Key Points
Filamin A associates with both inactive and active platelet integrin αIIbβ3 to differentially regulate bidirectional signaling.
Filamin A induces a structural transition in the αIIb CT to strongly link αIIbβ3 with actin to promote outside-in signaling.
Abstract
The communication of talin-activated integrin αIIbβ3 with the cytoskeleton (integrin outside-in signaling) is essential for platelet aggregation, wound healing, and hemostasis. Filamin, a large actin crosslinker and integrin binding partner critical for cell spreading and migration, is implicated as a key regulator of integrin outside-in signaling. However, the current dogma is that filamin, which stabilizes inactive αIIbβ3, is displaced from αIIbβ3 by talin to promote the integrin activation (inside-out signaling), and how filamin further functions remains unresolved. Here, we show that while associating with the inactive αIIbβ3, filamin also associates with the talin-bound active αIIbβ3 to mediate platelet spreading. Fluorescence resonance energy transfer–based analysis reveals that while associating with both αIIb and β3 cytoplasmic tails (CTs) to maintain the inactive αIIbβ3, filamin is spatiotemporally rearranged to associate with αIIb CT alone on activated αIIbβ3. Consistently, confocal cell imaging indicates that integrin α CT–linked filamin gradually delocalizes from the β CT–linked focal adhesion marker-vinculin likely because of the separation of integrin α/β CTs occurring during integrin activation. High-resolution crystal and nuclear magnetic resonance structure determinations unravel that the activated integrin αIIb CT binds to filamin via a striking α-helix→β-strand transition with a strengthened affinity that is dependent on the integrin-activating membrane environment containing enriched phosphatidylinositol 4,5-bisphosphate. These data suggest a novel integrin αIIb CT–filamin-actin linkage that promotes integrin outside-in signaling. Consistently, disruption of such linkage impairs the activation state of αIIbβ3, phosphorylation of focal adhesion kinase/proto-oncogene tyrosine kinase Src, and cell migration. Together, our findings advance the fundamental understanding of integrin outside-in signaling with broad implications in blood physiology and pathology.
Introduction
Integrin αIIbβ3 (GPIIb/IIIa) is a heterodimeric transmembrane receptor essential for regulating platelet function and hemostasis.1-7 Upon stimulation by thrombin or adenosine 5′-diphosphate, which elicits signals to the cytoplasmic face of αIIbβ3, the integrin is activated through a process called inside-out signaling.4,7-11 The activated αIIbβ3 binds to extracellular ligands and then sends signals back to the cytoplasm to alter the cytoskeleton, cell morphology, etc (outside-in signaling).4,7 Such bidirectional signaling enables platelets to swiftly alter their activity to control the dynamics of platelet function.12-14 A key part of the bidirectional signaling of αIIbβ3 is its cytoplasmic tail (CT) linkage to the actin cytoskeleton,4,7,15-17 which is poorly understood despite extensive studies. However, major advances have been made in understanding the integrin inside-out signaling pathway, in which extracellular domains of inactive αIIbβ3 and other integrins were found to exist predominantly in a bent shape as stabilized by the association of α/β transmembrane domain CTs4,18-21 (supplemental Figure 1, available on the Blood website) and talin, a major actin-binding protein (activated upon the platelet stimulation), which can bind to integrin β CTs to promote global conformational change and ligand binding of the receptors.4,7,22-24
Filamin is also a well-known actin-binding protein that has 3 family members, A, B, and C, which all contain 2 N-terminal actin–binding domains followed by 24 immunoglobulin (Ig) repeats.25-28 Interestingly, in contrast to talin that activates integrins, filamin A (FLNa) uses several homologous Ig repeats, notably Ig21, to strengthen the integrin α/β–CT association to stabilize the inactive state of the receptor.29 Such negative regulation on integrin activation was further confirmed by recent in vivo data in which FLNa was shown to associate with the resting state platelet integrin αIIbβ3,30 whereas its dissociation from the integrin because of a disease mutation in FLNa enhances the integrin αIIbβ3 activation.31 Upon cellular stimulation, a type 1 γ phosphatidylinositol phosphate kinase (PIPKIγ) is recruited by talin32,33 to locally produce PIP2 that in turn activates talin, which is otherwise autoinhibited.34-38 By strongly interacting with integrin β CT, activated talin then outcompetes filamin,39,40 facilitating the separation of α/β CTs and a global conformational change of the integrin to competently bind ECM ligands.39,41,42 The β CT–bound talin also connects to actin while triggering the formation of large protein aggregates, called focal adhesions (FAs; outside-in signaling), through direct interactions with proteins such as vinculin and paxillin that in turn recruit many other proteins.23,43,44 The current dogma (supplemental Figure 1) deems that filamin is displaced from integrin by talin to trigger integrin activation, but the mechanism by which filamin further functions remains highly elusive.30,45-48 Filamin is often found to localize with actin stress fibers at the cell cortex,49 cell leading edge,26,50 and trailing ends of FAs,51 but not within FAs. However, besides its role in the dynamic reorganization of the actin cytoskeleton,26,27,50 filamin is widely known to regulate integrin-mediated cell spreading and migration,25-28 which are critically dependent on integrin outside-in signaling.30,52-55 Donada et al56 suggested that an FLNa mutant that disrupts filamin/αIIbβ3 interaction can also impair integrin outside-in signaling due to impaired fibrinogen ligand binding to αIIbβ3. These observations challenge the model in supplemental Figure 1 and raise a question about how filamin mechanistically mediates integrin outside-in signaling. We address this issue in this study by focusing on αIIbβ3, the prototypic and extensively characterized integrin.4,39,41,42 Using a series of biochemical, cellular, and structural approaches, we show that while filamin enhances the αIIb/β3–CT association to stabilize the inactive integrin,29 it is spatiotemporally rearranged to bind αIIb CT alone of active integrin via a unique helix-strand transition with enhanced affinity, suggesting a novel mechanism of outside-in signaling via integrin αIIb–filamin–actin linkage. Our results not only advance the understanding of the platelet function involving integrin αIIbβ3 but also bear broad significance in understanding the signaling mechanism of other integrins.
Methods
All constructs for the study used human FLNa and αIIbβ3 sequences. FLNa fragments for biochemical and structural studies were purified using affinity columns followed by gel filtration. The structure of the FLNa-αIIb CT complex was determined using nuclear magnetic resonance (NMR), and the structure of the FLNa-αIIb CT chimera was determined using crystallography. FLNa localization to FAs was analyzed in mouse embryonic fibroblasts using Leica SP5 confocal microscope. Coimmunoprecipitation and colocalization of αIIbβ3 with FLNa or talin were performed using human platelets.
Details of the constructs, protein-binding assays, structure determination, and cell-based analyses are provided in the supplemental Methods on the Blood website.
