Key Points
Tumor-associated MNP are diverse and spatially polarized: cDC2 colocalize with malignant HRSCs; plasmacytoid and activated DCs are excluded.
DCs, monocytes, and macrophages exist in an inflammatory niche in close proximity to HRSCs, and all express inhibitory molecules.
Abstract
Classic Hodgkin lymphoma (cHL) has a rich immune infiltrate, which is an intrinsic component of the neoplastic process. Malignant Hodgkin Reed-Sternberg cells (HRSCs) create an immunosuppressive microenvironment by the expression of regulatory molecules, preventing T-cell activation. It has also been demonstrated that mononuclear phagocytes (MNPs) in the vicinity of HRSCs express similar regulatory mechanisms in parallel, and their presence in tissue is associated with inferior patient outcomes. MNPs in cHL have hitherto been identified by a small number of canonical markers and are usually described as tumor-associated macrophages. The organization of MNP networks and interactions with HRSCs remains unexplored at high resolution. Here, we defined the global immune-cell composition of cHL and nonlymphoma lymph nodes, integrating data across single-cell RNA sequencing, spatial transcriptomics, and multiplexed immunofluorescence. We observed that MNPs comprise multiple subsets of monocytes, macrophages, and dendritic cells (DCs). Classical monocytes, macrophages and conventional DC2s were enriched in the vicinity of HRSCs, but plasmacytoid DCs and activated DCs were excluded. Unexpectedly, cDCs and monocytes expressed immunoregulatory checkpoints PD-L1, TIM-3, and the tryptophan-catabolizing protein IDO, at the same level as macrophages. Expression of these molecules increased with age. We also found that classical monocytes are important signaling hubs, potentially controlling the retention of cDC2 and ThExh via CCR1-, CCR4-, CCR5-, and CXCR3-dependent signaling. Enrichment of the cDC2-monocyte-macrophage network in diagnostic biopsies is associated with early treatment failure. These results reveal unanticipated complexity and spatial polarization within the MNP compartment, further demonstrating their potential roles in immune evasion by cHL.
Introduction
Classic Hodgkin lymphoma (cHL) accounts for between 12% and 15% of lymphoma diagnoses and occurs most frequently in adolescents and young adults, with another peak of incidence in older adults. Multiagent cytotoxic chemotherapy (including selective use of radiotherapy) is curative in between 80% and 95% of patients; in adults the potential for cure is largely determined by the disease stage.1-3 However, for patients with relapsed or refractory disease, the prospect of cure or long-term disease control is significantly diminished.
A cHL tumor is defined by CD30+ Hodgkin Reed-Sternberg cells (HRSCs), comprising between 1% and 5% of cells, and derived from germinal center (GC) B cells.4 HRSC survive within a tumor microenvironment (TME) containing abundant T cells and a network of mononuclear phagocytes (MNPs).4 HRSCs establish an immunosuppressive niche through multiple mechanisms, including downregulation of major histocompatibility complex (MHC) proteins and enhanced expression of programmed cell death ligand-1 (PD-L1), resulting from 9p23-p24 copy number gain or amplification.5,6 PD-L1 is also abundantly expressed by myeloid cells surrounding HRSCs, leading to the notion that MNPs in cHL are largely tumor-associated macrophages (TAMs).7,8
Recent cross-tissue surveys of MNP diversity, using single-cell approaches, have characterized multiple states of monocytes, macrophages, and dendritic cells, identifying significant differences in functional properties and roles in pathology.8 Although MNPs are frequently observed in cHL, details of their functional specialization are not known.
Here, we report the use of complementary high-resolution techniques to map MNP diversity in the microenvironment of cHL. We found that tumor-associated MNPs comprise multiple cell types and occupy distinct and prognostically relevant niches, and that MNPs adjacent to HRSCs exhibit immunosuppressive phenotypes. These results reveal the complexity of MNP networks in cHL, and critical intercellular communication pathways that may be therapeutically tractable.
Methods
Tissue samples for scRNA-seq
Healthy lymph nodes, unaffected by neoplastic disease, were acquired from adult donors by donation after circulatory death at the time of organ retrieval for transplantation by the Cambridge Biorepository for Translational Medicine with ethical approval (15/EE/0152, East of England, Cambridge South research ethics committee) and consent from donor families (Table 1). Lymph node samples were collected from inguinal, mesenteric, and thoracic regions at the end of organ donation, within 1 to 2 hours of cessation of circulation under cold ischemic conditions. Lymph node specimens were transported in ice-cold 0.9% saline. Tissue was processed as previously described.9 Lymph node data (290b, 298c, and 302c) are shared with James et al and were processed with additional flow sorting.9 Patients who donated diseased tissue for single-cell RNA sequencing (scRNAseq) were enrolled in the “Investigating how childhood tumors and congenital disease develop” study (National Health Service [NHS] National Research Ethics Service [16/EE/0394]).
Healthy donors included in this study
| Donor . | Age, y . | Gender . | Ethnicity . | Cause of death . | Comorbidities . | Smoking history . | Alcohol intake . | BMI . |
|---|---|---|---|---|---|---|---|---|
| 284C | 59 | Male | White | Hypoxic brain damage | Inhalers for seasonal wheeze | 25 pack y, stopped 17 y before death | 3-6 units/d for 40 y | 25.8 |
| 290B | 61 | Female | South American | Drug overdose, hypoxic brain damage | Overdose of paracetamol (acetaminophen) 2 years before death, received 4 bags of NAC paracetamol; probable irritable bowel syndrome; anorexia nervosa, celiac disease, date diagnosed unknown | 5.25 pack y | — | 27.7 |
| 292B | 58 | Male | White | Intracranial hemorrhage | Squint correction aged 12 | 46 pack y | — | 27.5 |
| 295B | 79 | Female | White | Intracranial hemorrhage | Cerebral amyloid angiopathy, mild cognitive impairment and findings suggestive of Parkinson disease. Myocardial infarct 3 years before death. | 49 pack y | 1-2 units/d | 24 |
| 298C | 55 | Male | White | Intracranial hemorrhage | Epilepsy and depression | 15 pack y | — | 23.5 |
| 302C | 44 | Male | White | Suspected suicide | Knee sprain, previous tonsillitis | 25 pack y | Unknown | 34.5 |
| 337C | 25 | Female | White | Meningitis | Constipation, laceration to right heel | 4.5 pack y | Minimal | 24.5 |
| 414C | 52 | Male | White | Intracranial thrombosis | Steroid injection to wrist | — | 1-2 units/d | 27.2 |
| Donor . | Age, y . | Gender . | Ethnicity . | Cause of death . | Comorbidities . | Smoking history . | Alcohol intake . | BMI . |
|---|---|---|---|---|---|---|---|---|
| 284C | 59 | Male | White | Hypoxic brain damage | Inhalers for seasonal wheeze | 25 pack y, stopped 17 y before death | 3-6 units/d for 40 y | 25.8 |
| 290B | 61 | Female | South American | Drug overdose, hypoxic brain damage | Overdose of paracetamol (acetaminophen) 2 years before death, received 4 bags of NAC paracetamol; probable irritable bowel syndrome; anorexia nervosa, celiac disease, date diagnosed unknown | 5.25 pack y | — | 27.7 |
| 292B | 58 | Male | White | Intracranial hemorrhage | Squint correction aged 12 | 46 pack y | — | 27.5 |
| 295B | 79 | Female | White | Intracranial hemorrhage | Cerebral amyloid angiopathy, mild cognitive impairment and findings suggestive of Parkinson disease. Myocardial infarct 3 years before death. | 49 pack y | 1-2 units/d | 24 |
| 298C | 55 | Male | White | Intracranial hemorrhage | Epilepsy and depression | 15 pack y | — | 23.5 |
| 302C | 44 | Male | White | Suspected suicide | Knee sprain, previous tonsillitis | 25 pack y | Unknown | 34.5 |
| 337C | 25 | Female | White | Meningitis | Constipation, laceration to right heel | 4.5 pack y | Minimal | 24.5 |
| 414C | 52 | Male | White | Intracranial thrombosis | Steroid injection to wrist | — | 1-2 units/d | 27.2 |
BMI, body mass index.
scRNA-seq analysis
scRNA-seq data from cHL specimens were kindly shared by the laboratory of Christian Steidl (University of British Columbia) as a raw-counts matrix.10 Mapping and quantification of scRNA-seq data generated for this study (WSI data set) was performed using kallisto-bustools (version 0.24.4),11 against the GRCh38 human reference. The unfiltered count matrix was profiled using defaultDrops in the DropletUtils package (version 1.10.3),12 in R (version 4.0.4).
We performed data integration with batch correction (using donor as the batch key) and dimensionality reduction concurrently by training a single-cell variational inference model using the scvi-tools Python package (version 0.9.0a0).13 We completed quality control on the integrated data set (supplemental Methods; supplemental Figure 1A-B; available on the Blood website). A 15-dimensional latent representation was used as input for nearest-neighbor graph construction, uniform manifold and approximation (UMAP), and Leiden clustering in Scanpy. Marker genes were calculated using the tf-idf metric and genes were ranked by the tf-idf metric, and P values calculated by a hypergeometric test, corrected for multiple testing (Benjamini-Hochberg method).14 Cell type annotation was performed using marker genes and canonical marker expression (supplemental Figure 1C), supplemented with labeling using the celltypist Python package (0.1.15).15 Concordance between annotated cell-type labels and predicted reference annotations was assessed by calculating the Jaccard distance between cell label strings (supplemental Figure 1D-E). Cell-type predictions between the King et al B-cell atlas16 was calculated by training a multilayer perceptron classifier on the reference data and calculating predictions on unseen test data, using the scikit-learn package (0.24.2).17
Relative abundance analysis was performed using the MiloR package (0.99.1).18 The annotated data set was represented as a k = 30 nearest-neighbor graphs calculated in scVI latent space. This graph representation was partitioned into 22 573 overlapping neighborhoods, and cell counts per 10× channel was calculated per neighborhood. Differential abundance between disease and health was calculated using the testNhoods function, setting the design as ∼center + disease, and norm.method = “TMM.”
Ligand-receptor analysis was performed using CellPhoneDB (2.1.7).19 The count matrix was normalized to 1e4 total counts before input to the Python implementation of CellPhoneDB, which was run with --iterations = 1000, --threshold = 0.1, and --result-precision = 3.
