Key Points
The guanine nucleotide exchange factor Grab is a critical regulator of Tfrc recycling and iron metabolism in mammalian erythropoiesis.
Grab activates Rab8 to promote the endocytic recycling and exocytosis of Tfrc in differentiating erythroblasts.
Abstract
Developing erythroblasts acquire massive amounts of iron through the transferrin (Tf) cycle, which involves endocytosis, sorting, and recycling of the Tf-Tf receptor (Tfrc) complex. Previous studies on the hemoglobin-deficit (hbd) mouse have shown that the exocyst complex is indispensable for the Tfrc recycling; however, the precise mechanism underlying the efficient exocytosis and recycling of Tfrc in erythroblasts remains unclear. Here, we identify the guanine nucleotide exchange factor Grab as a critical regulator of the Tf cycle and iron metabolism during erythropoiesis. Grab is highly expressed in differentiating erythroblasts. Loss of Grab diminishes the Tfrc recycling and iron uptake, leading to hemoglobinization defects in mouse primary erythroblasts, mammalian erythroleukemia cells, and zebrafish embryos. These defects can be alleviated by supplementing iron together with hinokitiol, a small-molecule natural compound that can mediate iron transport independent of the Tf cycle. Mechanistically, Grab regulates the exocytosis of Tfrc-associated vesicles by activating the GTPase Rab8, which subsequently promotes the recruitment of the exocyst complex and vesicle exocytosis. Our results reveal a critical role for Grab in regulating the Tf cycle and provide new insights into iron homeostasis and erythropoiesis.
Introduction
Developing red blood cells acquire massive amounts of iron for the synthesis of hemoglobin. At the terminal stages of differentiation, erythroblasts upregulate the expression of transferrin receptor (Tfrc) and porphyrin synthesis genes to increase iron assimilation and heme production. Tfrc interacts with iron-loaded transferrin (Tf) on the cell surface, and the complex is internalized through clathrin-mediated endocytosis.1-3 Once iron is released in maturing endosomes, the iron-free Tf-Tfrc complex is recycled to the cell surface for the next round of iron uptake.1-3 We have previously demonstrated that sorting nexin 3 (Snx3), a component of the retromer complex, regulates the sorting of Tfrc into recycling endosomes to promote its recycling.4 Depletion of Snx3 leads to hemoglobin defects and anemias in mammalian erythroblasts and zebrafish embryos, respectively.4 Recent studies suggest that proteins in recycling endosomes may still be diverted to degradative pathways as recycling endosomes can serve as both membrane sources and platforms for autophagosome formation.5-7 Indeed, Tfrc has been shown to localize to autophagosomes and be degraded through the autophagy lysosomal pathway.5,8 Thus, the trafficking and fate of Tfrc at the recycling endosomes of differentiating erythroblasts must be strictly regulated to ensure that the vast majority of it is recycled to the cell surface.
Vesicular protein transport is controlled by small Rab GTPases as well as their regulatory proteins including guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins.9,10 Polarized exocytosis also requires the exocyst, a hetero-octameric protein complex that mediates the targeting and tethering of exocytic vesicles to the plasma membrane.11,12 The exocyst subunit Sec15 (also known as Sec15l1 or Exoc6) is responsible for interacting with Rab GTPases to facilitate the recruitment of the exocyst complex to secretory vesicles.13-16 Mice with a spontaneous mutation in Sec15 display hypochromic microcytic anemia due to impaired Tfrc recycling.17-20 These results support an essential role for the exocyst complex in the Tf cycle, but it remains unclear how developing erythroblasts upregulate the activity of the exocyst complex to meet the increased demand for Tfrc recycling.
Here, we identify a Rab GEF, Grab, that plays a critical role in regulating Tfrc recycling during erythropoiesis. Differentiating erythroblasts upregulate the expression of Grab to activate Rab8, which in its guanosine triphosphate (GTP)-bound state promotes the exocytosis of Tfrc, likely through recruiting the exocyst to recycling endosomes. The Grab-Rab8-exocyst cascade is crucial for erythropoiesis as its defects impairs Tfrc recycling, iron uptake, and hemoglobinization.
Methods
Knockdown of Grab in mouse primary fetal liver erythroblasts
Grab silencing and hemoglobin quantification in mouse primary fetal liver cells were carried out using the procedure described previously.21,22 Two short hairpin RNAs (shRNAs) targeting Grab (CCGAGTTCCTAAAGGAGGAGCTATA and CAACATGAAGCAGGCGACATCGGAA) were designed using Invitrogen BLOCK-iT RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress/design.do) and cloned into murine stem cell retroviral vector-U3-H1. After packaging in HEK293T cells, the shRNA retroviruses were transduced into primary fetal liver erythroblasts isolated from E14.5 mouse embryos. The knockdown efficiency was examined by quantitative real-time polymerase chain reaction (qRT-PCR). The hemoglobin content was quantified with the Drabkin’s reagent.
Knockdown of rab3il1 in zebrafish embryos
Rab3il1 was silenced in zebrafish embryos using a splice-blocking antisense morpholino (MO, Gene Tools) that targets the intron 2 - exon 3 splice acceptor sites in rab3il1 pre–messenger RNA (mRNA). The sequence of rab3il1 MO is 5′-TCGATCTAACACAGACAGAAAGGTT-3′. Zebrafish embryos were injected with 0.5 mM rab3il1 MO or 0.5 mM standard control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′). The knockdown efficiency was analyzed by RT-PCR with the following primers: F1, 5′-GACGAGGAGTGTGAAAGACTGTC-3′; F2, 5′-GGATGGCTAATCCAGAGTTTATGAGC-3′; R1, 5′-GTTCGGAGACGAGGGTGTAGAG-3′. The morphant embryos were analyzed at 24 to 72 hours postfertilization (hpf).
Knockout of Grab and Rab8a in mammalian cells
The knockout mammalian cells were generated following the method described previously.22 The pX330 plasmids containing guide RNAs and a pEF1α plasmid, which carries a puromycin-resistance gene, were electroporated into mouse erythroleukemia (MEL) and human erythroleukemia (K562) cells using a Gene Pulser Xcell system (Bio-Rad). The cells were selected with 5 µg/mL puromycin for MEL and 2 µg/mL puromycin for K562. After 10 days, single clones were isolated for PCR genotyping followed by sequencing. The guide RNA target sites are TCTCCATGGAGGAACTGCGC and GAGATCATGAGGCTTCGGAA for mouse Grab, TCCATGGAGATCCGAGAGAA and TTGCTGGATGTAGCGGATGT for human GRAB, and CAGCTTGAACAGGTAATCGT and GAGACTGCACCGGAAGAAGC for mouse Rab8a.