Results
Filamin associates with the resting as well as the activated state of integrin αIIbβ3 in spatiotemporally different manners
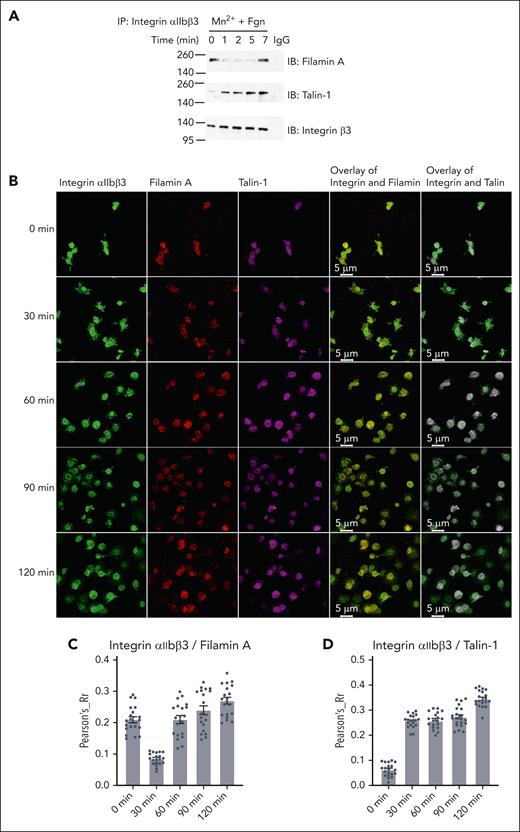
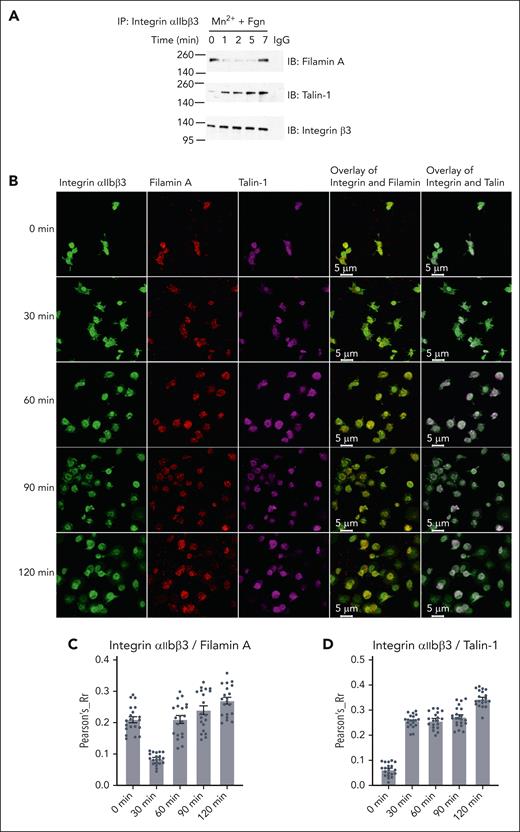
To examine how filamin engages integrin before and after the αIIbβ3 activation, we first performed the following filamin/αIIbβ3 association experiments using human platelets: (1) co-immunoprecipitation of αIIbβ3 with filamin within a very short period upon the platelet simulation. Figure 1A shows that although filamin associates with integrin αIIbβ3 in the resting state (0 minutes), the association drastically decreases and then surprisingly increases during 1 to 7 minutes period upon platelet stimulation. In comparison, talin does not associate with the resting state αIIbβ3 but associates with αIIbβ3 in a slightly enhanced manner from 1 to 7 minutes (Figure 1A). (2) Colocalization of αIIbβ3 with filamin using confocal imaging. We chose to spread platelets slowly on fibrinogen without stimulation to avoid rapid platelet aggregation. Figure 1B-D shows that filamin first delocalizes from αIIbβ3 (at a very early stage) and then reassociates with αIIbβ3 during the platelet spreading. In comparison, talin remains associated with αIIbβ3 during the same period. These data strongly indicate that upon the talin-mediated integrin activation, filamin first dissociates and then reassociates with integrin to regulate the integrin outside-in signaling.
Filamin A binds to activated integrin αIIbβ3 in stimulated human platelets. (A) Washed human platelets were stimulated with 0.5 mM Mn2+ and stirred in the presence of 25 μg/mL fibrinogen in suspension and lysed at indicated time points. Platelet lysates were immunoprecipitated with antibodies against integrin αIIbβ3 and then immunoblotted with antibodies against filamin A, talin-1, and integrin β3, respectively. (B) Representative confocal microscopic images colocalizing integrin αIIbβ3 with filamin A or talin-1 in human platelets spread on fibrinogen at the indicated time points. Experiments were repeated independently at least 3 times with similar results. Colocalization of integrin αIIbβ3 with filamin A (C) or talin-1 (D) was quantified using Pearson correlation coefficient calculation, using National Institutes of Health Image J JACoP plug-in57 with Costes automatic threshold. Twenty images from each time point were analyzed. A representative result from 2 independent experiments.
Filamin A binds to activated integrin αIIbβ3 in stimulated human platelets. (A) Washed human platelets were stimulated with 0.5 mM Mn2+ and stirred in the presence of 25 μg/mL fibrinogen in suspension and lysed at indicated time points. Platelet lysates were immunoprecipitated with antibodies against integrin αIIbβ3 and then immunoblotted with antibodies against filamin A, talin-1, and integrin β3, respectively. (B) Representative confocal microscopic images colocalizing integrin αIIbβ3 with filamin A or talin-1 in human platelets spread on fibrinogen at the indicated time points. Experiments were repeated independently at least 3 times with similar results. Colocalization of integrin αIIbβ3 with filamin A (C) or talin-1 (D) was quantified using Pearson correlation coefficient calculation, using National Institutes of Health Image J JACoP plug-in57 with Costes automatic threshold. Twenty images from each time point were analyzed. A representative result from 2 independent experiments.
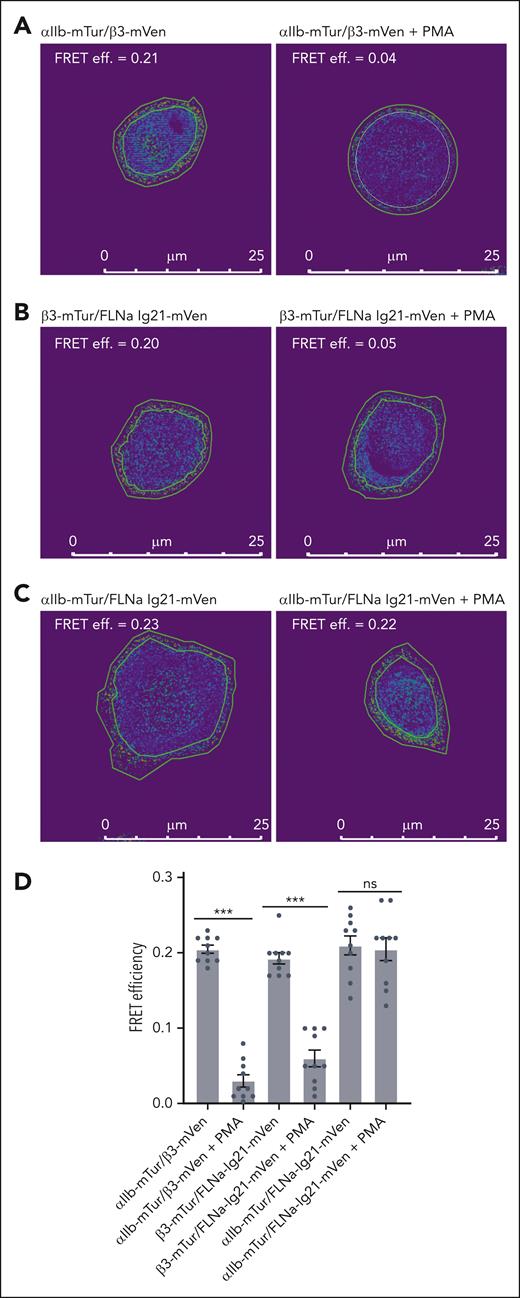
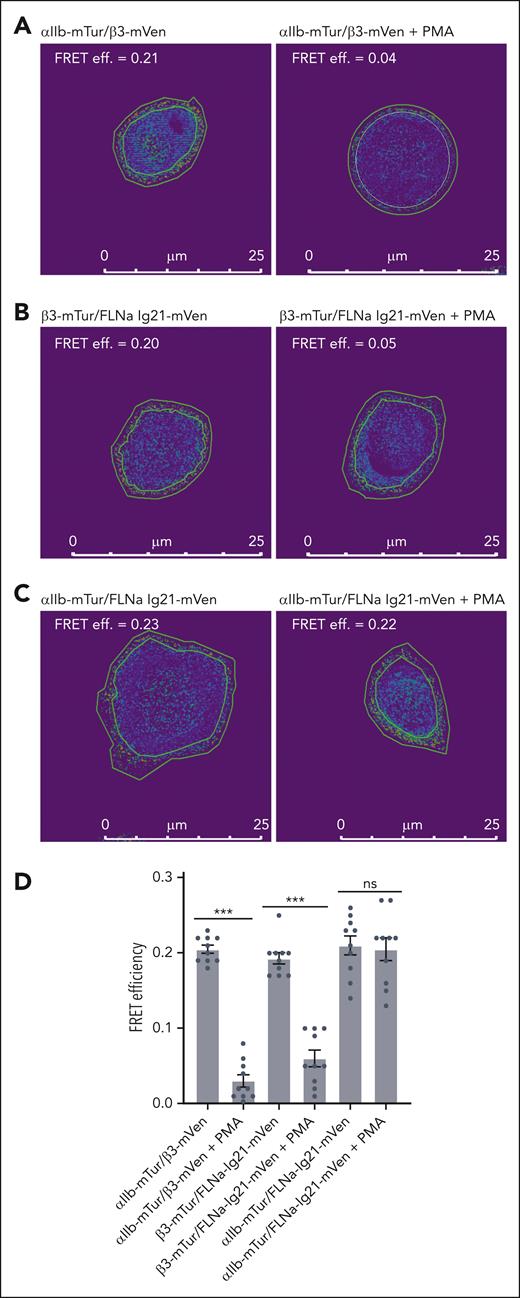
To understand how filamin associates with the active αIIbβ3, we then used fluorescence resonance energy transfer (FRET) microscopy, a powerful technique to study specific molecular interactions inside living cells.58 The monomeric Turquoise (mTur)/mVenus (mVen) fluorescence were used as a donor/acceptor pair. FRET efficiency was measured using the acceptor-photobleach method in which the donor (mTur) fluorescence intensity before and after specific photodestruction of acceptor (mVen) was compared. We cotransfected αIIb-mTur/β3-mVen, pcDNA3.1 (PC)-αIIb/β3-mTur/FLNa-Ig21-mVen, or αIIb-mTur/PC-β3/FLNa-Ig21-mVen as the experimental group with and without the treatment of PMA (phorbol myristate acetate), a potent activator of the protein kinase C pathway that is known to activate integrin αIIbβ3.59 We also cotransfected αIIb-mTur/PC-β3 or PC-αIIb/β3-mTur as control groups to ensure that the photobleaching of the acceptor (mVenus) has no direct effect on the donor (mTurquoise) emission intensity (data not shown). Firstly, we examined the integrin αIIb/β3–CT association/separation, a well-known step in integrin activation.39,40 Our results show that in αIIb-mTur/β3-mVen transfected cells with PMA treatment, FRET efficiency was significantly