Microarray data analysis
The GEO accessions GSE39133 and GSE17920 were downloaded using the GEOquery R package (version 2.60.0).20
For GSE39133, we performed differential expression between GC and HRSC samples using limma (version 3.48.1)21 and derived gene sets by retaining genes with an absolute log fold change of >3 and an adjusted P value <.01.
Transcription factor regulon and signaling network analysis
Transcription factor regulons were acquired using the DoRothEA R package (version 1.3.3).24 Regulons with confidence levels “A to C” were used to calculate transcription factor activities using run_viper.
We derived a network graph of transcription factors and targets, or ligands and receptors, using data from the OmnipathR R package (version 3.12),25 plotting the interaction networks using the ggraph R package (version 2.0.5).
Tissue samples for nanostring and multiplexed immunofluorescence
The study was approved by a UK NHS Health Research Authority Research Tissue Bank (Novopath Biobank, Newcastle upon Tyne Hospitals NHS Foundation Trust [17/NE0070]). Novopath released link-anonymized, formalin-fixed and paraffin embedded (FFPE) tissue, from patients with cHL. Tissue samples were surplus to diagnostic requirements and were released in accordance with the terms of the ethical approval.
Excised lymph node biopsies were selected from the pathology archives of Newcastle upon Tyne Hospitals NHS Foundation Trust, by members of Novopath, with appropriate ethical approval (17/NE0070). Limited, link-anonymized clinical data for patients in this study were collected by biobank staff. Hematoxylin and eosin (H&E)-stained and immunohistochemical (IHC) tissue sections were reviewed by a hematologist, together with original pathology reports. Fifty-four tumors were selected for the study, including biopsies from patients of all ages and histomorphological subtypes (Table 2). Patient biopsies were selected on the basis of high-quality, whole lymph node resection biopsies. Patients with coexisting malignant diagnoses were excluded from the study, even if these were not apparent in the tissue.
Clinical and pathological characteristics of cHL study cases for multiplexed immunofluorescence and nanostring GeoMx
| Study tumor reference . | Disease subtype∗ . | EBV status† . | GeoMx‡ . | Age at diagnosis (y) . | Sex . | Site of biopsy (excised node) . | Stage§ . | Clinical response|| . |
|---|---|---|---|---|---|---|---|---|
| CHL-1 | MCCHL | NEG | 80 | M | Inguinal | 2A | CR | |
| CHL-3 | MCCHL | NEG | 77 | F | Submandibular | NA | CR/relapse | |
| CHL-4 | LRCHL | POS | 75 | F | Neck | 2A | CR | |
| CHL-5 | NSCHL | NEG | 73 | F | Supraclavicular | 4B | CR | |
| CHL-6 | NSCHL | NEG | 73 | M | Neck | 1A | CR | |
| CHL-7 | NSCHL | NEG | 72 | M | Neck | 1A | CR | |
| CHL-9 | NSCHL | NEG | YES | 69 | M | Neck | 1A | PD/salvage CR |
| CHL-10 | NSCHL | NEG | YES | 69 | F | Neck | 3B | CR |
| CHL-11 | CHL-NOS | POS | 68 | F | Neck | 3B | CR/relapse | |
| CHL-12 | CHL-NOS | POS | 68 | F | Axilla | 1A | CR/relapse | |
| CHL-13 | MCCHL | NEG | 68 | M | Inguinal | 2A | CR | |
| CHL-14 | CHL-NOS | NEG | 64 | M | Mediastinum | NA | PD/salvage CR | |
| CHL-15 | MCCHL | POS | 64 | M | Neck | NA | NA | |
| CHL-16 | MCCHL | NEG | 63 | F | Neck | 2A | CR | |
| CHL-18 | MCCHL | POS | 60 | F | Supraclavicular | 4B | CR | |
| CHL-19 | MCCHL | NA | 53 | M | Inguinal | NA | PD | |
| CHL-20 | LRCHL | NA | 46 | M | Axilla | NA | NA | |
| CHL-21 | MCCHL | POS | 45 | M | Neck | 3S | PD | |
| CHL-22 | NSCHL | POS | 40 | M | Neck | 2A | CR/relapse | |
| CHL-23 | NSCHL | POS | 39 | M | Neck | 4A | PR/salvage CR | |
| CHL-24 | NSCHL | NA | 38 | M | Supraclavicular | 4A | PD | |
| CHL-25 | NSCHL | NEG | 33 | F | Neck | 3B | CR | |
| CHL-27 | NSCHL | POS | YES | 32 | M | Supraclavicular | 2A | PD |
| CHL-28 | MCCHL | POS | 32 | M | Neck | 3B | CR | |
| CHL-29 | NSCHL | NA | 32 | M | Neck | 4A | CR/relapse | |
| CHL-30 | MCCHL | POS | 28 | F | Neck | 2A | CR | |
| CHL-31 | NSCHL | NEG | 28 | M | Neck | 4B | CR | |
| CHL-32 | MCCHL | POS | YES | 26 | M | Neck | 2A | CR |
| CHL-33 | NSCHL | POS | 25 | M | Neck | 2B | CR | |
| CHL-34 | NSCHL | NEG | YES | 24 | M | Neck | NA | NA |
| CHL-36 | NSCHL | NEG | 23 | F | Neck | 2A | CR | |
| CHL-39 | MCCHL | NEG | 22 | F | Supraclavicular | NA | NA | |
| CHL-40 | NSCHL | NEG | YES | 20 | M | Supraclavicular | 3BS | CR |
| CHL-41 | NSCHL | NEG | 19 | F | Mediastinum | 2B | PD/salvage | |
| CHL-42 | NSCHL | NEG | 18 | F | Neck | 2B | PD | |
| CHL-43 | NSCHL | NEG | YES | 18 | M | Neck | 2A | CR |
| CHL-44 | NSCHL | POS | 18 | M | Postnasal space | 1A | CR | |
| CHL-45 | NSCHL | NEG | 17 | M | Neck | 2A | CR/relapsed and salvage | |
| CHL-48 | NSCHL | NEG | 16 | F | Axilla | 3A | PD/salvage | |
| CHL-49 | MCCHL | POS | 16 | M | Axilla | 1A | CR/relapsed and salvage | |
| CHL-50 | NSCHL | NEG | 16 | F | Neck | 2A | CR | |
| CHL-51 | NSCHL | POS | 16 | F | Supraclavicular | 3B | CR | |
| CHL-52 | NSCHL | NA | 15 | F | Neck | 2B | CR | |
| CHL-53 | CHL-NOS | NEG | 14 | F | Retroperitoneal | 3B | CR | |
| CHL-54 | NSCHL | NEG | YES | 14 | M | Supraclavicular | 2A | CR |
| CHL-55 | CHL-NOS | NEG | 14 | F | Supraclavicular | 3A | CR | |
| CHL-56 | NSCHL | NEG | 14 | F | Neck | 4A | CR | |
| CHL-57 | NSCHL | NEG | YES | 14 | M | Supraclavicular | 2A | CR |
| CHL-59 | NSCHL | NEG | YES | 13 | M | Neck | 4A | CR |
| CHL-60 | LR-CHL | POS | 12 | M | Neck | 1A | CR | |
| CHL-61 | NSCHL | POS | 12 | M | Neck | 2B | CR | |
| CHL-62 | MCCHL | POS | 8 | M | Hilar | 4A | CR/relapsed and salvage | |
| CHL-63 | CHL-NOS | POS | 7 | M | Neck | 4B | CR | |
| CHL-64 | MCCHL | POS | 6 | F | Neck | 3B | CR |
| Study tumor reference . | Disease subtype∗ . | EBV status† . | GeoMx‡ . | Age at diagnosis (y) . | Sex . | Site of biopsy (excised node) . | Stage§ . | Clinical response|| . |
|---|---|---|---|---|---|---|---|---|
| CHL-1 | MCCHL | NEG | 80 | M | Inguinal | 2A | CR | |
| CHL-3 | MCCHL | NEG | 77 | F | Submandibular | NA | CR/relapse | |
| CHL-4 | LRCHL | POS | 75 | F | Neck | 2A | CR | |
| CHL-5 | NSCHL | NEG | 73 | F | Supraclavicular | 4B | CR | |
| CHL-6 | NSCHL | NEG | 73 | M | Neck | 1A | CR | |
| CHL-7 | NSCHL | NEG | 72 | M | Neck | 1A | CR | |
| CHL-9 | NSCHL | NEG | YES | 69 | M | Neck | 1A | PD/salvage CR |
| CHL-10 | NSCHL | NEG | YES | 69 | F | Neck | 3B | CR |
| CHL-11 | CHL-NOS | POS | 68 | F | Neck | 3B | CR/relapse | |
| CHL-12 | CHL-NOS | POS | 68 | F | Axilla | 1A | CR/relapse | |
| CHL-13 | MCCHL | NEG | 68 | M | Inguinal | 2A | CR | |
| CHL-14 | CHL-NOS | NEG | 64 | M | Mediastinum | NA | PD/salvage CR | |
| CHL-15 | MCCHL | POS | 64 | M | Neck | NA | NA | |
| CHL-16 | MCCHL | NEG | 63 | F | Neck | 2A | CR | |
| CHL-18 | MCCHL | POS | 60 | F | Supraclavicular | 4B | CR | |
| CHL-19 | MCCHL | NA | 53 | M | Inguinal | NA | PD | |
| CHL-20 | LRCHL | NA | 46 | M | Axilla | NA | NA | |
| CHL-21 | MCCHL | POS | 45 | M | Neck | 3S | PD | |
| CHL-22 | NSCHL | POS | 40 | M | Neck | 2A | CR/relapse | |
| CHL-23 | NSCHL | POS | 39 | M | Neck | 4A | PR/salvage CR | |
| CHL-24 | NSCHL | NA | 38 | M | Supraclavicular | 4A | PD | |
| CHL-25 | NSCHL | NEG | 33 | F | Neck | 3B | CR | |
| CHL-27 | NSCHL | POS | YES | 32 | M | Supraclavicular | 2A | PD |
| CHL-28 | MCCHL | POS | 32 | M | Neck | 3B | CR | |
| CHL-29 | NSCHL | NA | 32 | M | Neck | 4A | CR/relapse | |
| CHL-30 | MCCHL | POS | 28 | F | Neck | 2A | CR | |
| CHL-31 | NSCHL | NEG | 28 | M | Neck | 4B | CR | |
| CHL-32 | MCCHL | POS | YES | 26 | M | Neck | 2A | CR |
| CHL-33 | NSCHL | POS | 25 | M | Neck | 2B | CR | |
| CHL-34 | NSCHL | NEG | YES | 24 | M | Neck | NA | NA |
| CHL-36 | NSCHL | NEG | 23 | F | Neck | 2A | CR | |
| CHL-39 | MCCHL | NEG | 22 | F | Supraclavicular | NA | NA | |
| CHL-40 | NSCHL | NEG | YES | 20 | M | Supraclavicular | 3BS | CR |
| CHL-41 | NSCHL | NEG | 19 | F | Mediastinum | 2B | PD/salvage | |
| CHL-42 | NSCHL | NEG | 18 | F | Neck | 2B | PD | |
| CHL-43 | NSCHL | NEG | YES | 18 | M | Neck | 2A | CR |
| CHL-44 | NSCHL | POS | 18 | M | Postnasal space | 1A | CR | |
| CHL-45 | NSCHL | NEG | 17 | M | Neck | 2A | CR/relapsed and salvage | |
| CHL-48 | NSCHL | NEG | 16 | F | Axilla | 3A | PD/salvage | |
| CHL-49 | MCCHL | POS | 16 | M | Axilla | 1A | CR/relapsed and salvage | |
| CHL-50 | NSCHL | NEG | 16 | F | Neck | 2A | CR | |
| CHL-51 | NSCHL | POS | 16 | F | Supraclavicular | 3B | CR | |
| CHL-52 | NSCHL | NA | 15 | F | Neck | 2B | CR | |
| CHL-53 | CHL-NOS | NEG | 14 | F | Retroperitoneal | 3B | CR | |
| CHL-54 | NSCHL | NEG | YES | 14 | M | Supraclavicular | 2A | CR |
| CHL-55 | CHL-NOS | NEG | 14 | F | Supraclavicular | 3A | CR | |
| CHL-56 | NSCHL | NEG | 14 | F | Neck | 4A | CR | |
| CHL-57 | NSCHL | NEG | YES | 14 | M | Supraclavicular | 2A | CR |
| CHL-59 | NSCHL | NEG | YES | 13 | M | Neck | 4A | CR |
| CHL-60 | LR-CHL | POS | 12 | M | Neck | 1A | CR | |
| CHL-61 | NSCHL | POS | 12 | M | Neck | 2B | CR | |
| CHL-62 | MCCHL | POS | 8 | M | Hilar | 4A | CR/relapsed and salvage | |
| CHL-63 | CHL-NOS | POS | 7 | M | Neck | 4B | CR | |
| CHL-64 | MCCHL | POS | 6 | F | Neck | 3B | CR |
CR, complete remission or complete metabolic response; F, female; M, male; NA, not available; NEG, negative; PD, progressive disease; POS, positive.