Uptake and exocytosis of Tf-Alexa 488
Chemically induced MEL cells were first incubated in Opti-MEM for 30 minutes at 37°C, followed by incubation with 30 μg/mL Alexa Fluor 488–labeled holo-transferrin (Tf-Alexa 488, Invitrogen) for 5 minutes on ice. For the Tf uptake assay, cells were further incubated in fresh growth medium at 37°C for 10 minutes. After washing the cells twice with an ice-cold acid wash buffer (50 mM glycine, 150 mM NaCl, pH 3.0), the fluorescence intensity of Tf-Alexa 488 in cells was quantified using Cytomics FC 500 Flow Cytometry System (Beckman). For the Tf exocytosis assay, cells were incubated in fresh growth medium for 2 minutes at 37°C following the incubation with Tf-Alexa 488. After washing twice with ice-cold phosphate-buffered saline, cells were transferred to fresh medium at 37°C and harvested at 0, 5, 10, 20, and 30 minutes. The mean fluorescence intensity of Tf-Alexa 488 was analyzed by flow cytometry (Cytomics FC 500, Beckman), and the values at different time points were normalized to the initial time point (0 minutes). Representative images of Tf-Alexa 488 at 0, 5, and 10 minutes were acquired by confocal microscopy (LSM710, Zeiss).
Identification of Grab-interacting proteins in MEL cells
The FLAG-Grab construct was electroporated into MEL cells, and the stable clones were derived by selecting the cells with 5 μg/mL puromycin for ∼10 days. The MEL cells stably expressing FLAG-Grab were chemically induced with dimethyl sulfoxide (DMSO) for 3 days. Proteins were extracted and crosslinked with 1 mM dithiobis (succinimidyl propionate) (Thermo Fisher). Affinity purification and mass spectrometry were performed according to the previously reported procedure.22 Briefly, FLAG-Grab and its interacting proteins were affinity purified using anti-FLAG M2 Affinity agarose (Sigma-Aldrich), and the purified proteins were subjected to liquid chromatography–tandem mass spectrometry analysis on a TripleTOF 5600 mass spectrometer (AB SCIEX).
Measurement of iron content by ICP-MS
The chemically induced MEL and K562 cells were digested in 65% HNO3 at 85°C until all HNO3 evaporated. The dried samples were dissolved in 2% HNO3, followed by measurement of iron concentration by inductively coupled plasma mass spectrometry (ICP-MS) using the Elan DRC-e ICP-mass spectrometer (Perkin-Elmer). For each sample, one-fifth of the cells were saved for measurement of the protein concentration with the Bradford reagent (Bio-Rad), and the iron concentration was normalized to the protein content.
Analysis of iron levels using iron-sensing dyes
The cellular and mitochondrial iron levels were determined with calcein green-AM (Thermo Fisher) and rhodamine B-[(1,10-phenanthrolin-5-yl)-aminocarbonyl] benzyl ester (RPA, Axxora), respectively, following a method described previously.23 Chemically induced MEL cells and K562 cells were incubated with 1 µM calcein green-AM and 1 µM RPA at 37°C for 15 minutes. After washing 2 times with phosphate-buffered saline, cells were photographed using a LSM710 microscope, and the fluorescence intensities were quantified using ImageJ software.
Heme staining and quantification
Heme in zebrafish embryos and chemically induced MEL and K562 cells was stained with o-dianisidine following the previously described method.4 The stained samples were analyzed on an Olympus IX73 microscope equipped with a Leica DFC310 FX camera.
Fluorescence porphyrin assays24 were performed to quantify heme in zebrafish embryos, chemically induced MEL, and K562 cells. Five volumes of 2 M oxalic acid were added to the cell lysates, and the samples were boiled for 30 minutes to release iron from heme. The fluorescence of porphyrin was measured on a Synergy H1 plate reader (excitation, 400 nm; emission, 608 nm; BioTek). The background porphyrin fluorescence of unboiled samples was subtracted, and the derived heme concentration was normalized to the protein content of the cell lysate.
Results
Grab expression is induced during erythroid differentiation
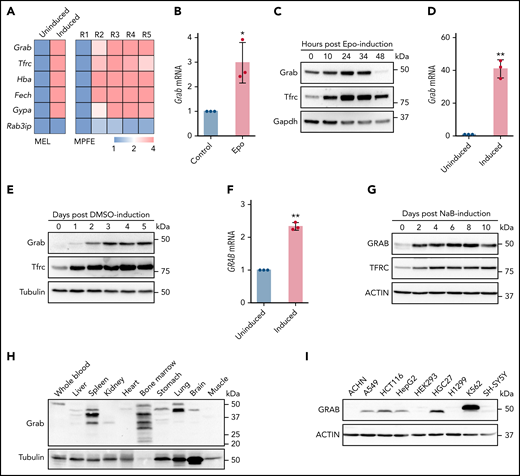
Differentiating erythroblasts acquire massive amounts of iron through the Tf cycle, which requires efficient recycling of Tfrc. To identify the molecular mechanisms responsible for the efficient Tfrc recycling, we screened for trafficking genes that are upregulated during erythropoiesis. Transcriptomics analyses of differentiating MEL and murine fetal liver erythroblasts22,25 revealed that Grab, a gene encoding a Rab GEF, was highly upregulated during erythroid differentiation (Figure 1A). In contrast, the expression of its closely related paralog, Rab3ip (also known as Rabin8), was not induced in maturing erythroblasts (Figure 1A). To confirm the erythroid-enriched expression of Grab, we isolated erythroid progenitors from the liver of E14.5 mouse embryos and induced their erythroid differentiation with Epo in vitro. Results from qRT-PCR and immunoblotting analyses both showed considerable upregulation of Grab following Epo-induced erythroid differentiation (Figure 1B-C). Consistently, the expression of Grab was substantially elevated in the MEL cell after DMSO-induced erythroid-like differentiation (Figure 1D-E). We found that the human erythroleukemia cell K562 also exhibited significantly increased GRAB expression during the erythroid-like differentiation (Figure 1F-G). This result is consistent with transcriptomics data on human erythroblasts derived from CD34+ hematopoietic stem and progenitor cells26,27 and suggests that the erythroid-induced expression of GRAB is conserved in mammals.