reduced (Figure 2A; supplemental Figure 2A-B), consistent with the separation of integrin αIIb/β3 CTs in integrin activation as previously observed using FRET for another integrin, αMβ2.55 Furthermore, with PMA treatment, FRET efficiency between β3-mTur/FLNa-Ig21-mVen was significantly reduced (Figure 2B; supplemental Figure 2C-D). This decrease is expected because the interaction between filamin and integrin β3-CT is predicted to be disrupted by talin to trigger the integrin activation.25,29 Surprisingly, the FRET efficiencies between αIIb-mTur and FLNa Ig21-mVen remain unexpectedly similar without and with the PMA treatment (Figure 2C; supplemental Figure 2E-F), indicating that filamin remains associated with αIIb CT after the integrin activation. Figure 2D provides a quantitative comparison of the FRET data (Figure 2A-C; supplemental Figure 2A-F). Note that previous FRET studies also suggested that filamin is associated with integrin αM CT.55 However, this study showed that αM CT binds filamin very weakly with dissociation constant KD in the millimolar range,55 which is similar to that of the the isolated αIIb CT/filamin interaction.29 Such weak affinity fits the previous model (supplemental Figure 1)39,40 in which filamin no longer effectively associated with the integrin after being displaced from β CT by talin. However, our FRET results strongly suggest that after being displaced from β3 CT by talin, filamin can reassociate with integrin αIIb CT.
Distinct associations of filamin with inactive and active integrin αIIbβ3 cytoplasmic face. Representative images of FRET efficiency using acceptor (mVenus) photobleaching of mTurquoise-mVenus pair transfected in CHO-K1 cells. Rings outlining the cell membrane were selected as the region of interest. Intensities within the region of interest before and after bleach were counted using Image Pro Plus 7 and used for calculating FRET efficiency (FRET effi.). Ten cells in each group were analyzed (see supplemental Figure 2A-F). A separation >100 Å between mTur and mVen would abolish FRET. (A) FRET of αIIb-mTur/β3-mVen pair without PMA treatment (left) shows good FRET efficiency, consistent with the association of αIIb/β3 CTs. Upon PMA treatment (right), the FRET efficiency is reduced, consistent with the separation of αIIb/β3 CTs upon integrin activation. (B) FRET of β3-mTur/FLNa Ig21-mVen pair without PMA treatment (left) shows good FRET efficiency, consistent with filamin association with β3 CT in the resting state of integrin. Upon PMA treatment (right), the FRET efficiency is reduced indicating the dissociation of filamin from β3 CT. (C) FRET of αIIb-mTur/FLNa Ig21-mVen pair with (right) and without (left) PMA treatment shows similar FRET efficiency, indicating that filamin remains associated with αIIb CT in both resting and active states of αIIbβ3. (D) Bar plot for FRET measured for αIIb-mTur/β3-mVen, or PC-αIIb/β3-mTur/FLNa Ig21-mVen, αIIb-mTur/PC-β3/FLNa Ig21-mVen cotransfected in CHO-K1 cells with or without PMA treatment. Data are average values ± SEM for 10 cells. ∗∗∗P < .001; ∗P > .05. SEM, standard error of mean; ns, nonsignificant.
Distinct associations of filamin with inactive and active integrin αIIbβ3 cytoplasmic face. Representative images of FRET efficiency using acceptor (mVenus) photobleaching of mTurquoise-mVenus pair transfected in CHO-K1 cells. Rings outlining the cell membrane were selected as the region of interest. Intensities within the region of interest before and after bleach were counted using Image Pro Plus 7 and used for calculating FRET efficiency (FRET effi.). Ten cells in each group were analyzed (see supplemental Figure 2A-F). A separation >100 Å between mTur and mVen would abolish FRET. (A) FRET of αIIb-mTur/β3-mVen pair without PMA treatment (left) shows good FRET efficiency, consistent with the association of αIIb/β3 CTs. Upon PMA treatment (right), the FRET efficiency is reduced, consistent with the separation of αIIb/β3 CTs upon integrin activation. (B) FRET of β3-mTur/FLNa Ig21-mVen pair without PMA treatment (left) shows good FRET efficiency, consistent with filamin association with β3 CT in the resting state of integrin. Upon PMA treatment (right), the FRET efficiency is reduced indicating the dissociation of filamin from β3 CT. (C) FRET of αIIb-mTur/FLNa Ig21-mVen pair with (right) and without (left) PMA treatment shows similar FRET efficiency, indicating that filamin remains associated with αIIb CT in both resting and active states of αIIbβ3. (D) Bar plot for FRET measured for αIIb-mTur/β3-mVen, or PC-αIIb/β3-mTur/FLNa Ig21-mVen, αIIb-mTur/PC-β3/FLNa Ig21-mVen cotransfected in CHO-K1 cells with or without PMA treatment. Data are average values ± SEM for 10 cells. ∗∗∗P < .001; ∗P > .05. SEM, standard error of mean; ns, nonsignificant.
