In this issue of Blood, Othman and colleagues compared clinical, pathologic, and molecular charactestics of nucleophosmin (NPM1) mutated treatment-related acute myeloid leukemia (T-AML) with de novo NPM1-mutated AML and T-AML with wild-type NPM1.1
As a clinician interested in acute myeloid leukemia (AML) and allogeneic hematopoietic cell transplantation (alloHCT), a potentially curative treatment modality for hematologic malignancies,2 I like to think through the lenses of my patients. For example, “Do I really need an alloHCT in first complete remission (CR1)? Are the antileukemic effects worth the increased risk of toxicity of the transplant?” In treatment-related AML (T-AML), mutations in the nucleophosmin (NPM1) help address this question. Fifteen years ago, the German–Austrian Acute Myeloid Leukemia Group3 showed that NPM1 mutations without FLT3-ITD mutations did not benefit from alloHCT in CR1. Subsequent studies have supported this conclusion, and thus AML with NPM1 mutations without other adverse-risk cytogenetic abnormalities has been placed in the favorable category of AML by European Leukemia network (ELN).4 Then what happens if relatively good prognostic mutational AML (NPM1 AML) occurs in a poor prognostic setting, such as T-AML? This intriguing question was tackled by Othman and colleagues by exploring the impact of a relatively good prognostic mutational AML (NPM1 AML) occurring in T-AML.1
NPM1 protein, encoded by NPM1 gene, is predominantly located in nucleolus and is involved in intracellular functions, including molecular chaperoning, nucleolar phase separation, ribosome biogenesis, centrosome replication, apoptosis, cell-cycle controlling, DNA repair and differentiation, tumor suppression (eg, TP53), and genome stability.5,6 However, when NPM1 is mutated, mostly in exon 12, alterations in location (predominantly in cytoplasm) and functions of nucleophosmin and nucleophosmin proteins occur. These aberrant forms play roles in leukemogenesis via not fully elucidated mechanisms, particularly in the presence of other mutations (eg, FLT3, DNMT3, and RAS).6NPM1 is the most common mutation in adults with AML (approximately 30% in all AMLs and 60% of those with normal karyotypes).6 It was also observed in approximately 15% of T-AML.7 T-AML results from genetic aberrations induced or selected by prior chemo- or radiotherapy (eg, TP53, PPM1D, chromosome 5 or 7 abnormalities, complex karyotypes, or clonal hematopoiesis). The ELN most recently described T-AML not as a disease entity, which is mostly defined by cytogenetic or molecular aberrations, but as a diagnostic qualifier.4 The reason is that T-AML (approximately 10%-15% of AML), in contrast to NMP1 AML, confers chemoresistance and thus offers a poor prognosis.
In the study by Othman and colleagues, 3 groups were included: T-NPM1 AML, de novo NPM1-mutated AML, and T-AML with wild-type NPM1. In this study, clinical, disease (molecular, cytogenetic), and patient characteristics were compared in these 3 groups. Major clinical outcomes were relapse, leukemia-free survival (LFS) and overall survival (OS) in patients who received intensive induction therapy (eg, “7 + 3”). T-AML was defined as a positive history of exposure to chemotherapy or radiotherapy. The major findings were as follows: T-NPM1 AML shared the common disease characteristics of de novo NPM1 AML (eg, aberrant localization of nucleophosmin in cytoplasm, predominance of normal karyotype, frequent coexistence with DNMT3A and TET2 mutations, upregulated HOX genes, and downregulated CD133 and CD34 expression). Regarding TP53 and PPM1D, T-NPM1 AML was again similar to de novo NPM1, and both had a much lower incidence compared with T-AML. Likewise, FLT3-ITD mutation was more common in both NPM1 AML groups (de novo or therapy related) compared with T-AML. FLT3 mutation seemed to have an adverse effect on survival in each of the NMP1 AML group. In terms of LFS and OS, T-NPM1 AML and de novo NPM1-mutated AML were similar to each other and were superior to T-AML. Approximately only 20% of each of the 3 groups received alloHCT in CR1, and alloHCT had no significant effect on survival by multivariate analysis.