Disease subtypes (n = 54): NSCHL, nodular sclerosis cHL (n = 30, 55.6%); MCCHL, mixed-cell cHL (n = 15, 27.8%); LRCHL, lymphocyte-rich cHL (n = 3, 5.6%); CHL-NOS, cHL, not otherwise specified (n = 6, 11.1%).
EBV status: Epstein-Barr virus (EBV), as assessed by EBV-encoded small RNAs (EBERs).
GeoMx: Tumor biopsies selected for Nanostring GeoMx assay.
Stage: Ann Arbor staging classification of lymphoma.
Clinical response: Best response with first-line treatment/any further event and treatment; CR; PD.
Nanostring spatial transcriptomics
Ten of 54 FFPE cHL tissue biopsies used in the multiplexed immunofluorescence analysis were selected for in situ transcriptomics (Table 2), using the Nanostring GeoMx platform (Nanostring, Seattle, WA). Biopsies were selected based upon year of fixation (2014-2019), high-quality multiplex staining, and successful RNAscope quality control (QC) tests (Advanced Cell Diagnostics Inc, Newark, CA). Five test slides were prepared, each arrayed with 2 unique, whole tissue biopsy sections; 1 slide also included reactive lymph node tissue. For each test slide, 4 consecutive 4-μm-thick sections were taken from each tissue block, prepared per manufacturer’s instructions: slide 1, H&E; slide 2, CD45 and PAX5 fluorescent immunostains; slide 3, CD274 (PD-L1) and PDCD1 (PD-1), RNAscope probes, DNA counterstain; slide 4, oligonucleotide RNA probe hybridization.
Circular regions of interest (ROIs; uniform diameter of 300 μm) were labeled upon the CD274/PDCD1 (PD-L1) RNAscope slides, with reference to H&E and IHC slides for histomorphological context. PD-L1 was used to identify HRSCs, rather than CD30, as fluorescent labeling with CD274 probes was more robust in optimization experiments. In all selected tumors, the HRSC-dense areas of tissue were known to be PD-L1+, and HRSCs were identified by 4′,6-diamidino-2-phenylindole (DAPI) signal and nuclear morphology. For each tumor, 3 ROIs were selected in PD-L1high areas and 3 ROIs in PD-L1low areas of tissue, unless otherwise stated. Assessments of PD-L1 intensity were made by visual inspection of normalized CD274 probe fluorescent signal within each tumor. Sclerotic bands, GCs, and areas of low cellularity were avoided in cHL tissues, with reference to DNA counterstaining and paired IHC. In the control reactive node tissue, 3 ROIs were labeled in germinal centers and 3 ROIs in interfollicular areas. RNA oligonucleotides from the Cancer Transcriptome Atlas (n = 1812 probes), coupled to photocleavable oligonucleotide tags (barcodes), were hybridized overnight. Hybridized barcodes were released from tissue by UV exposure from each ROI sequentially, before being counted and sequenced on the Illumina HiSeq 2500 platform.
Nanostring data analysis
QC excluded outlier probes, and data were normalized against the 75th percentile of signal from each tumor. After QC, 1372 (75.7%) probes were retained. The nanostring expression matrix (ROI by gene) was analyzed by principal component analysis using Scanpy, before constructing a shared–nearest neighbor graph representation. We identified k = 10 nearest neighbors in principal component analysis space using the FNN R package, before calculating the Jaccard distance between neighbor vectors per the approach of Levine et al.26 The graph was clustered using the Leiden algorithm27 with default settings.
Deconvolution was performed by first generating a matrix of 1-hot-encodings of marker genes per cell type, including HRSCs. We then used the rlm function in the MASS R package to perform robust linear regression. The resulting coefficients were treated as fractions to derive counts by multiplying by the number of nuclei in each ROI. These cell count estimates were then used to calculate differential abundance estimates for each cluster using DESeq2.28 Differential expression analysis between clusters was performed using limma (version 3.48.1).21 The top 10 differentially expressed genes per cluster were selected for plotting (supplemental Figure 4C).
Multiplexed immunofluorescence
Multiplexed immunofluorescence (IF) was performed by sequentially immunostaining 4-μm-thick tissue sections from selected FFPE biopsies, with primary antibodies, secondary reagents, and unique fluorochromes. Each of the 2 multiplexes comprised 6 primary antibodies, followed by a nuclear counterstain, 4',6-diamidino-2-phenylindole, as per published protocols.7,29,30 All immunostaining was performed on the automated Ventana Benchmark platform (Roche Diagnostics, Rotkreuz, Switzerland). Slides were air dried, mounted with Prolong Diamond antifade mounting medium (#P36965; Life Technologies), and stored at 4°C in a light-proof box until image acquisition. Target antigens, antibody clones, and dilutions for markers are listed in Table 3.
Antibodies and secondary reagents used for multiplex immunofluorescence panels
| . | Primary antibody . | Primary antibody dilution . | Clone . | Manufacturer . | Secondary antibody (Ventana) . | TSA-conjugated fluorochrome . |
|---|---|---|---|---|---|---|
| 1 | CD11c | 1:200 | 5D11 | Leica | Omnimap | Opal-520 |
| 2 | CD30 | 1:25 | BerH2 | Cell Marque | Ultramap | Opal-540 |
| 3 | CD68P∗ | 1:1500 | PGM1 | Dako | Omnimap | Opal-570 |
| 4 | IDO1∗ | 1:4000 | V1NC3IDO | Invitrogen | Omnimap | Opal-620 |
| 5 | PD-L1∗ | Predilute | SP263 | Ventana | Optiview | Opal-650 |
| 6 | TIM-3∗ | 1:200 | D5D5R | Cell Signaling Technology | Omnimap | Opal-690 |
| 7 | CADM1† | 1:10,000 | Polyclonal | Sigma-Aldrich | Omnimap | Opal-570 |
| 8 | CD1C† | 1:100 | OTI2F4 | Abcam | Omnimap | Opal-620 |
| 9 | LAMP3† | 1:100 | 1010E1 | Dendritics | Ultramap | Opal-650 |
| 10 | CD123† | 1:200 | BR4MS | Leica | Optiview | Opal-690 |
| . | Primary antibody . | Primary antibody dilution . | Clone . | Manufacturer . | Secondary antibody (Ventana) . | TSA-conjugated fluorochrome . |
|---|---|---|---|---|---|---|
| 1 | CD11c | 1:200 | 5D11 | Leica | Omnimap | Opal-520 |
| 2 | CD30 | 1:25 | BerH2 | Cell Marque | Ultramap | Opal-540 |
| 3 | CD68P∗ | 1:1500 | PGM1 | Dako | Omnimap | Opal-570 |
| 4 | IDO1∗ | 1:4000 | V1NC3IDO | Invitrogen | Omnimap | Opal-620 |
| 5 | PD-L1∗ | Predilute | SP263 | Ventana | Optiview | Opal-650 |
| 6 | TIM-3∗ | 1:200 | D5D5R | Cell Signaling Technology | Omnimap | Opal-690 |
| 7 | CADM1† | 1:10,000 | Polyclonal | Sigma-Aldrich | Omnimap | Opal-570 |
| 8 | CD1C† | 1:100 | OTI2F4 | Abcam | Omnimap | Opal-620 |
| 9 | LAMP3† | 1:100 | 1010E1 | Dendritics | Ultramap | Opal-650 |
| 10 | CD123† | 1:200 | BR4MS | Leica | Optiview | Opal-690 |
TSA, tyramide signal amplification.
CD11c and CD30 are used in both multiplexes. Other antibodies are annotated, as below:
Multiplex 1 only.
Multiplex 2 only.
Image acquisition
Test regions for multiplex IF analysis were initially identified for each tumor biopsy (n = 54) by reviewing H&E and immunofluorescence tissue sections (original magnification ×4). Three nonoverlapping ROIs were then acquired at original magnification ×20 for each multiplex panel. Each of these 3 test regions comprised 9 contiguous fields of view (FOVs), arranged as a 3 × 3 rectangular grid (measuring 2008 μm × 1508 μm in total). In total, there were 27 FOVs per tumor, for each multiplex.
