPCR for the quantification of MRD in CLL. However, the need for patient-tailored primer design remains an obstacle for the widespread implementation of a ddPCR-based approach.
In conclusion, we herein present an academically developed, IGHV leader-based NGS assay for the detection and quantification of MRD in CLL beyond MRD 10−5. The assay has high sensitivity and is quantitative and linear beyond MRD 10−5 when using 5 μg DNA input. Measurement to MRD 10−6 is possible, conditional on sufficient DNA input. Furthermore, as the assay employs primers that target the IGHV leader sequence, it is insensitive to SHM, as demonstrated by the successful stratification of 31 patients in whom ASO-qPCR MRD assay design was impossible because of SHM in primer-annealing sites. Implementation in trials and clinical care is feasible, as the generation of patient-specific primers is not required. The deeper MRD measurements enabled by the IGHV leader-based NGS assay resulted in improved stratification of patients with CLL following treatment.
Acknowledgment
The authors acknowledge Nikos Darzentas for his support in the immunoinformatic analysis.
Authorship
Contribution: P.J.H., M.-D.L. and A.W.L. contributed to the study design, data acquisition, analysis and interpretation, and manuscript writing; M.Y.v.d.K., F.G.K., and E.d.J. contributed to data acquisition; S.R. contributed to data analysis and interpretation; P.M.K., J.L.J.C.A., L.v.d.S., M.R., and P.E.W. contributed to data interpretation; F.D. designed and provided the IGHV leader primer pool and protocol; V.G., K.F., and M.H. designed and operated the CLL11 trial and provided access to the clinical samples and patient data; and all authors critically read and approved the final version of the article.
Conflict-of-interest disclosure: S.R. received honoraria from AstraZeneca. M.R. received research funding from and was an advisory board member for Roche. K.F. received travel grants from Roche. V.G. received consulting fees from Roche, was an advisory board member for Roche, Janssen, Gilead Sciences, and AbbVie, and received honoraria from Roche, Janssen, and Gilead Sciences. M.H. received honoraria and research funding from and was an advisory board member for AbbVie, Amgen, Celgene, Roche, Gilead Sciences, Janssen, and Mundipharma. A.W.L. received research funding via an unrestricted grant from Roche-Genentech and a speaker fee from Janssen. The remaining authors declare no competing financial interests.
Correspondence: Anton W. Langerak, Department of Immunology, Erasmus MC, University Medical Centre Rotterdam, Dr Molewaterplein 40, 3015 GD Rotterdam, The Netherlands; e-mail: a.langerak@erasmusmc.nl.
References
Author notes
Next-generation sequencing data discussed in this article have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus and are accessible through Gene Expression Omnibus series accession number GSE188587. All other original data can be obtained on reasonable request.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
M.-D.L. and A.W.L. contributed equally and share last authorship.