These types of studies are rare and difficult to complete because of the infrequency of each subset. Therefore, as done in this study, significant collaboration among centers from different countries with detailed, accurate data was used. The study has the drawbacks of being a retrospective study (eg, treatment choices will be subject to providers’ biases as well as to the period when patients were treated in this long-duration study). Some current era important data were lacking, such as minimal residual disease. No details of prior radio- or chemotherapy were available, and the definition of T-NPM1 AML was gross. Many factors—such as conditioning, graft-versus-host disease prophylaxis, and donor types—are unknown. However, it is impossible to compare these 3 groups in a prospective study. Moreover, to improve some of the limitations and thus improve the quality of the study, the authors performed molecular tests uniformly in a center in available samples (of note, the sets of patients for clinical and genetics studies are not exactly same).
This study enhances our knowledge in some of the following areas, including (1) the general characteristics and biology of T-NPM1 AML that are similar to de novo NPM1 AML. TNPM1 AML does not carry most of the unfavorable known factors (eg, TP53, complex cytogenetics) found in T-AML. A previous study also found that T-NPM1 AML was similar to de novo NPM1 AML rather than T-AML. In fact, the authors were so surprised by their findings that they suggested that perhaps the patients did not have T-NPM1 AML but had de novo NPM1 AML instead.7 (2) Regarding the response to treatment and survival, T-NPM1 AML again is similar to de novo NPM1 AML. This is very likely due to not having unfavorable factors in most patients with T-NPM1 AML in contrast to other T-AMLs. (3) DNMT3A and TET2 mutations are common in both NPM1 groups and likely represent premalignant clonal hematopoiesis rather than mutations selected by chemo- or radiotherapy.
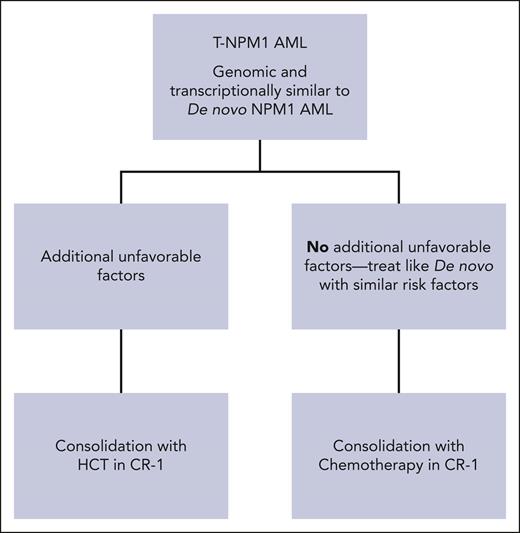
Now, to address the original question: Is alloHCT indicated in CR1 for T-NPM1 AML? Despite the limitations, especially for alloHCT in the study, at least one cannot say straightforwardly “yes, it is indicated” especially for those with no other poor prognostic factor (see figure). Lastly, NPM1 mutation, whether of de novo or therapy related, seems to present a good target for several treatments: avrainvillamide or XPO1 inhibitors (relocating cytoplasmic mutant NPM1 to nucleolus), actinomycin D (inducing nucleolar stress), small molecules interfering in NPM1 oligomerizations (NSC34888) or targeting HOX expression via inhibiting menin-MLL 1 (MI-3454), arsenic trioxide and ATRA (proteasome-dependent NPM1 mutant degradation), NPM1 mutant-specific engineered T cells. Some US Food and Drug Administration–approved drugs for AML mechanistically may also have a potential to be more effective due to disease characteristics (eg, antiCD33 agents gemtuzumab ozogamicin or intuzumab-Ac225 due to high expression of CD33, and bcl2 inhibitors with hypomethylating agents).6,8 These have potential to change positively our NPM1 AML treatment in the future.
The phrase “Gold is gold even in mud” in the title of this commentary is a Turkish proverb.
Conflict-of-interest disclosure: C.U. receives honoraria from Blueprint and Takeda for being on the speaker bureau and advisory board.