Regions were selected to exclude tissue artifacts, best represent the overall tissue, and to include CD30+ HRSCs. Regions were imaged and deconvoluted using the Vectra multispectral imaging platform (Vectra 3, Inform 2.4.8, Akoya Biosciences, Marlborough, MA), using specific spectral libraries.
Results
Single-cell transcriptomic profiling of lymphoma-affected and unaffected lymph nodes
To overview the cellular ecosystem of cHL, we compiled a census of 243 753 single-cell transcriptomes from lymphoma-affected and unaffected lymph nodes. We sourced data from Aoki et al10 incorporating droplet encapsulation single-cell transcriptomes (10× Genomics) from reactive and cHL nodes (Figure 1A). In addition, we performed scRNA-seq (10× Genomics) with cell suspensions from lymph nodes from deceased organ donors, and 2 lymphoma lymph nodes, 1 with nodular sclerosis cHL (NSCHL), and 1 with nodular lymphocyte–predominant Hodgkin lymphoma/nodular lymphocyte–predominant B-cell lymphoma (NLPHL/NLPBL), a biologically distinct and recently reclassified subtype.31 The combined data set comprises tissue from 13 donors who did not have lymphoma (8 deceased donors, 5 donors with reactive lymph node hyperplasia), 1 NLPHL/NLPBL, and 23 cHL lymph nodes (Figure 1A; Table 4).
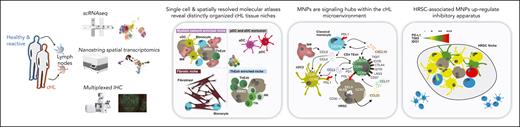
Study design and transcriptional profiling of immune and neoplastic cells in the cHL microenvironment. (A) Outline of the scRNA-seq data set. Two data sets (Aoki et al10 and “this study”/WSI data set) were merged, and contain data from lymph nodes unaffected by lymphoma (reactive lymph nodes and nondiseased lymph nodes from deceased donors) highlighted in blue, and lymphoma-infiltrated lymph nodes highlighted in red. The numbers of cells are given in dark gray, and the number of donors in light gray. (B) UMAP plot of 243 753 cells from an integrated scRNA-seq data set, colored by cell type and organized by compartment (T cells, B cells, innate lymphocytes, myeloid, and stroma). (C) UMAP plot of 22 573 graph neighborhoods, colored by the differential abundance (log fold change) in lymphoma-affected or nonlymphoma-affected (deceased-donor lymph nodes or reactive lymph node) samples. Dot size is proportional to neighborhood size (median neighborhood size = 50 cells). (D) Volcano plot showing differentially expressed genes between microdissected GCs and HRSCs. Significant genes for HRSCs are indicated (|LFC| > 3; adjusted P value <.01). (E) Heatmap showing transcription factor (TF) regulon normalized enrichment scores for GCs vs HRSCs calculated with DoRothEA. (F) Graph indicating predicted interactions between genes (red, transcription factors; gray, other genes) differentially expressed in HRSCs. Edges drawn where an interaction is documented by OmniPath. (G) UMAP plot of 2727 myeloid cells colored by cell type. (H) Heatmap showing mean normalized expression levels (color) and fraction of cells expressing (dot size) markers of myeloid cell subsets. (I) Heatmap showing the proportion of each cell type derived from lymphoma and nonlymphoma samples.
Study design and transcriptional profiling of immune and neoplastic cells in the cHL microenvironment. (A) Outline of the scRNA-seq data set. Two data sets (Aoki et al10 and “this study”/WSI data set) were merged, and contain data from lymph nodes unaffected by lymphoma (reactive lymph nodes and nondiseased lymph nodes from deceased donors) highlighted in blue, and lymphoma-infiltrated lymph nodes highlighted in red. The numbers of cells are given in dark gray, and the number of donors in light gray. (B) UMAP plot of 243 753 cells from an integrated scRNA-seq data set, colored by cell type and organized by compartment (T cells, B cells, innate lymphocytes, myeloid, and stroma). (C) UMAP plot of 22 573 graph neighborhoods, colored by the differential abundance (log fold change) in lymphoma-affected or nonlymphoma-affected (deceased-donor lymph nodes or reactive lymph node) samples. Dot size is proportional to neighborhood size (median neighborhood size = 50 cells). (D) Volcano plot showing differentially expressed genes between microdissected GCs and HRSCs. Significant genes for HRSCs are indicated (|LFC| > 3; adjusted P value <.01). (E) Heatmap showing transcription factor (TF) regulon normalized enrichment scores for GCs vs HRSCs calculated with DoRothEA. (F) Graph indicating predicted interactions between genes (red, transcription factors; gray, other genes) differentially expressed in HRSCs. Edges drawn where an interaction is documented by OmniPath. (G) UMAP plot of 2727 myeloid cells colored by cell type. (H) Heatmap showing mean normalized expression levels (color) and fraction of cells expressing (dot size) markers of myeloid cell subsets. (I) Heatmap showing the proportion of each cell type derived from lymphoma and nonlymphoma samples.
Outline of the merged scRNA-seq data set
| . | Histology . | Number of cases . | Cell number . | Data source . |
|---|---|---|---|---|
| Hodgkin lymphoma | NSCHL | 13 | 59 885 | This study and Aoki et al10 |
| MCCHL | 9 | 38 046 | Aoki et al10 | |
| LRCHL | 1 | 4 683 | Aoki et al10 | |
| NLPHL/NLPBL | 1 | 21 862 | This study | |
| Non-Hodgkin lymphoma | Healthy | 8 | 79 823 | This study |
| Reactive | 5 | 23 187 | Aoki et al10 |
| . | Histology . | Number of cases . | Cell number . | Data source . |
|---|---|---|---|---|
| Hodgkin lymphoma | NSCHL | 13 | 59 885 | This study and Aoki et al10 |
| MCCHL | 9 | 38 046 | Aoki et al10 | |
| LRCHL | 1 | 4 683 | Aoki et al10 | |
| NLPHL/NLPBL | 1 | 21 862 | This study | |
| Non-Hodgkin lymphoma | Healthy | 8 | 79 823 | This study |
| Reactive | 5 | 23 187 | Aoki et al10 |
Histological subtypes of HL: NSCHL, nodular sclerosis cHL; MCCHL, mixed-cell cHL; LRCHL, lymphocyte-rich cHL; NLPHL/NLPBL, nodular lymphocyte–predominant HL/nodular lymphocyte–predominant B-cell lymphoma.
After QC (supplemental Methods; supplemental Figure 1A-B), we performed data set integration and dimensionality reduction using single-cell variational inference13 and annotated cell types on the basis of marker genes and external data set validation (Figure 1B; supplemental Figure 1C-F).
The cellular ecosystem of cHL encompassed subsets of CD4+ and CD8+ T cells, including regulatory T cells (Treg), T-follicular helper cells (Tfh), and exhausted CD4+ (ThExh) and CD8+ T cells (CD8 TExh) (Figure 1B). ThExh matched the recently identified LAG3+ subset, and expressed checkpoint molecules and exhaustion markers including CD27, TNFRSF18, LAG3, and ICOS10,32 (supplemental Figure 1G). The B-cell compartment split into 2 clusters of memory and naive subsets, in addition to plasmablasts, and GC B cells (Figure 1B; supplemental Figure 1C-F).
The inclusion of nonlymphoma lymph nodes, including healthy, nonreactive tissue, allowed us to robustly test the differential abundance of cells between lymphoma and nonlymphoma, using a neighborhood partitioning approach (MiloR, see “Methods”).18 Lymphoma was enriched for cytotoxic lymphocyte subsets including effector memory CD8+ T cells and natural killer (NK) cells, in addition to CD4+ T-cell subsets expressing checkpoint apparatus, including ThExh and Tfh cells (Figure 1C; supplemental Figure 2B). Within the lymphoma samples, cHL was enriched for exhausted T-cell subsets, in contrast with the NLPHL/NLPBL sample, which was dominated by cytotoxic subsets (NK cells, effector memory CD8+ T cells, and CD8 central memory T cells) (supplemental Figure 2C).
HRSCs secrete chemokines to attract and retain exhausted T cells
We did not detect HRSCs, likely because of the rarity, size, and fragility of these cells; this was confirmed by assessing for canonical copy number alterations (supplemental Fig 3A-C). To probe their transcriptional programs, which potentially orchestrate this immunoregulatory milieu, we leveraged a microarray data set profiling microdissected HRSCs and GCs (see “Methods”).33 Differential expression between HRSCs and GCs identified an HRSC gene signature, including TNFRSF8 (CD30) (Figure 1D). Scoring of transcription factor regulons demonstrated nuclear factor κB (NF-κB) activation (NFKB1 and its activatory heterodimer partner RELA) in HRSCs (Figure 1E), consistent with previous reports.34 Next, we performed in silico identification of interactions between active transcription factors and potential targets within the HRSC gene set (see “Methods”). This demonstrated an NF-κB–centric network coordinating the upregulation of the chemokines CCL5, CCL17, and CCL22 capable of the positioning and retention of ThExh via CCR5 and CCR4 ligation (Figure 1F; supplemental Figure 1G).
Myeloid cells comprise a diverse repertoire of monocytes, macrophages, and DCs
Within the myeloid compartment, we identified macrophages coexpressing CD14, CD68, and resident macrophage markers FOLR2 and MRC1 (Figure 1G-H). These cells were transcriptionally distinct from classical monocytes, which expressed a characteristic signature including S100A9, CD14, VCAN, and FCN135 (Figure 1G-H). Among DCs, we identified cDC1 (key transcripts: CLEC9A, CADM1, and IDO1) and cDC2 (key transcripts: CD1C, CLEC10A, and FCER1A) (Figure 1G-H). We also identified LAMP3+ DCs in both healthy and some lymphoma samples, expressing the chemokine receptor CCR7 and the chemokines CCL17 and CCL19, which we termed “activated DC” (aDC), consistent with the nomenclature of transcriptionally similar cells described in human thymus and spleen,36,37 and in murine lung neoplasms38 (Figure 1G-H; supplemental Figure 1E). We found a population of plasmacytoid DCs (pDC) with a dominant contribution from lymphoma samples (Figure 1I; supplemental Figure 2A), expressing IL3RA (CD123) and CLEC4C (Figure 1G-H).