Filamin strongly binds to a highly conserved N-terminal region of activated αIIb CT that has an open conformation
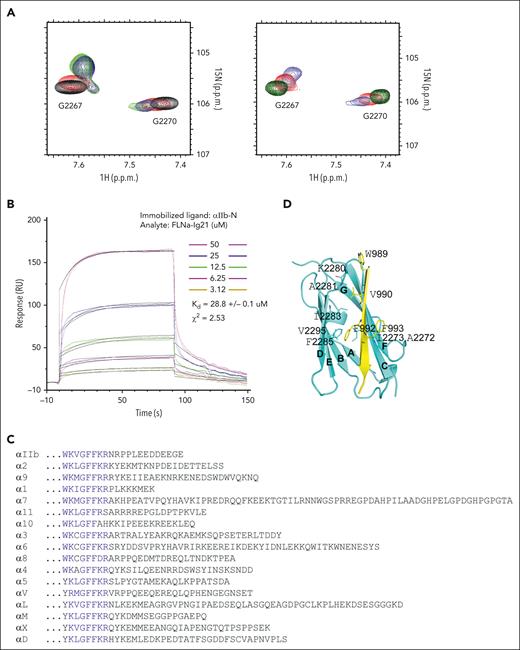
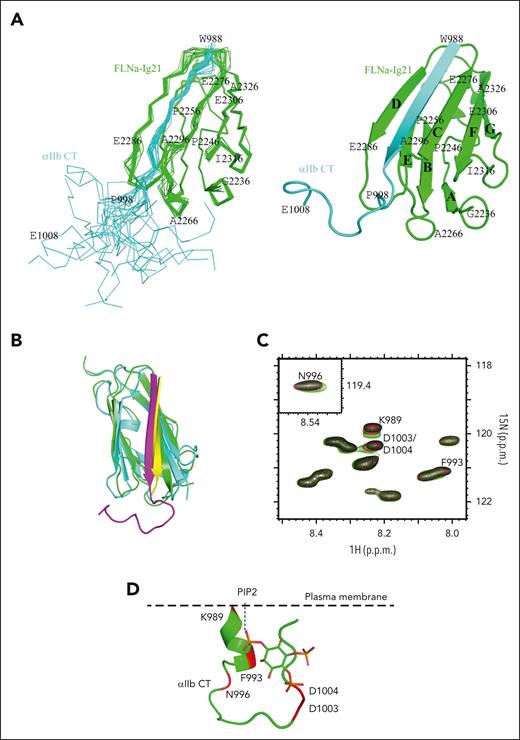
Next, we set to investigate the molecular basis for filamin association with αIIb CT of the activated αIIbβ3. Previous structural studies revealed that inactive integrin αIIb CT can adopt a closed conformation in which an N-terminal membrane-proximal helix and C-terminal loop pack against each other.19,29 Because the N-terminal helix of the inactive αIIb CT interacts with filamin very weakly,29 we wondered whether the C-terminus of αIIb CT, which forms the closed conformation by binding to the N-terminal helix, negatively regulates the αIIb CT binding to filamin. To test this hypothesis, we deleted the αIIb CT C–terminus (E1001-E1009) and examined the filamin binding only to αIIb N–terminus (αIIb-N, W988-L1000). Amazingly, αIIb-N induced much larger chemical shift changes of FLNa-Ig21 than full-length αIIb CT in a 2D NMR–based HSQC (heteronuclear single quantum coherence) experiment (Figure 3A), indicating that αIIb-N binds more strongly to FLNa-Ig21 than αIIb CT. Consistently, quantitative surface plasmon resonance experiments revealed a KD of ∼28 μM for αIIb-N/FLNa-Ig21 (Figure 3B) which is almost an order of magnitude stronger than the full-length integrin αIIb CT binding to FLNa-Ig21 (KD ∼0.23 mM).29 Because the binding interface between N- and C- termini of αIIb CT is largely electrostatic involving the positively charged N–terminus and the negatively charged C–terminus, we mutated the negatively charged residues D1003D1004E1005 at the C-terminus into the positively charged K1003K1004K1005, namely DDE/KKK mutant, so the interface would be disrupted based on the structure of full-length αIIb CT.19 Remarkably, the αIIb CT--DDE/KKK mutant acted like αIIb-N when bound to FLNa-Ig21 (Figure 3A). These data support our hypothesis that αIIb C–terminus conformationally restricts the binding of N-terminus to bind to filamin. We note that the N-terminal regions of integrin α CTs are highly conserved (Figure 3C), suggesting that filamin binds similarly to these α CT regions, but the interactions are likely negatively regulated by the C-terminus in a conformation-dependent manner. Overall, our results indicate that although filamin weakly associates with the closed/inactive conformation of integrin αIIb CT,29 the association becomes dramatically enhanced when αIIb CT is activated into an open conformation.
FLNa-Ig21 strongly binds to activated integrin αIIb CT that displays an open conformation. (A) (left) Selected region of HSQC of 0.1 mM 15N-labeled FLNa-Ig21 in the absence (black) and presence (red) of 0.2 mM αIIb CT, (blue) 0.2 mM αIIb-N, or (green) 0.2 mM αIIb CT DDE to KKK mutant; (right) selected region of HSQC of 0.1 mM 15N-labeled FLNa-Ig21 in 0% PIP2 vesicle (black), 0% PIP2 with 0.2 mM αIIb CT (red), 20% PIP2 to mimic locally enriched PIP2 (green), or 20% PIP2 with 0.2 mM αIIb CT (blue). (B) Representative real-time surface plasmon resonance sensorgrams of the binding between αIIb-N and FLNa-Ig21 (n = 2). The real-time binding curves (colored) were fitted (gray fitting curves) using a global fitting algorithm for a 1:1 binding model, resulting in the KD ∼28 uM. (C) Sequence alignment of α CTs of human integrins. Highly conserved residues are in blue. (D) The X-ray crystal structure of FLNa-Ig21-αIIb CT chimeric protein. β-strands are labeled from A to G based on the canonical immunoglobulin topology. FLNa-Ig21 is colored in cyan, and αIIb CT, in yellow, showing that αIIb CT N–terminus adopts the β-strand to bind the CD groove of FLNa-Ig21. Interface side chains are displayed and labeled.
FLNa-Ig21 strongly binds to activated integrin αIIb CT that displays an open conformation. (A) (left) Selected region of HSQC of 0.1 mM 15N-labeled FLNa-Ig21 in the absence (black) and presence (red) of 0.2 mM αIIb CT, (blue) 0.2 mM αIIb-N, or (green) 0.2 mM αIIb CT DDE to KKK mutant; (right) selected region of HSQC of 0.1 mM 15N-labeled FLNa-Ig21 in 0% PIP2 vesicle (black), 0% PIP2 with 0.2 mM αIIb CT (red), 20% PIP2 to mimic locally enriched PIP2 (green), or 20% PIP2 with 0.2 mM αIIb CT (blue). (B) Representative real-time surface plasmon resonance sensorgrams of the binding between αIIb-N and FLNa-Ig21 (n = 2). The real-time binding curves (colored) were fitted (gray fitting curves) using a global fitting algorithm for a 1:1 binding model, resulting in the KD ∼28 uM. (C) Sequence alignment of α CTs of human integrins. Highly conserved residues are in blue. (D) The X-ray crystal structure of FLNa-Ig21-αIIb CT chimeric protein. β-strands are labeled from A to G based on the canonical immunoglobulin topology. FLNa-Ig21 is colored in cyan, and αIIb CT, in yellow, showing that αIIb CT N–terminus adopts the β-strand to bind the CD groove of FLNa-Ig21. Interface side chains are displayed and labeled.
The drastic affinity enhancement of FLNa-Ig 21 binding to αIIb-N vs to αIIb-N (helix) of the full-length αIIb CT prompted us to investigate the structural basis of such affinity enhancement. We carefully analyzed the 2D 1H NOESY spectrum of αIIb-N (supplemental Figure 3A) or αIIb CT--DDE/KKK mutant (supplemental Figure 3B) bound to FLNa-Ig21 and found that the sequential amide-amide nuclear overhauser enhancements (NOEs), which are characteristic of the N-terminal helix, were lost in contrast to the full-length wild-type (WT) αIIb CT either in the absence19 or presence of FLNa-Ig21.29 On the other hand, sequential αHi-NHi+1 NOEs are strong in the same region of αIIb-N bound to FLNa-Ig21 (supplemental Figure 3C). The disappearance of sequential amide-amide NOEs and the appearance of strong αHi-NHi+1 NOEs are consistent with the formation of a β strand conformation, suggesting that the dramatically enhanced binding of FLNa-Ig21 to αIIb-N or αIIb CT--DDE/KKK mutant vs WT αIIb CT is due to a major secondary structural change at the N-terminal region of activated/open αIIb CT.
