In this issue of Blood, Shen et al from the Ghobrial group, describe a study using a multicolor fluorescent tag or a DNA barcode system to enable the tracking of individual multiple myeloma tumor cells in a sophisticated mouse xenograft model.1 The authors use this model to gain insight into the mechanisms that underpin tumor dissemination and clonal outgrowth in myeloma, features that are critical in multiple myeloma disease progression.
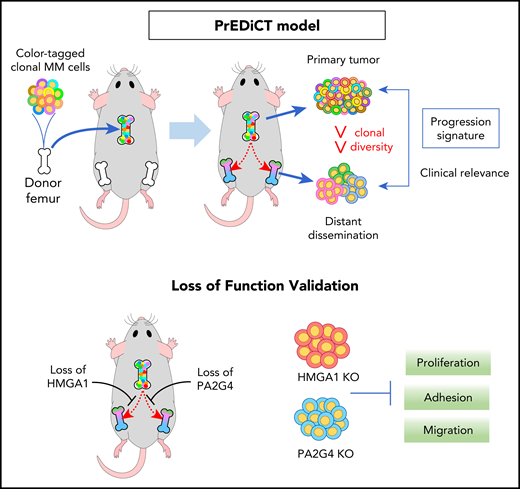
In the MM PrEDiCT (Progression through Evolution and Dissemination of Clonal Tumor Cells) model, human myeloma cell lines are injected into an isolated mouse bone, which is then implanted subcutaneously into an immunodeficient SCID-beige mouse. The implanted bone graft provides the crucial bone marrow microenvironment for the growth of the myeloma tumor cells in vivo. The labeling of individual myeloma tumor cells, with either a fluorescent tag system or via lentiviral DNA barcoding, enables tracking of the growth and dissemination of individual tumor cells and their progeny in an in vivo setting. Gene expression analysis of the primary and disseminated tumor in this model can reveal clinically relevant genes that may play an important role in the pathogenesis of myeloma. See the visual abstract in the article by Shen et al that begins on page 2360.
In the MM PrEDiCT (Progression through Evolution and Dissemination of Clonal Tumor Cells) model, human myeloma cell lines are injected into an isolated mouse bone, which is then implanted subcutaneously into an immunodeficient SCID-beige mouse. The implanted bone graft provides the crucial bone marrow microenvironment for the growth of the myeloma tumor cells in vivo. The labeling of individual myeloma tumor cells, with either a fluorescent tag system or via lentiviral DNA barcoding, enables tracking of the growth and dissemination of individual tumor cells and their progeny in an in vivo setting. Gene expression analysis of the primary and disseminated tumor in this model can reveal clinically relevant genes that may play an important role in the pathogenesis of myeloma. See the visual abstract in the article by Shen et al that begins on page 2360.
Tumor dissemination is a key process in the development of multiple myeloma (MM), with one of the defining diagnostic features of the disease being the presence of multiple tumors that have spread to sites throughout the skeleton. This dissemination is thought to occur through a process of hematogenous dissemination, with tumor cells that have become established at one site in the bone marrow migrating into the blood stream and homing and establishing at a distant bone marrow site. This process is important in both the initial development of myeloma and in disease relapse, with elevated circulating myeloma cell numbers being associated with more rapid progression to myeloma, accelerated disease relapse following therapy and poorer overall survival in patients.
The factors that facilitate the homing of myeloma tumor cells from the circulation into the bone marrow are relatively well-characterized. They include adhesion molecules, such as integrins, CD44 and N-cadherin, which enable binding to vascular endothelial cells, and the CXCL12/CXCR4 chemokine axis which drives migration from the blood stream into the bone marrow. However, a lack of well-established models for the investigation of spontaneous dissemination in myeloma has limited progress in the identification of factors that regulate the egress of myeloma tumor cells from the bone marrow. Notably, very few genes have been shown to regulate the spontaneous dissemination of myeloma tumor cells in vivo.2-4
In this study, Shen et al have used a novel tumor xenograft model to track the fate of individual tumor cells, or clones, in the primary tumor and during subsequent dissemination via the circulation to the host bone marrow (see figure). These experiments demonstrated clonal competition in the primary tumor that was microenvironment specific. Although the tumor cells had similar abilities to proliferate in vitro, the in vivo microenvironment applied selective pressures that enabled only some clones to grow and contribute to the subsequent tumor. In addition, it was evident that the subclonal diversity was further reduced, both in the circulation and at secondary bone marrow sites compared with the primary tumor. This reduction suggests multiple bottlenecks in the seeding and dissemination process, with mobilization of tumor cells to the peripheral circulation and seeding at secondary sites providing sequential selective pressures on the tumor. These results support previous findings of other groups, including our own, that show that the establishment of myeloma tumor cells in the bone marrow after IV injection is highly inefficient, with very few cells proliferating and contributing to the final tumor, despite a large number of single cells reaching and surviving in the bone marrow.5,6 Notably, the current study is the first to demonstrate in a spontaneous metastasis model that these bottlenecks in the tumor establishment and dissemination process also occur at the point of mobilization of the tumor cells from the bone marrow and their subsequent survival in the circulation.
A question that remains is whether the clonal selection observed by Shen et al reflects a cell intrinsic process, an adaptation to the bone marrow microenvironment, or a largely stochastic process. Notably, mobilization of the myeloma cells from the primary tumor in the model is specific to the implanted bone microenvironment, as subcutaneously injected MM.1S cells failed to disseminate despite being able to grow without the supportive niche. This therefore suggests that the bone niche provides a selective or supportive environment, critical for the dissemination process. Importantly, previous studies from the Ghobrial group suggest that bone marrow hypoxia is an important driver of MM tumor dissemination in vivo, through modulation of adhesion and response to chemokines.7
The distinct spatial distribution of growing clones observed at secondary sites suggests the existence of specific bone marrow niches that support outgrowth, a phenomenon previously described by Lawson et al.6 These studies demonstrated distinct differences in niches that support myeloma cell growth and dormancy, with dormant cells being largely localized close to endosteal bone surfaces. In patients, the immune microenvironment also plays a critical role in modulating myeloma tumor cells growth, a feature that is not modeled in the immunodeficient SCID-beige mice used by Shen et al. That said, previous studies using the syngeneic Vk*Myc and 5T/KaLwRij mouse models report similar bottlenecks in seeding, bone marrow growth, and dissemination in mouse models with an intact immune system.5,6
Shen et al performed a transcriptomic analysis of the primary and disseminated tumors, identifying a gene expression signature that was associated with dissemination and that may reflect adaptation to the tumor microenvironment. Identified genes included those with known roles in myeloma, such as MYC and CDKN2A, and potential novel contributors to myeloma pathogenesis, HMGA1 and PA2G4. Although Shen et al focused on gene signatures that are important for myeloma cell growth, these signatures provide an important proof of concept in the utility of this model for the identification of novel genes and pathways that may play a role in the dissemination process. Notably, the identified gene signatures were also validated in patient data sets as being associated with overall survival, demonstrating the potential for this model to identify clinically relevant genes that play a role in the pathophysiology of this disease.
Intratumoral subclonal heterogeneity is an established feature in patients with MM, with emerging data revealing both spatial8 and temporal9,10 variations in subclonal diversity. The PrEDiCT model provides a tool to validate candidate genetic or epigenetic variants associated with subclone expansion and dissemination and paves the way for future investigations of how the cells and factors that make up the bone marrow niches contribute to this process, potentially revealing novel therapeutic targets.
Conflict-of-interest disclosure: The author declares no competing financial interests.