Within the stromal cell compartment, we identified fibroblasts (key transcripts: LUM and DCN), and endothelial cells (key transcripts: CLDN5, PECAM1, and PLVAP [fenestrated endothelium]) (Figure 1B; supplemental Figure 1C).
We used this lymph node–wide account of the immune landscape of cHL as a reference to interrogate the cellular composition of cHL tissues, focusing on myeloid cells.
Spatial and bulk transcriptomics identifies microenvironments with divergent cell enrichments and prognostic significance
We first identified spatially distinct microenvironments using targeted spatial transcriptomic profiling (Nanostring GeoMx Cancer Transcriptome Atlas), defining 300-μm-diameter ROIs in both PD-L1high and PD-L1low regions of 9 NSCHL and 1 mixed-cell cHL (MCCHL) lymph nodes and follicular and interfollicular regions of 1 control reactive lymph node (supplemental Figure 4A-B). We represented these transcriptional profiles in a shared–nearest neighbor graph and identified 5 clusters (Figure 2A-B). We then deconvoluted the cell composition of each cluster using our scRNA-seq reference, followed by cell-type count estimation and differential cell-type abundance analysis (Figure 2C, see “Methods”). The clusters encompassed 2 “neoplastic” PD-L1high HRSC-enriched clusters (clusters 3 and 4), 2 “nonneoplastic” PD-L1low HRSC-depleted clusters (clusters 1 and 5), and 1 intermediate neighborhood (cluster 2).
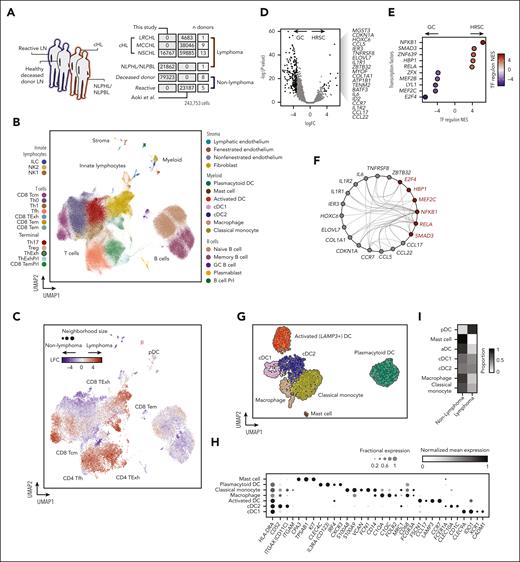
Heterogeneous immune cell networks occupy distinct tissue niches with divergent mononuclear phagocyte enrichment patterns and prognostic significance. (A) Shared–nearest neighbor graph embedding of transcriptional profiles of nanostring ROIs from reactive and cHL lymph nodes. Color indicates clusters identified using Leiden clustering with resolution = 1. (B) Shared–nearest neighbor graph embedding of transcriptional profiles of nanostring ROIs colored by PD-L1 expression status. (C) Differential abundance estimates of cell-types deconvolved from nanostring ROIs between clusters shown in (a), LFC estimates represent differential abundance of cell types in each cluster vs all other clusters. (MΦ, macrophage). (D) Upper panel: heatmap showing WGCNA module coexpression patterns for 6 modules (modules A-F) across 130 cHL gene expression profiles (microarray) split by treatment outcome. Lower panel: heatmap showing results for a hypergeometric enrichment test between module gene membership and cell type markers from the scRNA-seq data set for WGCNA modules (modules A-F). (MΦ, macrophage). (E) Representative micrographs from multiplexed IF imaging of cHL samples. One of 3 test regions from tumor CHL-27 is shown, cropped to 1662 μm by 1076 μm; CD11c (cyan), CD30 (orange), CADM1 (green), CD1c (yellow), LAMP3 (magenta), and CD123 (red) fluorescent signals are represented by unique pseudocolors. To improve clarity, DAPI is not shown. Selected areas are highlighted, each at original magnification ×100 (160 μm × 108 μm); CD30+ HRSC-dense area (region 1, inset left), internodular area with no CD30+ HRSCs (region 2, inset right), and area with CADM1+ cDC1 (region 3, inset top right). Individual pseudocolors are shown below the main image (second & third lines), with DAPI (blue) to identify cell nuclei. Each individual image refers to a multiplexed area above (inset), indicated by the corresponding number (top right). (F) HRSC and DC map, corresponding to the same region from panel E. Phenotyped cells are identified by colored nuclei, with each cell type represented by a different color; CD11c+ ONLY (CD11c+ CD1c−, cyan), CD30+ HRSC (light orange), cDC1 (green), cDC2 (yellow), LAMP3+ aDC (magenta), and pDCs (red) are shown. Cells with no assigned phenotype (“null”) and CD1c+ ONLY cells are excluded from this visualization. The “HRSC neighborhood” is shown as circles surrounding each CD30+ HRSC (dark orange). A selected area is highlighted, at original magnification ×200 (80 μm × 54 μm; inset, center). (G) Isobar plots show the location and density of each cell phenotype of interest, for the corresponding tumor region from panels E and F (note the uncropped 2008 μm by 1502 μm region is shown here). HRSCs (orange dots, left) and DC subsets (colors corresponding to each phenotype, right) are shown. (H) Summarized interaction plot across all study tumors in aggregate, displaying the ratio of the nearest neighbor distance between phenotype pairs (index cell type [gray] to test cell type [black]) compared with the expected baseline distance. Red indicates cell type cooccurrence, blue indicates cell type exclusion.
Heterogeneous immune cell networks occupy distinct tissue niches with divergent mononuclear phagocyte enrichment patterns and prognostic significance. (A) Shared–nearest neighbor graph embedding of transcriptional profiles of nanostring ROIs from reactive and cHL lymph nodes. Color indicates clusters identified using Leiden clustering with resolution = 1. (B) Shared–nearest neighbor graph embedding of transcriptional profiles of nanostring ROIs colored by PD-L1 expression status. (C) Differential abundance estimates of cell-types deconvolved from nanostring ROIs between clusters shown in (a), LFC estimates represent differential abundance of cell types in each cluster vs all other clusters. (MΦ, macrophage). (D) Upper panel: heatmap showing WGCNA module coexpression patterns for 6 modules (modules A-F) across 130 cHL gene expression profiles (microarray) split by treatment outcome. Lower panel: heatmap showing results for a hypergeometric enrichment test between module gene membership and cell type markers from the scRNA-seq data set for WGCNA modules (modules A-F). (MΦ, macrophage). (E) Representative micrographs from multiplexed IF imaging of cHL samples. One of 3 test regions from tumor CHL-27 is shown, cropped to 1662 μm by 1076 μm; CD11c (cyan), CD30 (orange), CADM1 (green), CD1c (yellow), LAMP3 (magenta), and CD123 (red) fluorescent signals are represented by unique pseudocolors. To improve clarity, DAPI is not shown. Selected areas are highlighted, each at original magnification ×100 (160 μm × 108 μm); CD30+ HRSC-dense area (region 1, inset left), internodular area with no CD30+ HRSCs (region 2, inset right), and area with CADM1+ cDC1 (region 3, inset top right). Individual pseudocolors are shown below the main image (second & third lines), with DAPI (blue) to identify cell nuclei. Each individual image refers to a multiplexed area above (inset), indicated by the corresponding number (top right). (F) HRSC and DC map, corresponding to the same region from panel E. Phenotyped cells are identified by colored nuclei, with each cell type represented by a different color; CD11c+ ONLY (CD11c+ CD1c−, cyan), CD30+ HRSC (light orange), cDC1 (green), cDC2 (yellow), LAMP3+ aDC (magenta), and pDCs (red) are shown. Cells with no assigned phenotype (“null”) and CD1c+ ONLY cells are excluded from this visualization. The “HRSC neighborhood” is shown as circles surrounding each CD30+ HRSC (dark orange). A selected area is highlighted, at original magnification ×200 (80 μm × 54 μm; inset, center). (G) Isobar plots show the location and density of each cell phenotype of interest, for the corresponding tumor region from panels E and F (note the uncropped 2008 μm by 1502 μm region is shown here). HRSCs (orange dots, left) and DC subsets (colors corresponding to each phenotype, right) are shown. (H) Summarized interaction plot across all study tumors in aggregate, displaying the ratio of the nearest neighbor distance between phenotype pairs (index cell type [gray] to test cell type [black]) compared with the expected baseline distance. Red indicates cell type cooccurrence, blue indicates cell type exclusion.
Clusters 1 and 5 were PD-L1low and represented fibrotic regions and healthy follicular microenvironments, respectively. Cluster 1 was enriched for stromal (fibroblast and endothelial) cells, classical monocytes, and pDCs but was depleted of cDCs (Figure 2C). Differentially expressed genes in this cluster included genes encoding signaling mediators of fibrosis FGFR4, TGFB2, and PTCH1 (supplemental Figure 4C). In contrast, cluster 5 was exclusively derived from control samples and enriched for GC B cells, Tfh, and plasmablasts (Figure 2A-C).
The PD-L1high neoplastic clusters exhibited divergent leukocyte enrichment. Cluster 3 was enriched for ThExh, Th, and NK cells. In contrast, cluster 4 represented a myeloid-enriched niche, characterized classical monocyte, macrophage, and cDC2 infiltration (Figure 2C). Marker genes for cluster 4 highlighted inflammatory signaling, with upregulation of chemokines associated with Th2 responses and CCR3-dependent eosinophil recruitment (CCL18, CCL13, CCL24, CCL26, and CCL23) and granulocyte-attracting chemokines CXCL1 and CXCL6 (supplemental Figure 4C).
Next, we sought to identify transcriptional correlates of these microenvironments in publicly available gene expression data. We performed weighted correlation network analysis (WGCNA) using data from 130 diagnostic cHL lymph nodes.22,39 This analysis yielded 6 modules of coexpressed genes (modules A-F) (Figure 2D). We performed enrichment analysis using both scRNA-seq references and Gene Ontology terms to annotate these modules and found that whereas module C corresponds to the cell cycle program (supplemental Figure 4D), the other modules strikingly mirror the tissue niche patterns identified in our spatially resolved (Nanostring) transcriptomics data (Figure 2D), suggesting a highly conserved organization of pathological niches in cHL.
Module A enriched for stromal cell signatures, similarly to PD-L1low nanostring cluster 1. In contrast, module B represented myeloid-skewed inflammation, enriching for monocyte, macrophage, and cDC2 signatures, aligning to nanostring cluster 4 (Figure 2D).