To further examine this structural change, we designed a chimeric construct in which αIIb CT was fused to the C-terminus of FLNa-Ig21 with a connecting linker, GASGSGASGSSGS so that αIIb CT is forced to be spatially close to FLNa-Ig21 to promote stronger binding to αIIb CT. This chimeric construct was successfully crystallized. The structure of the complex was solved (PDB ID 7SC4) (supplemental Table 1), and it revealed that αIIb N–terminus does indeed adopt β-strand conformation as part of a β-sheet by inserting into a C/D groove of FLNa-Ig 21, which is known to bind to canonical ligands, such as C-termini of integrin β CTs.25,29 In such a configuration, the C-terminus of αIIb CT no longer contacts with the N-terminus and becomes highly flexible and invisible in the electron density map, which is consistent with an open conformation of αIIb CT. The contacts in the interface of the structure include the following highly conserved hydrophobic interactions: αIIb CT W988/hydrophobic side chain of FLNa-Ig21 K2280, αIIb CT V990/FLNa-Ig21 A2281/I2283, αIIb CT F992/FLNa-Ig21 I2273/I2283/F2285/V2295, and αIIb CT F993/FLNa-Ig21 A2272 (Figure 3D). These hydrophobic contacts are much more extensive than those between the N-terminal helix of inactive αIIb CT and FLNa-Ig-21, which mostly involve some hydrophilic interactions,29 thus providing a basis for understanding that the drastic affinity enhancement of the αIIb CT binding to filamin is a result of a helix-strand transition.
PIP2-enriched integrin-activating environment promotes conformational activation of αIIb CT to strongly bind filamin
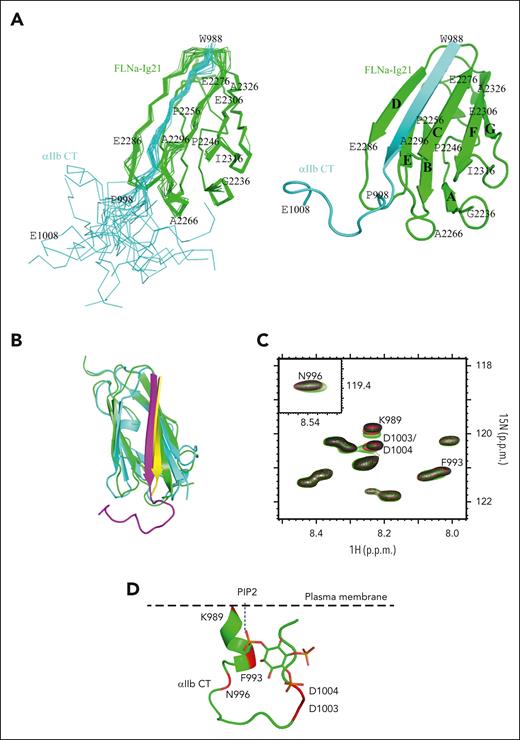
To further understand the αIIb CT binding to filamin, which was described earlier, we examined its binding in a membrane vesicle containing phosphatidylserine (PS) and phosphatidylcholine (PC) (PSPC vesicle; see details in “Methods”) enriched with PIP2. As mentioned in “Introduction,” PIP2, which is locally enriched by talin-bound PIPKIγ,32 is known to activate talin to trigger integrin activation,35-38 and, thus, a PIP2-enriched PSPC mimics the integrin-activating membrane environment. Interestingly, HSQC-based chemical shift perturbation experiments show that PIP2-enriched PSPC vesicle can also strengthen the interaction of WT αIIb CT with FLNa-Ig21 in a manner similar to αIIb-N or αIIb CT--DDE/KKK mutant with FLNa-Ig21 (Figure 3A). Further examination of the NOESY spectra of αIIb CT bound to FLNa-Ig21 (supplemental Figure 3D) revealed that sequential amide-amide NOE crosspeaks of αIIb CT disappeared in the presence of PIP2-enriched PSPC vesicle but not non-PIP2–enriched PSPC. Such NOE disappearance was also observed for αIIb-N or αIIb CT–DDE/KKK mutant bound to FLNa-Ig21 (supplemental Figure 3B) because of the α-helix–to–β-strand transition. To obtain a more definitive atomic view of this process, we determined the total NMR structure of αIIb CT bound to FLNa-Ig21 in PIP2-enriched PSPC vesicle (PDB ID 7SFT; see the representative intermolecular NOEs in supplemental Figure 3E, the ensemble superposition of the NMR structures in Figure 4A, and structural statistics in supplemental Table 2). Remarkably, the structure is essentially identical to the crystal structure of chimeric αIIb CT/FLNa-Ig complex (Figure 4B), with the αIIb N–terminus adopting β-strand conformation, whereas C-terminus is unstructured with no contact with the N-terminus (open conformation). These data reveal a novel structural transition mechanism of αIIb CT upon binding to filamin in the integrin-activating environment.
NMR structure of FLNa-Ig21 bound to αIIb CT in PIP2-enriched integrin-activating membrane environment. (A) (left) Superposition of 20 calculated complex structures of αIIb CT (cyan) and FLNa-Ig21 (green) with the lowest energies, showing a well-defined structure; (right) corresponding cartoon representation of the structure with the lowest energy. β-strands are labeled from A to G based on the canonical immunoglobulin topology. (B) Overlay of NMR solution structure of αIIb CT (magenta) in complex with FLNa-21 (green) and crystal structure of αIIb CT (yellow)–FLNa-21 (cyan) chimera, showing the structures are very similar (backbone root mean square deviation is 1.6 Å). (C) Selected region of HSQC of 0.05 mM 15N-labeled αIIb CT in the absence (black) and the presence of 0.25 mM (red) and 0.5 mM (green) IP3. IP3 significantly perturbed chemical shifts of N-terminal K989, F993, N996, and C-terminal D1003/D1004 of αIIb CT, consistent with an equilibrium shift from closed to an open conformation. (D) A model illustrating how negatively charged PIP2 embedded on the plasma membrane may open the closed conformation by interacting with the positively charged N–terminus while sterically and electrostatically repelling the negatively charged C–terminus. αIIb CT structure is modified from PDB 2MTP.
NMR structure of FLNa-Ig21 bound to αIIb CT in PIP2-enriched integrin-activating membrane environment. (A) (left) Superposition of 20 calculated complex structures of αIIb CT (cyan) and FLNa-Ig21 (green) with the lowest energies, showing a well-defined structure; (right) corresponding cartoon representation of the structure with the lowest energy. β-strands are labeled from A to G based on the canonical immunoglobulin topology. (B) Overlay of NMR solution structure of αIIb CT (magenta) in complex with FLNa-21 (green) and crystal structure of αIIb CT (yellow)–FLNa-21 (cyan) chimera, showing the structures are very similar (backbone root mean square deviation is 1.6 Å). (C) Selected region of HSQC of 0.05 mM 15N-labeled αIIb CT in the absence (black) and the presence of 0.25 mM (red) and 0.5 mM (green) IP3. IP3 significantly perturbed chemical shifts of N-terminal K989, F993, N996, and C-terminal D1003/D1004 of αIIb CT, consistent with an equilibrium shift from closed to an open conformation. (D) A model illustrating how negatively charged PIP2 embedded on the plasma membrane may open the closed conformation by interacting with the positively charged N–terminus while sterically and electrostatically repelling the negatively charged C–terminus. αIIb CT structure is modified from PDB 2MTP.
To understand how PIP2 promotes structural changes in αIIb CT to favorably bind to filamin, we performed the HSQC experiments of 15N-labeled αIIb CT with and without the presence of IP3 (inositol 1, 4, 5-trisphosphate), the head group of PIP2. Figure 4C shows that IP3 perturbed αIIb CT K989, F993, N996, D1003, and D1004, which are near or in the interface between N- and C-termini of the closed αΙIb CT.19,20 Considering the topology of αIIb CT in the membrane environment (Figure 4D), our results suggest that the negatively charged PIP2 head group favorably binds to the positively charged N–terminus while sterically and also electrostatically repelling the negatively charged C–terminus, thereby causing the closed-to-open conformational transition. Such transition releases the αIIb N–terminus to favor the binding to filamin, leading to the α-helix–to–β-strand transition with a substantial enhancement of the binding affinity (from KD ∼0.23 mM to ∼0.028 mM). Overall, our data strongly indicate that there is a spatiotemporal rearrangement of filamin association with integrin before and after the integrin activation. To gain more evidence of such rearrangement, we performed a series of NMR-based HSQC competition experiments. Consistent with the presented data, supplemental Figure 3F,G show that although FLNa-Ig21 forms a ternary complex with αIIb and β3 CTs representing the resting state integrin, FLNa-Ig21 is displaced from β3 CT by talin while becoming strongly associated with αIIb CT in the presence of PIP2-enriched PSPC membrane vesicle that simulates the integrin-activating membrane environment (see supplemental Figure 3F-G for a detailed description).
