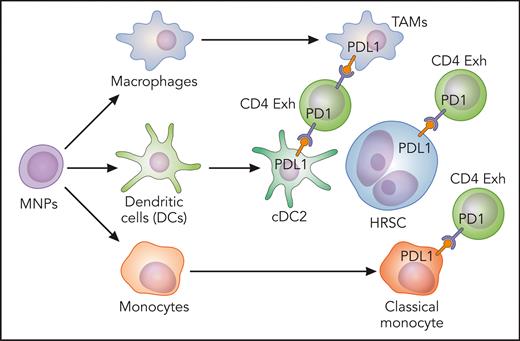
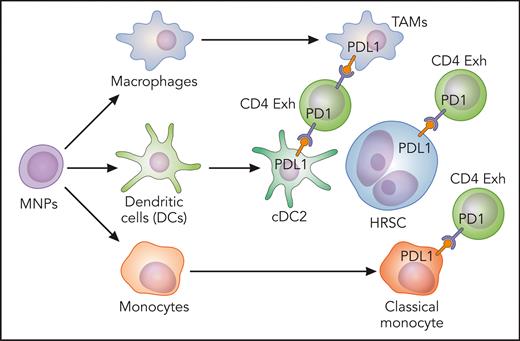
In this issue of Blood, Stewart et al1 describe mononuclear phagocyte (MNP) networks in the microenvironment of classic Hodgkin lymphoma (cHL), in which classical monocytes, macrophages, and conventional dendritic cells 2 (cDC2) expressing inhibitory molecules are enriched in the vicinity of Hodgkin Reed-Sternberg cells (HRSC).
cHL is unique among lymphoid malignancies, given that malignant HRSC comprise less than 1% of the total cell population. HRSC secrete chemokines and cytokines that drive the formation of a complex microenvironment of nontumor cells. This microenvironment consists of both innate as well as adaptive immune cells.2 The extensive but ineffective immune cell infiltrates found in cHL suggest that HRSC have developed mechanisms to escape immunosurveillance. Furthermore, HRSC depend on microenvironmental signals for survival and growth. Even though many cHL patients are cured with current treatment regimens, 20% to 30% fail to respond or experience a relapse after treatment. Moreover, especially in adolescents and young adults, treatment-related toxicity and long-term morbidity are persistent challenges requiring more basic research to identify new therapeutic targets, particularly within this unique microenvironment.3,4 Stewart et al provide evidence for the first time of the expression of immunoregulatory checkpoints and the secretion of chemokines from previously uncharacterized cells that contribute to the exhaustion of T cells and contribute to immune evasion in cHL.
HRSC have numerous mechanisms to elude the immune system, such as the overexpression of programmed cell death-1 ligand (PDL1) due to genetic alterations, that promote the interaction with PD1+ T cells that suppress antitumor immunity.5 The expression of PDL1 by tumor-associated macrophages (TAMs) also contributes to this immunosuppressive niche, and these TAMs physically colocalize with PDL1+ HRSC. PDL1+ macrophages are also enriched for contacts with T cells surrounding HRSC, implicating CD4 T cells as a target of PD1 blockade.6 In fact, in cHLs, concomitant expansion of PD1+ effector T helper 1 cells and regulatory PD1− T cells has been described, working as complementary mechanisms of immunosuppression.7 Even though the clinical success of PD1 blockade in some patients with cHL suggests that exhaustion of T effector cells could be reversible, it was demonstrated that T cells and TAMs positive for CTLA4 and its ligand CD86, respectively, also contribute to the HRSC immunosuppressive niche, raising the possibility that patients with cHL refractory to PD1 blockade may benefit from CTLA4 blockade.8 The expression of PDL1 was attributed to the HRSC and, in the microenvironment, to macrophages, even though diverse subsets of monocytes, macrophages, and dendritic cells are present. The aim of the study performed by Stewart et al was to characterize the complexity and diversity of MNP networks and explore their interactions with HRSC by means of complementary high-resolution techniques.
The study demonstrated by single cell transcriptomic profiling the recruitment of exhausted CD4 and CD8 T cells with specific markers such as CD27, TNFRSF18, LAG3, and ICOS in cHL. Enrichment of exhausted LAG3+ and CD4 T cells around HRSC has been previously reported.9 The analysis of microdissected HRSC revealed NF-κB activation, with an NF-κB–centric network coordinating the upregulation of the chemokines CCL5, CCL17, and CCL22 capable of the positioning and retention of exhausted T cells via CCR5 and CCR4 ligation. Targeted spatial transcriptomic profiling identified 2 “neoplastic” PDL1high HRSC-enriched clusters, one enriched for exhausted T cells and the other characterized by classical monocyte, macrophage, and cDC2 infiltration, showing, for the first time, that not only HRSC and macrophages are involved in PDL1 expression to create an immunosuppressive niche around tumor cells but other cells of the MNP network are also involved. Furthermore, the presence of the cDC2 monocyte-macrophage niche was associated with early relapse following treatment, providing evidence that other cells of the MNP network, not only macrophages,10 are associated with poor outcomes in advanced cHL.
The existence of this immune-privileged niche was subsequently confirmed by multiplexed immunofluorescence panels and revealed enrichment of cDC2 and CD11c+ monocytes in the immediate vicinity of HRSC. Both plasmacytoid DC and activated DC were excluded from the HRSC niche. Furthermore, a second panel was designed to confirm PDL1 expression. PDL1 was expressed by HRSC and by macrophages, as expected,6 and also by CD11c+/CD68− MNPs around HRSC, suggesting that monocytes and dendritic cells also contribute to the immunosuppressive niche by expressing PDL1 (see figure) and other coinhibitory receptors such as TIM3 and IDO1. The immunosuppressive signaling could be different in pediatric and young adult cHL from cHL in older adults, given the fact that the proportion of CD11c+/CD68− and CD11c+/CD68+ MNPs expressing inhibitory molecules increased with age at diagnosis. The analysis of ligand-receptor interactions predicted paracrine CCL3, CCL4, and CXCL10 signaling by macrophages and classical monocytes to CCR5-, CCR1-, and CXCR3-expressing cDC2 and exhausted T cells.
Classical monocytes, macrophages, and cDC2-expressing inhibitory molecules enriched in the vicinity of HRSC. Exh, exhausted. Professional illustration by Patrick Lane, ScEYEnce Studios.
Classical monocytes, macrophages, and cDC2-expressing inhibitory molecules enriched in the vicinity of HRSC. Exh, exhausted. Professional illustration by Patrick Lane, ScEYEnce Studios.
Stewart et al provide evidence of distinct and recurring tissue niches within cHL lymph nodes. Another important strength of this study is that MNPs, in particular classical monocytes and conventional DCs, have a key role in directing and amplifying immunosuppressive niche signaling, as previously reported for macrophages. In addition, even though recent reports suggested the presence of alternative immune checkpoints in cHL,8,9 the coexpression of inhibitory molecules and chemokines by MNPs, as shown in this study, suggests extensive redundancy in immunosuppressive signaling. Therefore, the MNPs adjacent to HRSC exhibit critical intercellular communication pathways that may be therapeutically targetable and provide the basis to design future strategies towards new inhibitory molecules or an MNP-depleted phenotype.
Stewart et al performed a high-resolution analysis of the content of cHL lymph nodes and confirmed the expression of specific inhibitory molecules by means of customized multiplexed immunofluorescence panels. However, no functional experiments to validate their conclusions were included. Therefore, it will be important to continue the functional characterization of novel and existing checkpoint molecules for the development of rational combination therapeutic strategies in cHL and to increase our knowledge in the complexity and spatial polarization within the MNP compartment and their potential roles in immune evasion.
Conflict-of-interest disclosure: The author declares no competing financial interests.