We then interrogated the relationship between module gene expression and treatment outcomes. High expression of the fibrosis/stroma enriched module was associated with treatment success, whereas high expression of the myeloid enriched module was associated with early treatment failure. Expression of module F (B-cell follicle) was not associated with differences in outcome (Figure 2D).
Identification and quantification of specific MNP subtypes in cHL biopsies
Given that variation in microenvironmental myeloid infiltration appears to be a conserved and prognostically important feature of cHL pathobiology, we sought to define MNPs within the HRSC niche at single-cell resolution. Using marker gene coexpression patterns in the scRNA-seq data (supplemental Figure 4E), we designed multiplexed immunofluorescence panels to identify MNPs in cHL tissue sections (n = 54 tumors, representative images: Figure 2E-G). We identified CADM1+/CD11c+ cDC1, CD1c+/CD11c+ cDC2, LAMP3+ aDCs with distinctive dendritic morphology, CD123+ pDC, CD11c+ monocytes and macrophages (CD11c+ ONLY), and CD30+ HRSCs (Figure 2E). We then phenotyped segmented cells and performed neighborhood analyses, taking a 25-μm-radius neighborhood around each CD30+ HRSC and measuring the relative enrichment of MNP subsets in aggregated neighborhoods, compared with “nonneighborhood” regions (Figure 2F). This revealed enrichment of cDC2 and CD11c+ monocytes in the immediate vicinity of HRSCs. In contrast, both pDC and aDC were excluded from the HRSC niche and occupy regions with a low density of CD11c+ cells (Figure 2E-H; Figure 3A-H).
pDCs and aDCs are excluded from the HRSc microenvironment but immunosurveillant cDC1s and cDC2s are enriched. (A) In total, 54 unique tumors were examined by multiplexed, immunofluorescence image analysis. A representative region from CHL-13 is shown, which measures 2008 μm (x-axis) by 1502 μm (y-axis). The centroid of each cell is identified with a colored circle. The CD30+ HRSCs are indicated by a magenta circle. The cell nuclei, which lie within a 10-to-25-μm radius from each HRSC centroid (HRSC neighborhood) are indicated by orange circles and those nuclei beyond the 25-μm areas (nonneighborhood) are indicated by blue circles. Blood vessels/other endothelial structures are indicated (white) and all cells encompassed within these areas are excluded from analysis. (B-D) Spatial distribution of DC subtypes that cooccur with HRSCs. For each phenotype a representative tumor region is identified and shown in all plots: (B) cDC2 (CD11c+ CD1c+) = represented by CHL-13. (C) CD11c+ ONLY cells = CHL-43. (D) cDC1 = CHL-31. (E-F) Spatial distribution of DC subtypes that are “excluded” from HRSCs. For each phenotype a representative tumor region is identified and shown in all plots: (E) pDC = CHL-11. (F) aDC / LAMP3+ = CHL-13. A cropped, illustrative area from each tumor region (measuring 1004 μm by 751 μm, left) is shown as a cell map; phenotyped cells are identified by colored nuclei, with each cell type represented by a different color. CD11c+ ONLY (cyan), CD30+ HRSCs (light orange), cDC1 (green), cDC2 (yellow), LAMP3+ aDCs (magenta), and pDCs (red) are shown. Cells with no assigned phenotype (null) and CD1c+ ONLY cells are excluded from this visualization. The HRSC neighborhood is shown as circles surrounding each CD30+ HRSC (dark orange), each circle extending to a radius of 25 μm around each HRSC. Topological maps of the corresponding full ROI (2008 μm by 1502 μm) of HRSCs (orange, upper right), immune cells of interest (colored based on cell phenotype, lower right). Representative immunofluorescence images (original magnification ×80), showing cells of interest (second line). CD11c (cyan), CD30 (orange), CADM1 (green), CD1c (yellow), LAMP3 (magenta), and CD123 (red) fluorescent signals are represented by unique pseudocolors. To improve visualization, certain signals may not be shown in each particular image (these excluded colors are shown in gray in each respective key). (G) Violin plots comparing the observed cell proportions for each tumor sample (n = 54) for the CD30+ HRSC neighborhood (orange) and across the entire examined tissue area (nonneighborhood; blue), for each DC subtype. Statistical significance: QQ plots showed that the differences in cell proportions between the CD30 neighborhood and the entire sample were not normally distributed, therefore a Wilcoxon signed-rank test was used to assess statistical significance for each phenotype. (H) Histograms showing the distance from each HRSC to its nearest neighbor of an immune-phenotype (n = 54). Distance is shown as a ratio to the expected distance if the immune cell was distributed randomly within the tissue section, with values less than 1 (left of dotted line) indicating that cells cooccur with HRSCs and values >1 (right of dotted line) indicating that cells are excluded from HRSC.s
pDCs and aDCs are excluded from the HRSc microenvironment but immunosurveillant cDC1s and cDC2s are enriched. (A) In total, 54 unique tumors were examined by multiplexed, immunofluorescence image analysis. A representative region from CHL-13 is shown, which measures 2008 μm (x-axis) by 1502 μm (y-axis). The centroid of each cell is identified with a colored circle. The CD30+ HRSCs are indicated by a magenta circle. The cell nuclei, which lie within a 10-to-25-μm radius from each HRSC centroid (HRSC neighborhood) are indicated by orange circles and those nuclei beyond the 25-μm areas (nonneighborhood) are indicated by blue circles. Blood vessels/other endothelial structures are indicated (white) and all cells encompassed within these areas are excluded from analysis. (B-D) Spatial distribution of DC subtypes that cooccur with HRSCs. For each phenotype a representative tumor region is identified and shown in all plots: (B) cDC2 (CD11c+ CD1c+) = represented by CHL-13. (C) CD11c+ ONLY cells = CHL-43. (D) cDC1 = CHL-31. (E-F) Spatial distribution of DC subtypes that are “excluded” from HRSCs. For each phenotype a representative tumor region is identified and shown in all plots: (E) pDC = CHL-11. (F) aDC / LAMP3+ = CHL-13. A cropped, illustrative area from each tumor region (measuring 1004 μm by 751 μm, left) is shown as a cell map; phenotyped cells are identified by colored nuclei, with each cell type represented by a different color. CD11c+ ONLY (cyan), CD30+ HRSCs (light orange), cDC1 (green), cDC2 (yellow), LAMP3+ aDCs (magenta), and pDCs (red) are shown. Cells with no assigned phenotype (null) and CD1c+ ONLY cells are excluded from this visualization. The HRSC neighborhood is shown as circles surrounding each CD30+ HRSC (dark orange), each circle extending to a radius of 25 μm around each HRSC. Topological maps of the corresponding full ROI (2008 μm by 1502 μm) of HRSCs (orange, upper right), immune cells of interest (colored based on cell phenotype, lower right). Representative immunofluorescence images (original magnification ×80), showing cells of interest (second line). CD11c (cyan), CD30 (orange), CADM1 (green), CD1c (yellow), LAMP3 (magenta), and CD123 (red) fluorescent signals are represented by unique pseudocolors. To improve visualization, certain signals may not be shown in each particular image (these excluded colors are shown in gray in each respective key). (G) Violin plots comparing the observed cell proportions for each tumor sample (n = 54) for the CD30+ HRSC neighborhood (orange) and across the entire examined tissue area (nonneighborhood; blue), for each DC subtype. Statistical significance: QQ plots showed that the differences in cell proportions between the CD30 neighborhood and the entire sample were not normally distributed, therefore a Wilcoxon signed-rank test was used to assess statistical significance for each phenotype. (H) Histograms showing the distance from each HRSC to its nearest neighbor of an immune-phenotype (n = 54). Distance is shown as a ratio to the expected distance if the immune cell was distributed randomly within the tissue section, with values less than 1 (left of dotted line) indicating that cells cooccur with HRSCs and values >1 (right of dotted line) indicating that cells are excluded from HRSC.s
Ligand-receptor interactions between MNPs, T cells, and HRSCs
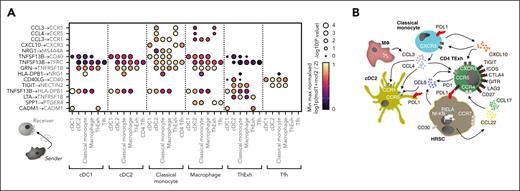
Next, we investigated which signals might coordinate the positioning of MNP subsets and T cells found in close association with HRSCs. To interrogate ligand-receptor interactions (LRIs), we calculated the statistical enrichment of candidate LRIs between MNPs and T- cells in our scRNA-seq data, using the CellPhoneDB tool.19 This analysis predicted paracrine CCL3 and CCL4 signaling by macrophages and classical monocytes to CCR5- and CCR1-expressing cDC2 and ThExh (Figure 4A). Furthermore, classical monocyte–derived CXCL10 was predicted to signal to cDC1, ThExh, and Tfh via CXCR3. Nominated inhibitory interactions from ThExh included TIGIT signaling via NECTIN2 expressed by cDC2, classical monocytes, and macrophages (Figure 4A). Analysis of putative LRIs inferred from Nanostring cluster 4 and WGCNA module B also highlighted CCR1- and CCL5-mediated signaling (supplemental Figure 5A). A proposed schematic illustrates potential interactions between HRSCs, cDC2s, classical monocytes, and TExh (Figure 4B); NF-κB–driven expression of CCL22, CCL17, and CCL5 by HRSCs coordinates recruitment of TExh and cDCs; the niche is further coordinated by CCL3, CCL4, and CXCL10 signaling from monocytes and macrophages.
Reciprocal mononuclear phagocyte chemokine signaling shapes the immunosuppressive microenvironment in cHL. (A) Heatmap of reciprocal ligand-receptor interactions between myeloid subsets and T cells implicated in the cHL TME. Point size indicates permutation P value (CellPhoneDB). Color indicates the min-max normalized aggregate mean expression level of ligand and receptor. (B) Model for intercellular interactions between MNP, dysfunctional T cells, and HRSCs in the cHL TME. HRSCs produce CCL22, CCL17, and CCL5 downstream of NF-κB activity, orchestrating T-cell recruitment via indicated ligand-receptor interactions. CCL3, CCL4, and CXCL10 produced by monocytes and macrophages direct positioning of dysfunctional T cells and cDC2s. Widespread inhibitory molecule expression is seen on HRSCs, MNPs, and T cells.