Grab is upregulated in differentiating erythroblasts. (A) Relative expression of Grab and control genes from RNA sequencing analyses on differentiating MEL cells (left)22 and mouse primary fetal liver erythroblasts (MPFE, right).25 MEL cells before (uninduced) and after DMSO-induced differentiation were analyzed. R1 to R5 represent the progressive stages of differentiating erythroblasts. (B-C) qRT-PCR (B) and immunoblotting (C) analyses of Grab in differentiating mouse fetal liver erythroblasts. Erythropoietin (Epo) was supplemented to the erythroid progenitors isolated from the livers of E14.5 mouse embryos to induce the erythroid differentiation. Tfrc and Gapdh were used as controls. *P < .05. (D-E) qRT-PCR (D) and immunoblotting (E) analyses of Grab in MEL cells before and after DMSO-induced erythroid-like differentiation. Tfrc and Tubulin were used as controls. **P < .01. (F-G) qRT-PCR (F) and immunoblotting (G) analyses of GRAB in K562 cells before and after sodium butyrate (NaB)-induced erythroid-like differentiation. TFRC and ACTIN were used as controls. **P < .01. (H-I) Western analyses of Grab in multiple mouse tissues (H) and human cell lines (I).
Grab is upregulated in differentiating erythroblasts. (A) Relative expression of Grab and control genes from RNA sequencing analyses on differentiating MEL cells (left)22 and mouse primary fetal liver erythroblasts (MPFE, right).25 MEL cells before (uninduced) and after DMSO-induced differentiation were analyzed. R1 to R5 represent the progressive stages of differentiating erythroblasts. (B-C) qRT-PCR (B) and immunoblotting (C) analyses of Grab in differentiating mouse fetal liver erythroblasts. Erythropoietin (Epo) was supplemented to the erythroid progenitors isolated from the livers of E14.5 mouse embryos to induce the erythroid differentiation. Tfrc and Gapdh were used as controls. *P < .05. (D-E) qRT-PCR (D) and immunoblotting (E) analyses of Grab in MEL cells before and after DMSO-induced erythroid-like differentiation. Tfrc and Tubulin were used as controls. **P < .01. (F-G) qRT-PCR (F) and immunoblotting (G) analyses of GRAB in K562 cells before and after sodium butyrate (NaB)-induced erythroid-like differentiation. TFRC and ACTIN were used as controls. **P < .01. (H-I) Western analyses of Grab in multiple mouse tissues (H) and human cell lines (I).
Next, we examined the expression of Grab in various mouse tissues and human cell lines. Consistent with its enhanced expression in erythroblasts, Grab displayed the most abundant expression in the hematopoietic tissue bone marrow (Figure 1H). Spleen and lung also expressed large amounts of Grab, whereas other tissues, such as liver, kidney, heart, and muscle, exhibited low expression of Grab (Figure 1H). Among all the human cell lines examined, the hematopoietic cell line K562 showed the highest GRAB expression (Figure 1I). Together, these results demonstrate that Grab is highly expressed during mammalian erythropoiesis.
Immunoblotting analysis revealed several immunoreactive bands of distinct sizes in mouse tissues (Figure 1H). RT-PCR detected at least 3 different Grab transcripts in bone marrow (supplemental Figure 1A-B, available on the Blood Web site), indicating that some of these bands are protein isoforms. Besides, other events such as posttranslational modification and protein degradation may contribute to the generation of these protein products.
Loss of Grab leads to impaired hemoglobin production
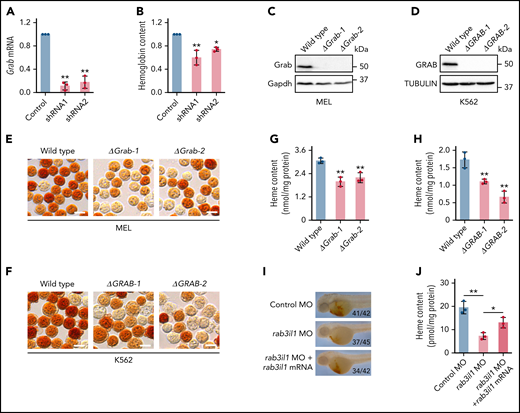
Grab was first identified as a GEF for Rab3a, a Ras-like GTPase that regulates vesicle exocytosis in neurons.28 Recent studies revealed that Grab also served as a GEF for another Rab protein, Rab8.29-31 We found that Grab expression is induced in differentiating erythroblasts, suggesting that it may play an important role in erythropoiesis. To investigate its erythroid function, we knocked down Grab in MPFE using 2 shRNAs (Figure 2A). Drabkin’s assay showed that silencing of Grab in differentiating MPFE led to a significant reduction in the hemoglobin production (Figure 2B). To corroborate this result, we knocked out Grab in both mouse and human hematopoietic cell models using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system(supplemental Figure 2A-B). Deletion of Grab in MEL and K562 cells was validated by both PCR genotyping and western analysis (Figure 2C-D; supplemental Figure 2C-D). Consistent with the data in mouse primary erythroblasts, o-dianisidine staining demonstrated that loss of Grab in MEL and K562 cells both caused diminished hemoglobin production (Figure 2E-F).
Grab is required for heme synthesis during erythropoiesis. (A) qRT-PCR validating knockdown of Grab in mouse primary fetal liver erythroblasts by 2 shRNAs. **P < .01. (B) Quantification of the hemoglobin content in control and Grab-silencing mouse primary fetal liver erythroblasts. *P < .05; **P < .01. (C-D) Immunoblotting analyses validating the knockout of Grab in MEL cells (C) and K562 cells (D). (E-F) o-Dianisidine staining of hemoglobin in wild-type and Grab knockout MEL cells (E) and K562 cells (F) post–erythroid-like induction. Scale bars, 10 μm. (G-H) Quantification of heme levels in MEL cells (G) and K562 cells (H) by fluorescence porphyrin assays. **P < .01. (I-J) Analysis of heme content in control zebrafish embryos, rab3il1 morphants, and rab3il1 morphants coinjected with rab3il1 mRNA. The zebrafish embryos were analyzed at 48 hours postfertilization. (I) and (J) are the representative images of o-dianisidine staining and quantification of total heme, respectively. The number of embryos with reduced o-dianisidine staining over the total number analyzed is shown in each image. *P < .05; **P < .01.