Cellular localization of filamin in relation to FAs
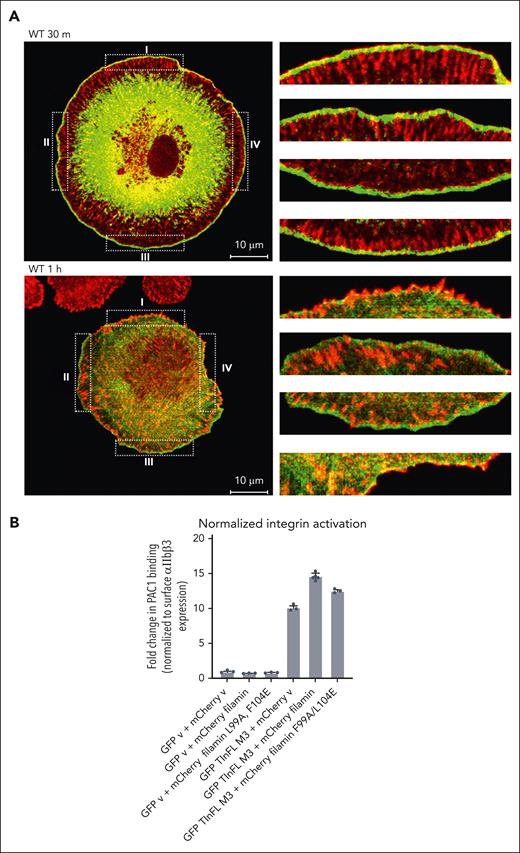
Next, because the filamin/integrin–α CT association is strengthened upon integrin activation, we questioned whether the filamin is localized to nascent FAs that form right after the integrin activation. FAs are defined using common markers such as vinculin or paxillin that indirectly link to integrin β CT upon binding to talin.60 However, although filamin may localize to nascent FAs, it may not be readily detected by these β CT–linked markers because filamin bound to integrin α CT may be distant from β CT–linked FA markers owing to the long-distance separation (>100 Å61) of α/β CTs after integrin activation. To explore this possibility, we allowed mouse embryonic fibroblasts to only spread for 30 or 60 minutes before fixation. Consistent with previous findings,51 our confocal microscopy images (Figure 5A; supplemental Figure 4) show that filamin is enriched along actin filaments stained with phalloidin in 2 separate locations: one is in the axial direction trailing the vinculin-marked FA, and the other is in the lateral direction at the cell cortex. Intriguingly, in 30-minute–spread cells, whenever the axial vinculin (red) crosses the filamin in cell cortex (green), they overlay (by changing colors to yellow) on the tip of vinculin, but in 60-minute–spread cells, this colocalization is significantly reduced. These results are consistent with our speculation that at the very early stage of the integrin activation, α CT–filamin complex is still in close proximity to β CT–linked FA protein complexes, but at the later stage of integrin activation, α CT–filamin complex delocalizes far away from β CT–linked FA complexes because of the long-distance separation of α/β CTs. Such separation during the formation of the activated integrin appears to also depend on the α CT–filamin linkage to actin because actin-binding defective filamin mutant (F99A/L104E)62 disrupts the colocalization between filamin and vinculin even in the 30-minute– or 60-minute–spread cells (supplemental Figure 4).
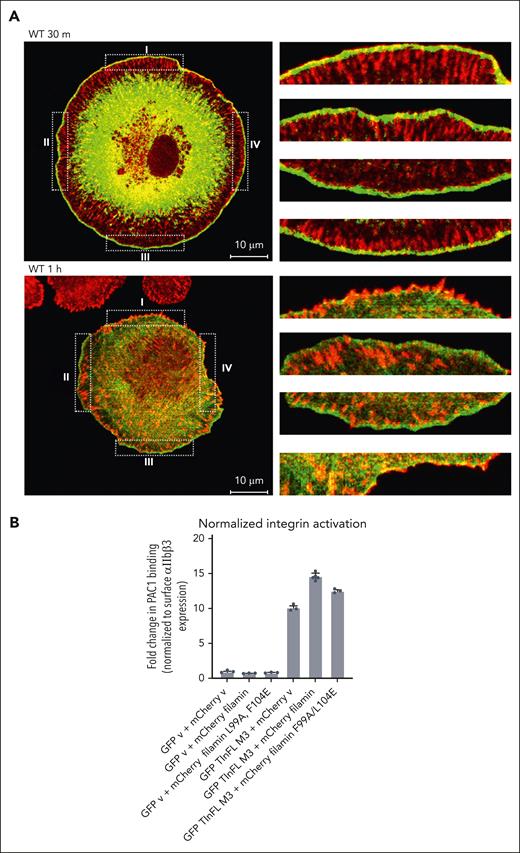
Cellular location of filamin and its regulation on integrin-ECM ligand binding and wound healing. (A) Filamin localizes to nascent FAs upon integrin activation. Merged confocal sections of EGFP (enhanced green fluorescent protein)-filamin WT were fixed after plating on FN-coated glass for 30 minutes or 1 hour. Four randomly selected regions (dotted boxes labeled I-IV, left panel) were enlarged (right) to show the colocalization at the cell cortex. The enlarged boxes on the right correspond as from I to IV from top to bottom, respectively. Note that the resolving power of a confocal microscope is, on average, from 100 to 200 nm laterally (x-y–axes), and 500 nm axially (z-axis). Given that α/β CTs are separated by >100Å61 and that extended forms of both filamin26 and talin63 are >100 nm, filamin (bound to α CT) and vinculin (bound to talin-β CT) can be easily separated beyond the resolving power of a confocal microscope. Scale, 10 μm; green, EGFP-filamin; red, vinculin. Green fluorescent protein–expressing cells were considered filamin-transfected and therefore selected for imaging. See supplemental Figure 4 for individual stains. (B) Filamin promotes active talin-triggered integrin activation. Bar graph generated from flow cytometry data using PAC-1 antibody to analyze the level of activated integrins on cells transfected with indicated vectors. Data are averages ± SEM for 3 independent experiments.
Cellular location of filamin and its regulation on integrin-ECM ligand binding and wound healing. (A) Filamin localizes to nascent FAs upon integrin activation. Merged confocal sections of EGFP (enhanced green fluorescent protein)-filamin WT were fixed after plating on FN-coated glass for 30 minutes or 1 hour. Four randomly selected regions (dotted boxes labeled I-IV, left panel) were enlarged (right) to show the colocalization at the cell cortex. The enlarged boxes on the right correspond as from I to IV from top to bottom, respectively. Note that the resolving power of a confocal microscope is, on average, from 100 to 200 nm laterally (x-y–axes), and 500 nm axially (z-axis). Given that α/β CTs are separated by >100Å61 and that extended forms of both filamin26 and talin63 are >100 nm, filamin (bound to α CT) and vinculin (bound to talin-β CT) can be easily separated beyond the resolving power of a confocal microscope. Scale, 10 μm; green, EGFP-filamin; red, vinculin. Green fluorescent protein–expressing cells were considered filamin-transfected and therefore selected for imaging. See supplemental Figure 4 for individual stains. (B) Filamin promotes active talin-triggered integrin activation. Bar graph generated from flow cytometry data using PAC-1 antibody to analyze the level of activated integrins on cells transfected with indicated vectors. Data are averages ± SEM for 3 independent experiments.
Distinct associations of filamin with inactive vs active integrin regulate the transition from integrin inside-out to outside-in signaling
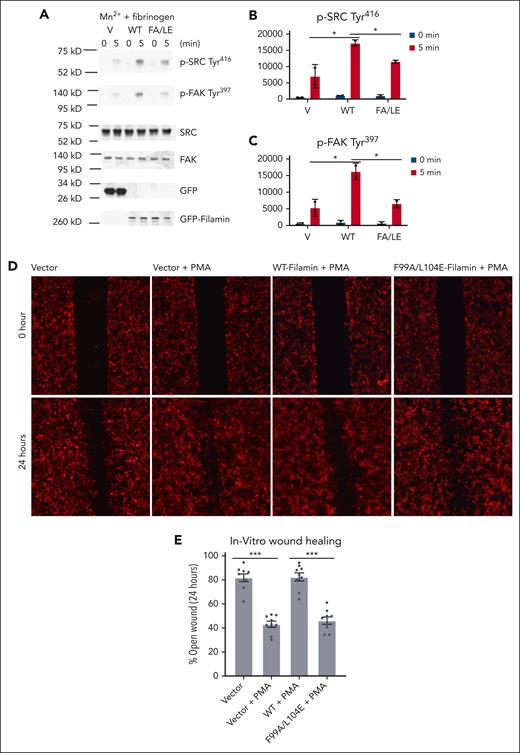
Based on the presented data and also the fact that both talin and filamin bind to actin filaments, it is conceivable that the spatiotemporally rearranged filamin-αIIb association would lead to a major change of the integrin cytoplasmic face with respect to the actin cytoskeleton (outside-in signaling); that is, from the inactive state (state 1) in which the clasped α/β CTs of inactive integrin are bound to filamin (connected to actin) to the active state (state 2) in which separated α CT of active integrin is strongly bound to filamin (connected to actin), whereas separated β CT is strongly bound to talin (also connected to actin). Filamin in state 2 would cooperate with activated talin to stabilize or strengthen the active state of integrin. Indeed, when we coexpressed FLNa and a constitutively active talin mutant (M319A, T1767L, E1770K, and tln-M3; see Lu et al24), PAC-1, an antibody specific to active integrin αIIbβ3, had increased binding to αIIbβ3 vs tln-M3 alone. Yet the actin-binding defective filamin mutant (F99A/L104E),62 when coexpressed with tln-M3, decreased the ligand binding (Figure 5B). Consistently, the actin-binding defective filamin mutation (F99A/L104E) also decreased phosphorylation of steroid receptor coactivator and focal adhesion kinases (Figure 6A-C), common integrin outside-in signaling markers. These data provide evidence that filamin-mediated state 2 promotes integrin outside-in signaling.
Actin-binding defective mutation in filamin A impairs integrin outside-in signaling. (A) Analysis of αIIbβ3 outside-in signaling in HEL cells transfected with EGFP vector (V), EGFP-filamin (WT), and EGFP-filamin F99A/L104E (FA/LE) and simulated with 0.5 mM Mn2+ in the presence of 25 μg/mL fibrinogen in suspension. At indicated time points, HEL cells were lysed and analyzed using western blotting, with antibodies recognizing phosphorylated Src Tyr416, phosphorylated FAK Tyr397, Src, FAK, and GFP. Band intensity of phosphorylated Src (B) and phosphorylated FAK (C) was quantified using imageJ. Data are averages ± SEM for 2 independent experiments. ∗P < .05. (D) Regulation of cell migration by filamin. Representative images of HEK293T cells transfected with pmCherry, pmCherry-WT-Filamin, pmCherry-F99A/L104E-Filamin respectively at time 0 and 24 hours after a scratch was introduced in the monolayer with a 20 μl pipette tip. (E) The average area of the open wound of the indicated conditions at 24 hours after a scratch was introduced. The area was shown as the percentage of wound area at time 0. Data are averages ± SEM for 6 independent experiments. ∗∗∗P < .001. FAK, focal adhesion kinase; HEL, human erythroleukemia cell; Src, proto-oncogene tyrosine kinase.
Actin-binding defective mutation in filamin A impairs integrin outside-in signaling. (A) Analysis of αIIbβ3 outside-in signaling in HEL cells transfected with EGFP vector (V), EGFP-filamin (WT), and EGFP-filamin F99A/L104E (FA/LE) and simulated with 0.5 mM Mn2+ in the presence of 25 μg/mL fibrinogen in suspension. At indicated time points, HEL cells were lysed and analyzed using western blotting, with antibodies recognizing phosphorylated Src Tyr416, phosphorylated FAK Tyr397, Src, FAK, and GFP. Band intensity of phosphorylated Src (B) and phosphorylated FAK (C) was quantified using imageJ. Data are averages ± SEM for 2 independent experiments. ∗P < .05. (D) Regulation of cell migration by filamin. Representative images of HEK293T cells transfected with pmCherry, pmCherry-WT-Filamin, pmCherry-F99A/L104E-Filamin respectively at time 0 and 24 hours after a scratch was introduced in the monolayer with a 20 μl pipette tip. (E) The average area of the open wound of the indicated conditions at 24 hours after a scratch was introduced. The area was shown as the percentage of wound area at time 0. Data are averages ± SEM for 6 independent experiments. ∗∗∗P < .001. FAK, focal adhesion kinase; HEL, human erythroleukemia cell; Src, proto-oncogene tyrosine kinase.
Using a wound healing assay, a well-established assay to study cell migration, Haage et al64 previously showed that increased integrin activation as well as abnormal traction force induced by a talin autoinhibition–disrupted mutant significantly delay the wound closure because of defective cell migration. To further functionally illustrate the importance of filamin-integrin–α CT interaction in integrin outside-in signaling, we performed the same wound healing assay in which we compared the cell migration kinetics by expressing WT filamin vs the actin-binding defective filamin mutant (F99A/L104E)62 upon PMA-induced integrin activation. Figure 6D-E shows that compared with the vector control, PMA, which is expected to induce the integrin activation, significantly stimulated cell migration. However, the overexpression of WT FLNa, but not FLNa (F99A/L104E), significantly delayed PMA-induced cell migration (Figure 6D-E). These data suggest that the overexpression of filamin inhibits dynamic cell movement likely because of excessively enhanced integrin outside-in signaling or increased cell adhesion and abnormal traction force,51 which is induced by filamin overexpression–enhanced α CT–actin linkage. These results are consistent with our structural, biochemical, and FRET-based cellular data and strongly support the mechanism that upon integrin activation, filamin further associates with active integrin to regulate outside-in signaling and promote cytoskeleton reassembly and cell adhesion.
Discussion
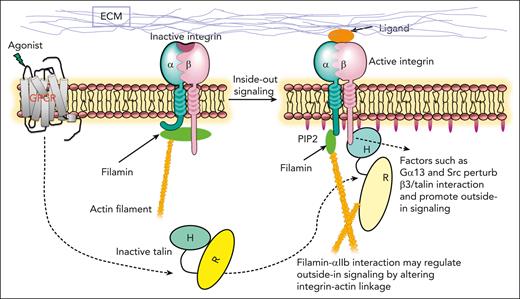
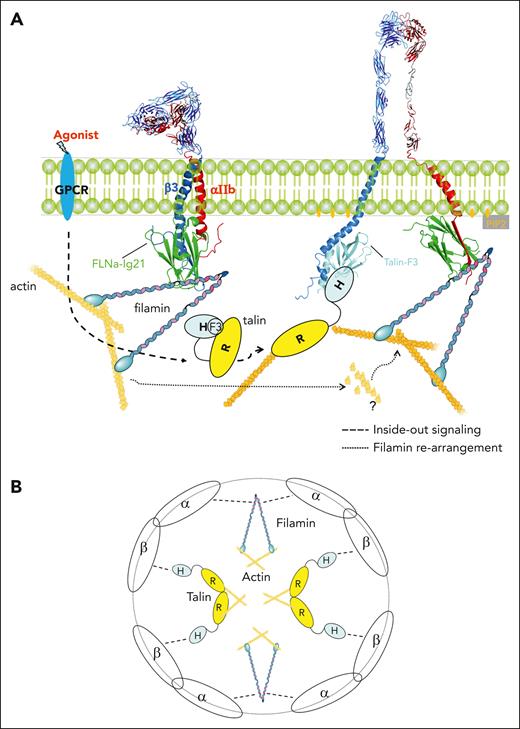
Upon the activation of talin and its binding to integrin β CT to sterically disrupt the integrin α/β CT complex and trigger the inside-out signaling of the receptor, talin is known to further initiate the formation of FAs that are linked to actin filaments (outside-in signaling), thereby promoting stress fiber formation and firm cell adhesion.23,44,65 This β CT–talin (FAs)-actin linkage is then disrupted by a distinct regulator such as Gα13 that competes with talin for binding to β3 CT66 or calpain that cleaves talin;67 thereby, further mediating the integrin outside-in signaling events, such as cytoskeleton reassembly and cyclic adhesion processes, such as cell migration and spreading. Although this integrin β CT–mediated integrin outside-in signaling has been extensively characterized, surprisingly little is known about how α CT of activated integrin functions during the integrin outside-in signaling. In fact, integrin α CT is typically drawn as a “naked” tail in literature without engaging any partners after integrin activation. The results obtained from our study indicate that upon activation, αIIb CT of integrin αIIbβ3 undergoes a distinct α-helix–β-strand structural transition to strongly bind to filamin, thereby inducing a novel outside-in signaling pathway linking ECM (via integrin αIIb CT–filamin) to the actin cytoskeleton. Because filamin was previously shown to associate with the αIIb/β3 CT complex to stabilize the resting state integrin,1 the spatial rearrangement of filamin to strongly associate with integrin αIIb CT suggests a positive role of filamin in promoting integrin outside-in signaling, which appears to occur immediately after talin-induced αIIb/β3 CT separation. This previously unrecognized role of filamin after integrin activation is supported by our extensive functional and structural data. Thus, filamin is a unique dual regulator that can spatiotemporally control both integrin inside-out (inhibitor) and outside-in (promoter) signaling. Based on this presented information, we propose a new model to illustrate 2 key steps of how filamin modulates bidirectional signaling of αIIbβ3 and likely other integrins: step 1, filamin strengthens the integrin α/β CT complex stabilizing inactive integrin; and step 2, filamin is displaced from β CT by talin and then rapidly rearranged to strongly associate with integrin α CT (Figure 7A). Such rearrangement in the integrin cytoplasmic face leads to a new integrin-actin linkage, that is, active integrin α–filamin-actin and active integrin β–talin-actin linkages, which differ from the inactive integrin–filamin-actin linkage. Note that because both talin and filamin are dimers, our model can be expanded to suggest that talin-integrin β/filamin-integrin α linkages may further promote the clustering of active integrins (Figure 7B) and their affinity for multivalent ligands (Figure 5B); see recent talin-based studies24). Disruption of filamin-mediated integrin-actin linkage may destabilize the activation state of integrin and induce dynamic events regulated by integrin outside-in signaling, such as cytoskeleton reassembly, cell-ECM adhesion, and cell migration, as shown in this study (Figure 6D-E; supplemental Figure 4). Such disruption may be mediated by distinct regulators, such as migfilin that strongly binds to canonical C/D groove of filamin Ig2148 and other Ig repeats69 to trigger dynamic cell adhesion. Interestingly, a recent study showed that the knockout of migfilin in platelets prolongs the integrin-filamin association and controls the dynamics of integrin outside-in signaling after the activation of integrin αIIbβ3.30 Data obtained in our study indicate that the prolonged integrin-filamin association in migfilin null mice30 involves integrin αIIb CT because integrin β3 CT is already occupied by talin upon the integrin activation. Consistently, the administration of a filamin-binding migfilin peptide was shown to dissociate filamin from activated integrin αIIbβ3, triggering cytoskeleton reorganization and platelet aggregation.30
Diagrammatic illustrations of spatiotemporal regulation of integrin signaling by filamin. (A) At the resting state, integrin is stabilized by filamin that forms a ternary complex with both integrin α and β CTs.29 Agonist stimulation triggers a cascade of cellular signals that lead to the recruitment/activation of talin to bind integrin. The activation of talin is mediated by PIP2 that is generated by PIPKIγ recruited by talin (not drawn; see the text). The talin-activating PIP2 also opens up the closed conformation of α CT (Figure 4D), which drastically enhances its binding to filamin accompanied by a major conformational change of the N-terminal of α CT from α-helix to β-strand. Note the dramatic change of the integrin cytoplasmic face where filamin initially links α/β CTs of inactive integrin with actin but is dissociated by talin (bound to β CT and actin) and then strongly reassociated with α CT of active integrin and actin to promote the outside-in signaling. The question mark indicates the complex actin cytoskeleton reassembly process as regulated by the spatial rearrangement of filamin. Note that in addition to filamin and talin drawn in this diagram, many molecules23 including other integrin CT binding proteins68 may be involved in mediating the outside-in signaling in a spatiotemporal manner, which remains to be investigated. Structures displayed in the diagram were modified from FLNa-Ig21/αIIb (7SC4) and protein data bank--integrin αVβ3 ectodomain (1JV2) and αIIbβ3 TMCT (2KNC), talin (2H7E), FLNa-Ig21/αIIb CT/β3 CT (2MTP). (B) Clustering of activated integrins as mediated by the dimerization of talin and filamin. Note that clustering may be also mediated by additional regulators, such as kindlin and paxillin.24
Diagrammatic illustrations of spatiotemporal regulation of integrin signaling by filamin. (A) At the resting state, integrin is stabilized by filamin that forms a ternary complex with both integrin α and β CTs.29 Agonist stimulation triggers a cascade of cellular signals that lead to the recruitment/activation of talin to bind integrin. The activation of talin is mediated by PIP2 that is generated by PIPKIγ recruited by talin (not drawn; see the text). The talin-activating PIP2 also opens up the closed conformation of α CT (Figure 4D), which drastically enhances its binding to filamin accompanied by a major conformational change of the N-terminal of α CT from α-helix to β-strand. Note the dramatic change of the integrin cytoplasmic face where filamin initially links α/β CTs of inactive integrin with actin but is dissociated by talin (bound to β CT and actin) and then strongly reassociated with α CT of active integrin and actin to promote the outside-in signaling. The question mark indicates the complex actin cytoskeleton reassembly process as regulated by the spatial rearrangement of filamin. Note that in addition to filamin and talin drawn in this diagram, many molecules23 including other integrin CT binding proteins68 may be involved in mediating the outside-in signaling in a spatiotemporal manner, which remains to be investigated. Structures displayed in the diagram were modified from FLNa-Ig21/αIIb (7SC4) and protein data bank--integrin αVβ3 ectodomain (1JV2) and αIIbβ3 TMCT (2KNC), talin (2H7E), FLNa-Ig21/αIIb CT/β3 CT (2MTP). (B) Clustering of activated integrins as mediated by the dimerization of talin and filamin. Note that clustering may be also mediated by additional regulators, such as kindlin and paxillin.24
In summary, our studies have suggested a crucial dual structural mechanism for filamin to effectively control integrin αIIbβ3 bidirectional signaling for dynamic regulation of cytoskeleton reassembly and platelet aggregation. Given that filamin can engage with other integrins and transmembrane receptors via conserved binding modes,25-28 our findings may help explain how different pools of filamin may mediate signaling of these receptors in a similar manner, thereby providing differential regulation of diverse cellular and physiological processes.
Acknowledgments
The authors thank Jun Yang for the technical assistance with the vesicle membrane preparation, Kemin Tang for crystallographic data collection at Argonne National Laboratory, Kasia Bialkowska for assistance in transfection using HEL cells, and Rui Chen and Thomas McIntyre for providing human platelets.
The work was supported by National Institutes of Health (NIH), National Heart, Lung, and Blood Institute grants R01 HL58758, P01 HL73311, and P01 HL154811, NIH National Institute of Diabetes and Digestive and Kidney Diseases grant R01DK127589, American Heart Association Career Development grant 18CDA34110364, and NIH Office of the Director NMR instrumentation grant S10OD023432. The work also used the Leica SP8 confocal microscope that was purchased with funding from NIH Scientific Interest Group grant 1S10OD019972-01.
Authorship
Contribution: J.L. was involved in conceptual development, performing the experiments, and writing the draft; F.L. was involved in imaging, focal adhesion in analysis, integrin-ligand binding experiments, and data interpretation; S.S.I. was involved in conceptual development and data interpretation; M.A. was involved in crystal structure determination; M.D. was involved in initial cell spreading studies and data interpretation; G.D. was involved in confocal imaging experiments and data interpretation; E.F.P. was involved in the data interpretation and manuscript writing; and J.Q. was involved in the entire process, including coordinating/supervising the work, conceptual development, designing the studies, data interpretation, and manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jun Qin, Department of Cardiovascular and Metabolic Sciences, NB20, Lerner Research Institute, Cleveland Clinic, 9500 Euclid Ave, Cleveland, OH 44195; e-mail: qinj@ccf.org.
References
Author notes
All data sets and protocols are available on reasonable request from the corresponding author, Jun Qin (qinj@ccf.org).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.