Reciprocal mononuclear phagocyte chemokine signaling shapes the immunosuppressive microenvironment in cHL. (A) Heatmap of reciprocal ligand-receptor interactions between myeloid subsets and T cells implicated in the cHL TME. Point size indicates permutation P value (CellPhoneDB). Color indicates the min-max normalized aggregate mean expression level of ligand and receptor. (B) Model for intercellular interactions between MNP, dysfunctional T cells, and HRSCs in the cHL TME. HRSCs produce CCL22, CCL17, and CCL5 downstream of NF-κB activity, orchestrating T-cell recruitment via indicated ligand-receptor interactions. CCL3, CCL4, and CXCL10 produced by monocytes and macrophages direct positioning of dysfunctional T cells and cDC2s. Widespread inhibitory molecule expression is seen on HRSCs, MNPs, and T cells.
Coexpression of inhibitory proteins by MNPs in cHL biopsies
We were surprised that the HRSC-associated MNP network includes cDC2 and occasional cDC1, which play key roles in immunosurveillance. Although macrophage expression of PD-L1 is established, upregulation of inhibitory ligands and receptors by DCs is less well described in cHL. To test this, we designed a second IF panel to examine expression of PD-L1, IDO1, and TIM-3 by CD11c+/CD68− cDC1 and cDC2, CD11c+/CD68+ monocytes, and CD11c−/CD68+ macrophages, and to assess their spatial relationships to CD30+ HRSC (Table 3).
HRSCs exhibited extensive PD-L1 expression, as expected. We also found enrichment of PD-L1+ CD11c+/CD68+ and CD11c+/CD68− MNPs in CD30+ HRSC neighborhoods (Figure 5A-D). Similarly, we found variable coexpression of the coinhibitory receptor TIM-3, and IDO1, an immunomodulating tryptophan catabolizing enzyme, on CD11c+/CD68+ and CD11c+/CD68− MNPs in close proximity to HRSCs (Figure 5A-E; supplemental Figure 6). MNPs expressing ≥1 inhibitory molecules are enriched around HRSCs and associated with other such MNPs; CD68+ CD11c− MNPs are distributed evenly across the tissue, with no enrichment around HRSCs (Figure 5D-E).
Mononuclear phagocytes expressing ≥1 immunosuppressive molecules are enriched around HRSCs in cHL. (A) Representative micrographs from multiplexed IF imaging of cHL samples, showing expression of inhibitory molecules (IDO1, PD-L1, and TIM-3), cell type–defining markers for MNPs (CD11c & CD68), and HRSCs (CD30). One of 3 test regions from CHL-7 is shown (2008 μm × 1502 μm), with CD11c (cyan), CD30 (orange), CD68 (magenta), PD-L1 (green), IDO1 (yellow), and TIM-3 (red) fluorescent signal represented by unique pseudocolors (to improve clarity DAPI is not shown). Inset 1 (original magnification ×100 [160 μm × 108 μm]; left) shows CD30 and IDO1 (top), CD30 and PD-L1 (middle), and CD30 and TIM-3 (bottom), with DAPI (blue) to identify cell nuclei. Inset 2 (original magnification ×100 [160 μm × 108 μm]; right) shows representative CD30+ HRSCs, surrounded by mononuclear phagocytes in closer detail. Individual pseudocolors from area 2, corresponding to each antibody, are shown below the main image (second and third lines). (B) Expression of PD-L1, IDO1, and TIM-3 by MNPs. Representative images at original magnification ×200 (54 μm × 54 μm) show PD-L1 expression (green, top line) and corresponding CD11c (cyan) and CD68 (magenta, second line) for the same areas. Images are separated to improve clarity, as colocalized. IDO1 expression (yellow, cytoplasmic localization) with CD11c and CD68 (third line). TIM-3 expression (red, cytoplasmic localization) with CD11c and CD68 (bottom line). (C) Proportion of mononuclear phagocytes expressing immuno-regulatory molecules. Left panel: lower key represents the combinations of inhibitory molecules (black, expressed; white, unexpressed) corresponding to the proportions shown in boxplots, individual points correspond to cHL cases. Right upper panel: proportions of each cell type expressing, 0, 1, 2, or 3 inhibitory molecules in aggregate across all images. Right lower panel: Proportion of single- or double-positive cells expressing each indicated combination of inhibitory molecules. (D) Summarized interaction plot across all study tumors in aggregate, displaying the ratio of the nearest neighbor distance between phenotype pairs (index phenotype (gray) to test phenotype (black)) compared with the expected baseline distance. Red indicates cell type cooccurrence, and blue indicates cell type exclusion. (E) Violin plots comparing the observed cell proportions for each tumor sample for the CD30+ neighborhood (orange) and across the entire examined tissue area (nonneighborhood; blue), for each MNP subtype. Wilcoxon signed-rank test used to assess statistical significance.
Mononuclear phagocytes expressing ≥1 immunosuppressive molecules are enriched around HRSCs in cHL. (A) Representative micrographs from multiplexed IF imaging of cHL samples, showing expression of inhibitory molecules (IDO1, PD-L1, and TIM-3), cell type–defining markers for MNPs (CD11c & CD68), and HRSCs (CD30). One of 3 test regions from CHL-7 is shown (2008 μm × 1502 μm), with CD11c (cyan), CD30 (orange), CD68 (magenta), PD-L1 (green), IDO1 (yellow), and TIM-3 (red) fluorescent signal represented by unique pseudocolors (to improve clarity DAPI is not shown). Inset 1 (original magnification ×100 [160 μm × 108 μm]; left) shows CD30 and IDO1 (top), CD30 and PD-L1 (middle), and CD30 and TIM-3 (bottom), with DAPI (blue) to identify cell nuclei. Inset 2 (original magnification ×100 [160 μm × 108 μm]; right) shows representative CD30+ HRSCs, surrounded by mononuclear phagocytes in closer detail. Individual pseudocolors from area 2, corresponding to each antibody, are shown below the main image (second and third lines). (B) Expression of PD-L1, IDO1, and TIM-3 by MNPs. Representative images at original magnification ×200 (54 μm × 54 μm) show PD-L1 expression (green, top line) and corresponding CD11c (cyan) and CD68 (magenta, second line) for the same areas. Images are separated to improve clarity, as colocalized. IDO1 expression (yellow, cytoplasmic localization) with CD11c and CD68 (third line). TIM-3 expression (red, cytoplasmic localization) with CD11c and CD68 (bottom line). (C) Proportion of mononuclear phagocytes expressing immuno-regulatory molecules. Left panel: lower key represents the combinations of inhibitory molecules (black, expressed; white, unexpressed) corresponding to the proportions shown in boxplots, individual points correspond to cHL cases. Right upper panel: proportions of each cell type expressing, 0, 1, 2, or 3 inhibitory molecules in aggregate across all images. Right lower panel: Proportion of single- or double-positive cells expressing each indicated combination of inhibitory molecules. (D) Summarized interaction plot across all study tumors in aggregate, displaying the ratio of the nearest neighbor distance between phenotype pairs (index phenotype (gray) to test phenotype (black)) compared with the expected baseline distance. Red indicates cell type cooccurrence, and blue indicates cell type exclusion. (E) Violin plots comparing the observed cell proportions for each tumor sample for the CD30+ neighborhood (orange) and across the entire examined tissue area (nonneighborhood; blue), for each MNP subtype. Wilcoxon signed-rank test used to assess statistical significance.
Expression of inhibitory molecules by age and histological characteristics
The proportion of CD11c+/CD68− and CD11c+/CD68+ MNPs expressing inhibitory molecules increased with age at diagnosis (P < .05), but there were no significant differences in the proportion of MNPs expressing inhibitory molecules based on sex, histological subtype, or Epstein-Barr virus status (supplemental Figure 7A-D). This finding suggests that immunosuppressive signaling in pediatric and young-adult cHL may differ from cHL in older adults.
Discussion
HRSCs have the remarkable ability to cohere an immunosuppressive microenvironment, including the recruitment of MNPs expressing immunoregulatory molecules. Through high-resolution analysis, we found that MNPs comprise diverse subsets of monocytes, macrophages, and DCs. We highlight several new features of this MNP network: the exclusion of pDCs and aDCs from the vicinity of HRSC; that immunoregulatory molecules are present on DCs close to HRSCs, at equivalent levels to macrophages; that multiple soluble and cell-to-cell interactions underpin this structure, communicating with HRSCs and lymphocytes. This work complements recent detailed analysis of lymphocytes in cHL, showing enrichment of exhausted LAG3+ CD4+ T cells around MHC-II− HRSCs,10 and elaboration of CXCL13 by Tfh-like cells in lymphocyte-rich cHL.32
We have shown that conventional DCs are closely associated with PD-L1+ HRSCs. These cells are usually involved in immunosurveillance and presentation of neoantigens to T cells, however, this mechanism appears to be stalled in cHL, likely as a result of immunosuppressive niche signaling. Colocalization and LRI analyses suggest that MNPs have proximal roles in directing and amplifying this signaling. These MNPs cooperate with HRSC-derived and NF-κB–directed chemokine expression, providing chemoattractant and inhibitory signals that recruit and instruct immunosuppressive T cells. Our analysis identifies classical monocytes, in particular, as important signaling hubs, controlling retention of cDC2 and ThExh via CCR1-, CCR4-, CCR5-, and CXCR3-dependent signaling (Figure 4A-B).
We propose that there are distinct and recurring tissue niches within cHL lymph nodes. Enrichment for the inflammatory cDC2-monocyte-macrophage niche is associated with early relapse following treatment, expanding upon previous reports that CD68+ TAM infiltration and macrophage gene expression is associated with poor outcomes in advanced cHL.39,40 Conversely, a fibrosis/stroma-rich tissue niche is associated with favorable outcomes. Computational pathology approaches and high-throughput multiplexed technologies may permit the characterization of these diverging cell networks in diagnostic tissue sections, before treatment.41,42 Future strategies might aim to shift the dominant pathology toward a fibrotic or MNP-depleted phenotype (eg, CCL2 inhibition or CSF-CSF1R blockade), and away from an inflammatory and immunosuppressive phenotype.43-45 DCs may be augmented by Flt3-ligand,46 CD40 agonists, or lentivirally-transduced chimeric antigen receptors.47
Therapeutic efforts to reverse tumor-associated immune suppression have targeted T cells, with recent studies suggesting alternative immune checkpoints, in addition to PD-1.10,48 The coexpression of inhibitory molecules and chemokines across MNPs suggests extensive redundancy in immunosuppressive signaling.49,50 This may partially explain resistance to PD-1 blockade, and offer opportunities for tailoring therapies on the basis of molecular profiling. In a murine model, it has been shown that TAMs rapidly transfer anti–PD-1 complexes away from T cells, through a FcɣR-mediated process.51 Intriguingly, a recent report demonstrated that HRSC eradication, after induction therapy with Nivolumab, was associated with a reduction in PD-L1+ TAM, rather than detectable cytotoxic responses.52 This suggests initial vulnerability of MNP networks to PD-1 blockade, such as dependence on tonic signaling by T cells or HRSCs, and may support the hypothesis of reverse signaling on HRSCs.53
Overall, these atlas-scale molecular and spatially resolved data provide evidence for distinctly organized tissue niches defining the pathology of cHL. Our results highlight a dominant, interacting MNP network associated with HRSCs with immunosuppressive phenotypes, the rational targeting of which may present valuable therapeutic opportunities.