Grab is required for heme synthesis during erythropoiesis. (A) qRT-PCR validating knockdown of Grab in mouse primary fetal liver erythroblasts by 2 shRNAs. **P < .01. (B) Quantification of the hemoglobin content in control and Grab-silencing mouse primary fetal liver erythroblasts. *P < .05; **P < .01. (C-D) Immunoblotting analyses validating the knockout of Grab in MEL cells (C) and K562 cells (D). (E-F) o-Dianisidine staining of hemoglobin in wild-type and Grab knockout MEL cells (E) and K562 cells (F) post–erythroid-like induction. Scale bars, 10 μm. (G-H) Quantification of heme levels in MEL cells (G) and K562 cells (H) by fluorescence porphyrin assays. **P < .01. (I-J) Analysis of heme content in control zebrafish embryos, rab3il1 morphants, and rab3il1 morphants coinjected with rab3il1 mRNA. The zebrafish embryos were analyzed at 48 hours postfertilization. (I) and (J) are the representative images of o-dianisidine staining and quantification of total heme, respectively. The number of embryos with reduced o-dianisidine staining over the total number analyzed is shown in each image. *P < .05; **P < .01.
Hemoglobin is composed of globin polypeptide chains and heme groups. Immunoblotting assay did not reveal notable difference in the globin expression between the Grab knockout and wild-type clones (supplemental Figure 2E-F). In contrast, loss of Grab induced a significant reduction in the heme content (Figure 2G-H), implying that the hemoglobinization defects in these cells are mainly attributed to impaired heme synthesis.
To evaluate whether Grab regulates erythropoiesis in vivo, we silenced its homolog, rab3il1, in zebrafish embryos by injecting a splice-blocking antisense MO (supplemental Figure 3A-B). Compared with the control, the rab3il1 morphant embryos displayed diminished o-dianisidine staining, a defect that was largely rescued by coinjection of the synthetic rab3il1 mRNA (Figure 2I; supplemental Figure 3C). This result was further verified by measuring the total heme content (Figure 2J). To find out whether rab3il1 depletion impairs erythroid lineage specification, we analyzed the expression of erythroid markers gata1, band 3, and αe3 globin. The expression of these markers was not reduced in rab3il1-morphant embryos (supplemental Figure 3D). Similarly, silencing of rab3il1 did not impair the expression of the Tg(globin-LCR:eGFP) transgene in zebrafish embryos (supplemental Figure 3E). These observations are consistent with the results from mammalian erythroid cells and suggest that Grab plays a major role in heme synthesis during erythropoiesis.
Grab is required for erythroid iron assimilation
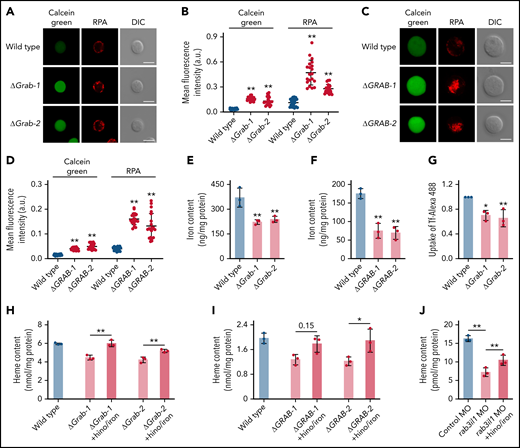
To determine whether the reduced heme production in Grab-deficient cells is due to insufficient iron supply, we assessed iron levels using calcein green and RPA, 2 fluorescent dyes that have been used as indicators of the cytosolic and mitochondrial iron, respectively.23 The fluorescence signals of these dyes inversely correlate with the iron levels. We found that knockout of Grab in MEL cells resulted in substantially increased fluorescence of both calcein green and RPA, indicating iron deficiency at both cellular and mitochondrial levels (Figure 3A-B). Consistently, the GRAB-deficient K562 cells also displayed enhanced fluorescence of calcein green and RPA (Figure 3C-D). The cellular iron deficiency observed in Grab knockout erythroblasts was further verified by quantitative analyses using ICP-MS (Figure 3E-F).
The heme defect exhibited by Grab-deficient erythroblasts is due to reduced iron uptake. (A-B) Representative fluorescence images (A) and quantification (B) of calcein green and RPA in wild-type and Grab knockout MEL cells. N = 30 for each group; **P < .01. (C-D) Representative fluorescence images (C) and quantification (D) of calcein green and RPA in wild-type and GRAB knockout K562 cells. N = 30 for each group; **P < .01. (E-F) Quantification of iron content in differentiating MEL cells (E) and K562 cells (F) by ICP-MS. **P < .01. (G) Flow cytometric analysis showing reduced uptake of Alexa Fluor 488–labeled holo-transferrin (Tf-Alexa 488) in differentiating Grab-deficient MEL cells. *P < .05; **P < .01. (H-I) Supplementation of iron citrate and hinokitiol restored heme synthesis in Grab-deficient MEL cells (H) and K562 cells (I) following chemically induced differentiation. *P < .05; **P < .01. (J) Supplementation of iron citrate and hinokitiol significantly rescued the heme defect induced by rab3il1 depletion. Zebrafish embryos at 72 hours postfertilization were analyzed. **P < .01. DIC, differential interference contrast.
The heme defect exhibited by Grab-deficient erythroblasts is due to reduced iron uptake. (A-B) Representative fluorescence images (A) and quantification (B) of calcein green and RPA in wild-type and Grab knockout MEL cells. N = 30 for each group; **P < .01. (C-D) Representative fluorescence images (C) and quantification (D) of calcein green and RPA in wild-type and GRAB knockout K562 cells. N = 30 for each group; **P < .01. (E-F) Quantification of iron content in differentiating MEL cells (E) and K562 cells (F) by ICP-MS. **P < .01. (G) Flow cytometric analysis showing reduced uptake of Alexa Fluor 488–labeled holo-transferrin (Tf-Alexa 488) in differentiating Grab-deficient MEL cells. *P < .05; **P < .01. (H-I) Supplementation of iron citrate and hinokitiol restored heme synthesis in Grab-deficient MEL cells (H) and K562 cells (I) following chemically induced differentiation. *P < .05; **P < .01. (J) Supplementation of iron citrate and hinokitiol significantly rescued the heme defect induced by rab3il1 depletion. Zebrafish embryos at 72 hours postfertilization were analyzed. **P < .01. DIC, differential interference contrast.
Given that erythroblasts acquire iron primarily though the Tf cycle, we measured the uptake of Tf-iron by incubating cells with the holo-Tf conjugated to the fluorescent dye Alexa 488 (Tf-Alexa 488). We found that Grab knockout MEL cells assimilated significantly less holo-Tf than the wild-type control (Figure 3G). To ascertain that the heme defects observed in Grab-deficient erythroblasts are attributed to the diminished iron uptake, we supplemented the knockout clones with iron and hinokitiol, a small-molecule natural compound known to mediate iron transport independent of the Tf cycle.23 Supplementation of hinokitiol-iron substantially increased the heme production in Grab knockout MEL and K562 cells (Figure 3H-I). Consistently, this hinokitiol-iron supplement significantly rescued the heme defect in rab3il1-morphant zebrafish embryos (Figure 3J). These data suggest that loss of Grab in erythroblasts caused a primary defect in iron assimilation.
Grab regulates the recycling of Tfrc
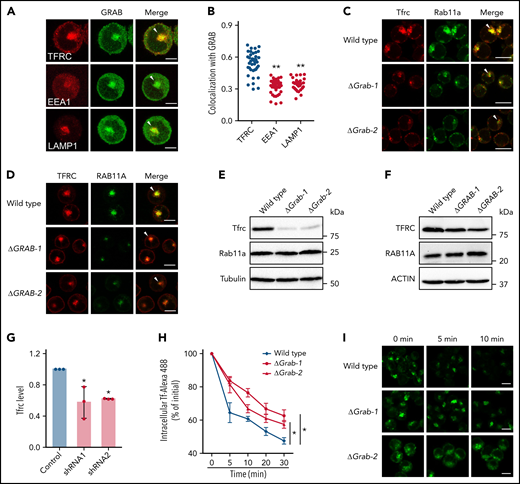
In K562 cells, the receptor responsible for the uptake of Tf-iron, TFRC, mainly localizes to the perinuclear recycling endosomes.32-34 We found that GRAB was also enriched in this juxtanuclear region (Figure 4A). Because the endocytic organelles including early and recycling endosomes and lysosomes are highly clustered in K562 cells,35 we investigated the colocalization between GRAB and the organelle markers TFRC, EEA1, and LAMP1 (Figure 4A). The highest colocalization was observed between GRAB and TFRC, indicating that GRAB preferentially localizes to recycling endosomes in differentiating K562 cells (Figure 4B).
Loss of Grab leads to impaired endocytic recycling of Tfrc. (A) Representative fluorescence images of GRAB and the indicated organelle markers in differentiating K562 cells. Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (B) Quantification of the colocalization between GRAB and the markers in differentiating K562 cells. Pearson's correlation coefficients are shown. N = 30 for each group. **P < .01 compared with the TFRC group. (C-D) Immunofluorescence assays of Tfrc and Rab11a in wild-type and Grab knockout MEL cells (C) and K562 cells (D). Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (E-F) Immunoblotting analyses of Tfrc and Rab11a in wild-type and Grab-deficient MEL cells (E) and K562 cells (F). (G) Quantification of the Tfrc protein abundance in the control and Grab-silencing mouse primary fetal liver erythroblasts. *P < .05. (H-I) Flow cytometric analyses (H) and fluorescence assays (I) showing delayed clearance of internalized Tf-Alexa 488 in Grab knockout cells. Following the internalization of Tf-Alexa 488 for 2 minutes, the cells were transferred to fresh medium without Tf-Alexa 488 and analyzed at indicated time points. *P < .05. Scale bars, 10 μm.
Loss of Grab leads to impaired endocytic recycling of Tfrc. (A) Representative fluorescence images of GRAB and the indicated organelle markers in differentiating K562 cells. Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (B) Quantification of the colocalization between GRAB and the markers in differentiating K562 cells. Pearson's correlation coefficients are shown. N = 30 for each group. **P < .01 compared with the TFRC group. (C-D) Immunofluorescence assays of Tfrc and Rab11a in wild-type and Grab knockout MEL cells (C) and K562 cells (D). Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (E-F) Immunoblotting analyses of Tfrc and Rab11a in wild-type and Grab-deficient MEL cells (E) and K562 cells (F). (G) Quantification of the Tfrc protein abundance in the control and Grab-silencing mouse primary fetal liver erythroblasts. *P < .05. (H-I) Flow cytometric analyses (H) and fluorescence assays (I) showing delayed clearance of internalized Tf-Alexa 488 in Grab knockout cells. Following the internalization of Tf-Alexa 488 for 2 minutes, the cells were transferred to fresh medium without Tf-Alexa 488 and analyzed at indicated time points. *P < .05. Scale bars, 10 μm.
Taking into consideration the role of Grab as a Rab GEF, the predominant localization of Grab to recycling endosomes suggests that it may play a role in endocytic recycling. We therefore examined the localization of Tfrc and found that deletion of Grab resulted in reduced abundance of Tfrc in recycling endosomes (Figure 4C-D). Additionally, the steady-state levels of Tfrc were decreased in both Grab knockout MEL and K562 cells (Figure 4E-F; supplemental Figure 4A-B). This observation was further verified in Grab-deficient mouse primary erythroblasts (Figure 4G). The Tfrc mRNA level was unchanged in Grab knockout cells (supplemental Figure 4C-D), indicating that the reduced Tfrc protein abundance was not attributed to its decreased expression.
To discern whether Grab deficiency caused a specific defect in Tfrc trafficking or a more general defect in endocytic recycling, we examined the localization of 2 recycling endosome markers, Rab11a and Rab35. Rab11a also exhibited diminished localization to the recycling endosomes in Grab knockout cells (Figure 4C-D), but its protein abundance was not reduced (Figure 4E-F). In wild-type MEL and K562 cells, the signal of Rab35 was less intensely concentrated in the perinuclear region than that of Rab11a and Tfrc (supplemental Figure 4E-F), suggesting that it localizes to a distinct population of recycling endosomes. Although GRAB depletion induced a moderate reduction in perinuclear Rab35 staining in K562 cells, it did not alter Rab35 distribution in MEL cells (supplemental Figure 4E-F). These results imply that Grab may regulate the protein trafficking at Rab11+ recycling endosomes.
Analysis of another recycling cargo, LDL receptor, further substantiated the important function of Grab in endocytic trafficking (supplemental Figure 4G-H). To find out whether Grab indeed regulates Tfrc recycling, we measured the exocytosis of the Tf-Tfrc complex by chasing the internalized Tf-Alexa 488. The Grab knockout cells cleared out the internalized Tf-Alexa 488 at much slower rates than the wild-type MEL control (Figure 4H-I). These results suggest that Grab regulates the endosomal trafficking and homeostasis of Tfrc.
Grab promotes the Tfrc recycling via activating Rab8a
To explore the mechanism by which Grab regulates Tfrc trafficking, we characterized the Grab interactome in differentiating MEL cells by affinity purification and mass spectrometry. Grab and its interacting proteins were pulled down from a MEL clone that stably expresses FLAG-tagged Grab using anti-FLAG M2 agarose beads (Figure 5A). Mass spectrometry analysis identified 44 proteins that copurified with FLAG-Grab (supplemental Table 1). Among the list, we identified the small GTPase Rab8a, which has been previously demonstrated to be a substrate for Grab.29-31 Coimmunoprecipitation experiments validated that FLAG-Grab was able to pull down endogenous Rab8a from MEL cell lysates (Figure 5B). To determine which nucleotide-bound state of Rab8a interacts with Grab, we constructed the GTP-locked Rab8a (Rab8a [Q67L]) andthe guanosine diphosphate (GDP)-locked Rab8a (Rab8a [T22N]) with N-terminal hemagglutinin (HA) epitopes. Because the transfection efficiencies of MEL and K562 cells are too low for biochemical assays on exogenous proteins, we transiently expressed these constructs and FLAG-Grab in the HEK293 cell for immunoprecipitation studies. Despite its low expression level in HEK293 cells, Rab8a (T22N) pulled down a considerably higher amount of Grab than that of Rab8a (Q67L) (Figure 5C). This binding preference is consistent with the role of Grab as a Rab8a GEF that binds the GDP-bound Rab8a and catalyzes its conversion into the active GTP-bound form.
Grab regulates the recycling of Tfrc through Rab8a. (A) Schematic of protein purification and mass spectrometry to identify Grab-interacting proteins. (B) Validation of the interaction between FLAG-Grab and Rab8a in MEL cells by coimmunoprecipitation assays. *Indicates immunoglobulin G (IgG). (C) Coimmunoprecipitation assays showing stronger interaction between HA-Rab8a (T22N) and FLAG-Grab in HEK293 cells. Rab8a (Q67L) and Rab8a (T22N) are constitutively GTP-locked and GDP-locked mutant forms of Rab8a, respectively. (D-E) Representative fluorescence images of Rab8a and Tfrc in wild-type and Grab-deficient MEL cells (D) and K562 cells (E). Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (F) Quantification of the colocalization between Rab8a and Tfrc in wild-type and Grab knockout cells. Pearson's correlation coefficients are shown. The numbers of cells analyzed are 23 (wild-type) and 21 (both Grab knockout clones) for MEL and 30 for each of the K562 clones. **P < .01. (G) Immunoblotting analyses of Rab8a and Tfrc in wild-type and Rab8a knockout MEL cells. (H) Quantification of heme content in wild-type and Rab8a-deficient MEL cells following DMSO-induced erythroid-like differentiation. *P < .05. (I) Immunofluorescence analyses of Tfrc in Grab knockout MEL cells transfected with the wild-type (HA-Rab8a) or the constitutively active form (HA-Rab8a [Q67L]) of Rab8a. Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (J) Total heme content in control zebrafish embryos, rab3il1 morphants, and rab3il1 morphants coinjected with rab8a (Q67L) mRNA. The zebrafish embryos were analyzed at 72 hours postfertilization. *P < .05; **P < .01. IB, immunoblotting; IP, immunoprecipitation.
Grab regulates the recycling of Tfrc through Rab8a. (A) Schematic of protein purification and mass spectrometry to identify Grab-interacting proteins. (B) Validation of the interaction between FLAG-Grab and Rab8a in MEL cells by coimmunoprecipitation assays. *Indicates immunoglobulin G (IgG). (C) Coimmunoprecipitation assays showing stronger interaction between HA-Rab8a (T22N) and FLAG-Grab in HEK293 cells. Rab8a (Q67L) and Rab8a (T22N) are constitutively GTP-locked and GDP-locked mutant forms of Rab8a, respectively. (D-E) Representative fluorescence images of Rab8a and Tfrc in wild-type and Grab-deficient MEL cells (D) and K562 cells (E). Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (F) Quantification of the colocalization between Rab8a and Tfrc in wild-type and Grab knockout cells. Pearson's correlation coefficients are shown. The numbers of cells analyzed are 23 (wild-type) and 21 (both Grab knockout clones) for MEL and 30 for each of the K562 clones. **P < .01. (G) Immunoblotting analyses of Rab8a and Tfrc in wild-type and Rab8a knockout MEL cells. (H) Quantification of heme content in wild-type and Rab8a-deficient MEL cells following DMSO-induced erythroid-like differentiation. *P < .05. (I) Immunofluorescence analyses of Tfrc in Grab knockout MEL cells transfected with the wild-type (HA-Rab8a) or the constitutively active form (HA-Rab8a [Q67L]) of Rab8a. Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (J) Total heme content in control zebrafish embryos, rab3il1 morphants, and rab3il1 morphants coinjected with rab8a (Q67L) mRNA. The zebrafish embryos were analyzed at 72 hours postfertilization. *P < .05; **P < .01. IB, immunoblotting; IP, immunoprecipitation.
Given that the GDP-to-GTP switch controls the association of Rab proteins with target membranes, we investigated the impact of Grab deficiency on the cellular localization of Rab8a. In both murine and human erythroleukemia cells, Rab8a strongly colocalized with Tfrc in recycling endosomes (Figure 5D-E). Knockout of Grab resulted in reduced colocalization between Rab8a and Tfrc (Figure 5D-F). Furthermore, quantitative analyses revealed that the enriched localization of Rab8a in the perinuclear recycling endosomes was abolished by Grab depletion (Figure 5D-E; supplemental Figure 5A-B), suggesting that Grab is required for the recruitment of Rab8a to recycling endosomes.
To substantiate the hypothesis that Grab regulates Tfrc recycling through Rab8a, we knocked out Rab8a in the MEL cell (Figure 5G; supplemental Figure 5C-D). Similar to the defects observed in Grab knockout cells, deletion of Rab8a also resulted in reduced Tfrc abundance and heme production (Figure 5G-H; supplemental Figure 5E-F). Additionally, we found that expression of the constitutively active form of Rab8a but not the wild-type Rab8a rescued the Tfrc localization defect in Grab knockout cells (Figure 5I). This rescue was further confirmed in zebrafish embryos in vivo (Figure 5J). These data support a model that Grab regulates Tfrc recycling via activating Rab8a.
Grab is required for the recruitment of Sec15 to recycling endosomes
The exocyst subunit Sec15 has been reported to be a downstream effector for Rab8a.36,37 Because Sec15 is also required for the exocytosis of Tfrc in maturing erythroblasts,17-20 we investigated whether it functions in the same pathway as Grab and Rab8a. In wild-type MEL cells, Sec15 predominantly localizes to recycling endosomes (Figure 6A-B). This localization pattern is largely impaired in both Rab8a- and Grab-deficient cells (Figure 6A-B), indicating that the activation of Rab8a by Grab is required for the recruitment of Sec15 to recycling endosomes.
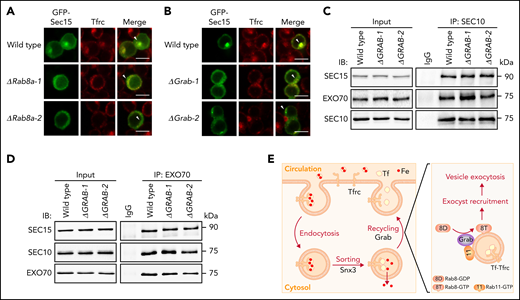
Grab and Rab8a are required for the recruitment of Sec15 to recycling endosomes. (A-B) Representative fluorescence images of GFP-tagged Sec15 and Tfrc in differentiating wild-type, Rab8a knockout (A), and Grab knockout (B) MEL cells. Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (C-D) Analyses of the interaction among the exocyst subunits SEC10, SEC15, and EXO70 in wild-type and GRAB knockout K562 cells by coimmunoprecipitation assays. SEC10 and EXO70 were immunoprecipitated in (C) and (D), respectively. (E) The proposed role of Grab in regulating the exocytosis and recycling of Tfrc in differentiating erythroblasts. IB, immunoblotting; IgG, immunoglobulin G; IP, immunoprecipitation.
Grab and Rab8a are required for the recruitment of Sec15 to recycling endosomes. (A-B) Representative fluorescence images of GFP-tagged Sec15 and Tfrc in differentiating wild-type, Rab8a knockout (A), and Grab knockout (B) MEL cells. Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (C-D) Analyses of the interaction among the exocyst subunits SEC10, SEC15, and EXO70 in wild-type and GRAB knockout K562 cells by coimmunoprecipitation assays. SEC10 and EXO70 were immunoprecipitated in (C) and (D), respectively. (E) The proposed role of Grab in regulating the exocytosis and recycling of Tfrc in differentiating erythroblasts. IB, immunoblotting; IgG, immunoglobulin G; IP, immunoprecipitation.
Within the exocyst complex, Sec15 forms a stable pair with another subunit Sec10, and this pair associates with the Exo70-Exo84 pair to assemble into a subcomplex.14-16 To find out whether the activation of Rab8a promotes the association of Sec15 with other exocyst subunits, we investigated its interactions with Sec 10 and Exo70. Knockout of GRAB did not interfere with the interactions among SEC15, SEC10, and EXO70 in K562 cells (Figure 6C-D; supplemental Figure 6A-B). We were also able to analyze the interaction between Sec10 and Exo70 in MEL cells, and similarly, we found that their interaction was not perturbed by Grab depletion (supplemental Figure 6C-F). Accordingly, it is more likely that Grab regulates the recruitment of the exocyst complex to recycling endosomes in erythroblasts.
Discussion
In this study, we demonstrate that Grab is a key regulator of Tfrc trafficking and iron metabolism in mammalian erythropoiesis. The loss of Grab impairs Tfrc recycling, leading to iron deficiency and hemoglobinization defects. In developing erythroblasts, Grab functions as a GEF to activate Rab8a, which in turn promotes the exocytosis and recycling of Tfrc (Figure 6E).
Differentiating erythroblasts rely on the Tf cycle to acquire iron for the synthesis of large amounts of heme and hemoglobin. This iron uptake cycle requires the efficient recycling of Tfrc, which involves endosomal sorting and exocytosis. We have previously demonstrated that the retromer protein Snx3 regulates the sorting of Tfrc into recycling endosomes.4 Previous studies on the hemoglobin deficit (hbd) mouse have revealed that the rapid recycling of Tfrc also requires the exocyst protein Sec15.17-20 Here, we demonstrate that developing red cells upregulate the expression of Grab, which acts as a GEF to activate Rab8a. The active, GTP-bound Rab8a promotes the recruitment of the exocyst to recycling endosomes, leading to enhanced exocytosis of Tfrc (Figure 6E). Thus, the induction of Grab is crucial for enhancing Tfrc recycling to meet the increased demand for iron assimilation during erythroid differentiation.
Grab has been reported to be a GEF for Rab3 and Rab8.28-31,38 Here, we demonstrate that in developing erythroblasts, Grab regulates the Tfrc recycling via activating Rab8. During the exocyst-mediated exocytosis, Rab8-GTP is known to recruit the exocyst by interacting with its effector Sec15.36,37,39,40 Our data suggest that the activation of Rab8a by Grab may facilitate Tfrc exocytosis by recruiting Sec15 to the perinuclear recycling endosomes. Mutations of the exocyst genes are known to cause accumulation of secretory vesicles within cells.13,41,42 Interestingly, we observed a decreased population of perinuclear recycling endosomes in Grab knockout erythroid cells. A similar phenomenon has also been previously observed in RAB8-depleted HeLa cells.43 Because Rab8 acts downstream of Rab11,31,44,45 it may regulate the maintenance, but not the biogenesis, of recycling endosomes. Overexpression of Sec15 has been shown to enhance Rab11+ recycling endosomes in the perinuclear region,37,46 supporting a notion that Sec15 may be involved in Rab8-mediated regulation of recycling endosomes. However, there is also a possibility that Grab regulates recycling endosomes through an exocyst-independent mechanism. In that case, the impaired localization of Sec15 to the perinuclear region in Grab-deficient cells could be an indirect outcome of fewer perinuclear recycling endosomes. More research is needed to investigate how Grab regulates the population of recycling endosomes.
Mammals have 2 GEFs for Rab8, Grab and Rabin8, but only the expression of Grab is induced during erythropoiesis. Analysis of chromatin immunoprecipitation sequencing data revealed that Grab is a target of the hematopoietic transcription factors Gata1 and Tal1 (supplemental Figure 7A), both of which bind to the promoter region of Grab in erythroid cells.47,48 Within the cell, Grab protein predominantly localizes to recycling endosomes (Figure 4A). This localization may be orchestrated by its interaction with the recycling endosome protein Rab11.31,49 Additionally, we found that Grab also interacts with Tfrc (supplemental Figure 7B), suggesting that Tfrc may assist in the recruitment of Grab to the recycling endosomes in differentiating erythroblasts.
The regulation of Tfrc recycling by the Grab-Rab8-exocyst cascade is critical for the hemoglobin synthesis and red cell maturation. The loss of Grab in mammalian hematopoietic cells results in reduced iron uptake and hemoglobin production. In humans, genome-wide association studies have revealed strong association between the erythrocyte hemoglobin level and the single nucleotide polymorphisms of both GRAB (rs1151139) and RAB8A (rs1043407, rs17722795, rs200560717, rs61630159, rs62117911, rs73928741) (supplemental Table 2).50-53 Additionally, the most evident phenotype observed in hbd mice was hypochromic microcytic anemia.17,54 According to these observations, defects in any of the genes in this trafficking cascade, including GRAB, RAB8A, SEC15, and SEC10, are likely to cause hematologic disorders in humans. Thus, the findings of this work not only offer new insights into mammalian iron metabolism and erythropoiesis but may also provide a genetic basis for understanding related blood disorders such as congenital hypochromic anemias.
Acknowledgments
This work was supported by funding from the National Key Research and Development Program of China (2018YFA0507802 to C.C.), the Zhejiang Natural Science Foundation (LR17C110001 to C.C.), and the National Natural Science Foundation of China (31871200, 31930003, and 32071155 to C.C.).
Authorship
Contribution: C.C. conceived the project; M.C. and C.C. wrote the manuscript; M.C. performed most of the experiments and analyzed the data; Y.Z. and K.J. contributed to immunoprecipitation, immunofluorescence, qPCR, and zebrafish experiments; W.W., R.Z., and C.M. performed the primary erythroblast experiments; H.F. contributed to zebrafish experiments; Z.Z. generated the Rab8a knockout cells; J.S. contributed to the primary erythroblast experiments and edited the manuscript; and all authors discussed the results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Caiyong Chen, College of Life Sciences, Zhejiang University, 866 Yuhangtang Rd, Hangzhou 310058, China; e-mail: chency@zju.edu.cn.
Send data sharing requests via e-mail to the corresponding author.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
REFERENCES
Author notes
M.C., Y.Z., and K.J. contributed equally to this study.


![Grab regulates the recycling of Tfrc through Rab8a. (A) Schematic of protein purification and mass spectrometry to identify Grab-interacting proteins. (B) Validation of the interaction between FLAG-Grab and Rab8a in MEL cells by coimmunoprecipitation assays. *Indicates immunoglobulin G (IgG). (C) Coimmunoprecipitation assays showing stronger interaction between HA-Rab8a (T22N) and FLAG-Grab in HEK293 cells. Rab8a (Q67L) and Rab8a (T22N) are constitutively GTP-locked and GDP-locked mutant forms of Rab8a, respectively. (D-E) Representative fluorescence images of Rab8a and Tfrc in wild-type and Grab-deficient MEL cells (D) and K562 cells (E). Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (F) Quantification of the colocalization between Rab8a and Tfrc in wild-type and Grab knockout cells. Pearson's correlation coefficients are shown. The numbers of cells analyzed are 23 (wild-type) and 21 (both Grab knockout clones) for MEL and 30 for each of the K562 clones. **P < .01. (G) Immunoblotting analyses of Rab8a and Tfrc in wild-type and Rab8a knockout MEL cells. (H) Quantification of heme content in wild-type and Rab8a-deficient MEL cells following DMSO-induced erythroid-like differentiation. *P < .05. (I) Immunofluorescence analyses of Tfrc in Grab knockout MEL cells transfected with the wild-type (HA-Rab8a) or the constitutively active form (HA-Rab8a [Q67L]) of Rab8a. Arrowheads indicate recycling endosomes. Scale bars, 10 μm. (J) Total heme content in control zebrafish embryos, rab3il1 morphants, and rab3il1 morphants coinjected with rab8a (Q67L) mRNA. The zebrafish embryos were analyzed at 72 hours postfertilization. *P < .05; **P < .01. IB, immunoblotting; IP, immunoprecipitation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/10/10.1182_blood.2021015189/3/m_bloodbld2021015189f5.png?Expires=1765970490&Signature=ZIRCanT7uVHiZYjgUcQGOIqRvSnFuPjxWgRukbtj41mNMuQfjWOFQg39iEQ1P37qBfRFifwCf~Du8K5MU2N6eMFfnl5CoPpgrOC5v~C7MSNSS3Xnh4lgNHd-KoYnBQ95x87EJsXfvkEZKuARpnfuy9tqMdItUcxrzbEyyS8y1k1FU4UrcDwtd8sCoX-jgMVNGxa0C3CmQLQe7BANqm3qVDW3T77qz5qnLhnsN3i79SKWIX~ouHJetnx5hueIAeK~17MRrNGdURPol6z1tqH4~QczTnBUQOjx76jbUsAq9nbIZcI6O-YcKxKo93ewi07za9TJoOs7AgyHQv-nVrGdzA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)