Acknowledgments
The authors thank Christian Steidl (The University of British Columbia) for generous sharing of scRNA-seq data. The authors thank the Newcastle University/Newcastle upon Tyne NHS Foundation Trust Molecular diagnostics laboratory (NovoPath; https://www.novopath.co.uk/) for tissue handling and technical assistance.
B.J.S. is supported by a Clinical Doctoral Fellowship from the Wellcome Trust (216366/Z/19/Z). C.D.C. is supported by grant funding from the JGW Patterson Foundation (https://jgwpattersonfoundation.co.uk), Bright Red (https://brightred.org.uk), the Lymphoma Research Trust (https://www.lymphoma-research-trust.org.uk), a Medical Research Council Confidence in Concepts award, and a Wellcome Trust Small project grant award. S.B. is supported by the Wellcome Trust (206194).
Authorship
Contribution: B.J.S. performed data analysis and wrote the manuscript; M.F. performed data analysis and contributed to the manuscript; M.D.Y. contributed to data curation; C.J., A.S., A.B., C.M.B., V.R., J.R.F., K.R.J., and N.C. performed scRNA-seq experiments; K.T.M., L.H., N.J., and K.S.-P. acquired deceased-donor lymph node tissue; M.C., M.R.C., and S.B. supervised the project; and C.D.C. performed microscopy experiments, analyzed data, wrote the manuscript, and supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sam Behjati, Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, Saffron Walden CB10 1RQ, United Kingdom; e-mail: sb31@sanger.ac.uk; and Christopher D. Carey, Translational and Clinical Research Institute, Newcastle University, Newcastle upon Tyne, NE1 7RU, United Kingdom; e-mail: christopher.carey@newcastle.ac.uk.
References
Author notes
Code used to analyze data in the project is uploaded to Github at https://github.com/bjstewart1/hodgkin_lymphoma_analysis.
Data are available for download at https://hodgkinlymphoma.cellgeni.sanger.ac.uk as annotated data objects (.h5ad file) and interactive cellxgene portal.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Heterogeneous immune cell networks occupy distinct tissue niches with divergent mononuclear phagocyte enrichment patterns and prognostic significance. (A) Shared–nearest neighbor graph embedding of transcriptional profiles of nanostring ROIs from reactive and cHL lymph nodes. Color indicates clusters identified using Leiden clustering with resolution = 1. (B) Shared–nearest neighbor graph embedding of transcriptional profiles of nanostring ROIs colored by PD-L1 expression status. (C) Differential abundance estimates of cell-types deconvolved from nanostring ROIs between clusters shown in (a), LFC estimates represent differential abundance of cell types in each cluster vs all other clusters. (MΦ, macrophage). (D) Upper panel: heatmap showing WGCNA module coexpression patterns for 6 modules (modules A-F) across 130 cHL gene expression profiles (microarray) split by treatment outcome. Lower panel: heatmap showing results for a hypergeometric enrichment test between module gene membership and cell type markers from the scRNA-seq data set for WGCNA modules (modules A-F). (MΦ, macrophage). (E) Representative micrographs from multiplexed IF imaging of cHL samples. One of 3 test regions from tumor CHL-27 is shown, cropped to 1662 μm by 1076 μm; CD11c (cyan), CD30 (orange), CADM1 (green), CD1c (yellow), LAMP3 (magenta), and CD123 (red) fluorescent signals are represented by unique pseudocolors. To improve clarity, DAPI is not shown. Selected areas are highlighted, each at original magnification ×100 (160 μm × 108 μm); CD30+ HRSC-dense area (region 1, inset left), internodular area with no CD30+ HRSCs (region 2, inset right), and area with CADM1+ cDC1 (region 3, inset top right). Individual pseudocolors are shown below the main image (second & third lines), with DAPI (blue) to identify cell nuclei. Each individual image refers to a multiplexed area above (inset), indicated by the corresponding number (top right). (F) HRSC and DC map, corresponding to the same region from panel E. Phenotyped cells are identified by colored nuclei, with each cell type represented by a different color; CD11c+ ONLY (CD11c+ CD1c−, cyan), CD30+ HRSC (light orange), cDC1 (green), cDC2 (yellow), LAMP3+ aDC (magenta), and pDCs (red) are shown. Cells with no assigned phenotype (“null”) and CD1c+ ONLY cells are excluded from this visualization. The “HRSC neighborhood” is shown as circles surrounding each CD30+ HRSC (dark orange). A selected area is highlighted, at original magnification ×200 (80 μm × 54 μm; inset, center). (G) Isobar plots show the location and density of each cell phenotype of interest, for the corresponding tumor region from panels E and F (note the uncropped 2008 μm by 1502 μm region is shown here). HRSCs (orange dots, left) and DC subsets (colors corresponding to each phenotype, right) are shown. (H) Summarized interaction plot across all study tumors in aggregate, displaying the ratio of the nearest neighbor distance between phenotype pairs (index cell type [gray] to test cell type [black]) compared with the expected baseline distance. Red indicates cell type cooccurrence, blue indicates cell type exclusion.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/19/10.1182_blood.2022015575/2/m_blood_bld-2022-015575-gr2.jpeg?Expires=1769113368&Signature=Cz7hAC~XT97WD2QMXOmTzHiglE51CdwV-g4vF9EW4PBLj2GSOf0uRaLsqiexNDKgXTBEOvJEiuk~B61qPuMhsxx0D09ciQD5lVSVkP5JtAcZRGKtdnJ98d5wVtFqs4lYm0sAIzPjoO4NnWJEI8FS6njg8~dtJZ2OJBBJak9fAmCltzZIQSfnVlY~M4mZ3x-8CcKj6wEY72MVSg-7~PO25ovX~BfyAkZNTvkXY-Q2bQ2LwVw55bnx56V5y~o8UqAvUcHfoZapGk4O2q8iuOnfpC1BMxIfka0qDBMs2YYX~Cbzzl7ixJGm~53JHP8~HCk2JJmfPd4j5pFCTSn5OLTx4Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Mononuclear phagocytes expressing ≥1 immunosuppressive molecules are enriched around HRSCs in cHL. (A) Representative micrographs from multiplexed IF imaging of cHL samples, showing expression of inhibitory molecules (IDO1, PD-L1, and TIM-3), cell type–defining markers for MNPs (CD11c & CD68), and HRSCs (CD30). One of 3 test regions from CHL-7 is shown (2008 μm × 1502 μm), with CD11c (cyan), CD30 (orange), CD68 (magenta), PD-L1 (green), IDO1 (yellow), and TIM-3 (red) fluorescent signal represented by unique pseudocolors (to improve clarity DAPI is not shown). Inset 1 (original magnification ×100 [160 μm × 108 μm]; left) shows CD30 and IDO1 (top), CD30 and PD-L1 (middle), and CD30 and TIM-3 (bottom), with DAPI (blue) to identify cell nuclei. Inset 2 (original magnification ×100 [160 μm × 108 μm]; right) shows representative CD30+ HRSCs, surrounded by mononuclear phagocytes in closer detail. Individual pseudocolors from area 2, corresponding to each antibody, are shown below the main image (second and third lines). (B) Expression of PD-L1, IDO1, and TIM-3 by MNPs. Representative images at original magnification ×200 (54 μm × 54 μm) show PD-L1 expression (green, top line) and corresponding CD11c (cyan) and CD68 (magenta, second line) for the same areas. Images are separated to improve clarity, as colocalized. IDO1 expression (yellow, cytoplasmic localization) with CD11c and CD68 (third line). TIM-3 expression (red, cytoplasmic localization) with CD11c and CD68 (bottom line). (C) Proportion of mononuclear phagocytes expressing immuno-regulatory molecules. Left panel: lower key represents the combinations of inhibitory molecules (black, expressed; white, unexpressed) corresponding to the proportions shown in boxplots, individual points correspond to cHL cases. Right upper panel: proportions of each cell type expressing, 0, 1, 2, or 3 inhibitory molecules in aggregate across all images. Right lower panel: Proportion of single- or double-positive cells expressing each indicated combination of inhibitory molecules. (D) Summarized interaction plot across all study tumors in aggregate, displaying the ratio of the nearest neighbor distance between phenotype pairs (index phenotype (gray) to test phenotype (black)) compared with the expected baseline distance. Red indicates cell type cooccurrence, and blue indicates cell type exclusion. (E) Violin plots comparing the observed cell proportions for each tumor sample for the CD30+ neighborhood (orange) and across the entire examined tissue area (nonneighborhood; blue), for each MNP subtype. Wilcoxon signed-rank test used to assess statistical significance.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/19/10.1182_blood.2022015575/2/m_blood_bld-2022-015575-gr5.jpeg?Expires=1769113368&Signature=dVlEzRn7buH0zrJ7cXeFKokP0BJUBK57NIMzYaSe9zc1z77-ArQ3t8jYGrCZxkJtMfA1AmG4ihNClpgTZXYj2igvY94x4GFr3CVBS1VDDOPH7S2j7YNTyG4pD7cklAPNpTaXpvDKtgWy-bmILfn80J-lMZcpwtRUZ1XKKtFPv5bGp7-oM06BCz~~257rSLGmflZfMhheArJPsMHuhUnU2LPlpc1kVzf~xS1RtKutNIF12oDacNO8p7hZ~Z-z~TbIwtrp8P~bqzs0iGBwfWyZ58ELQOC4gdnr2FMb3Sbh6hBrwUK-8QpebKZCPamrFHrZvxtpmXVjXG2gOCxzL3S2Cw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal