Key Points
The HLA-B leader dimorphism informs survival after unrelated donor HCT.
The risks associated with HLA mismatching depend on the HLA-B leader genotype.
Abstract
Hematopoietic cell transplantation (HCT) from HLA-mismatched unrelated donors can cure life-threatening blood disorders, but its success is limited by graft-versus-host disease (GVHD). HLA-B leaders encode methionine (M) or threonine (T) at position 2 and give rise to TT, MT, or MM genotypes. The dimorphic HLA-B leader informs GVHD risk in HLA-B–mismatched HCT. If the leader influences outcome in other HLA-mismatched transplant settings, the success of HCT could be improved for future patients. We determined leader genotypes for 10 415 patients receiving a transplant between 1988 and 2016 from unrelated donors with one HLA-A, HLA-B, HLA-C, HLA-DRB1, or HLA-DQB1 mismatch. Multivariate regression methods were used to evaluate risks associated with patient leader genotype according to the mismatched HLA locus and with HLA-A, HLA-B, HLA-C, HLA-DRB1, or HLA-DQB1 mismatching according to patient leader genotype. The impact of the patient leader genotype on acute GVHD and mortality varied across different mismatched HLA loci. Nonrelapse mortality was higher among HLA-DQB1–mismatched MM patients compared with HLA-DQB1–mismatched TT patients (hazard ratio, 1.35; P = .01). Grades III to IV GVHD risk was higher among HLA-DRB1–mismatched MM or MT patients compared with HLA-DRB1–mismatched TT patients (odds ratio, 2.52 and 1.51, respectively). Patients tolerated a single HLA-DQB1 mismatch better than mismatches at other loci. Outcome after HLA-mismatched transplantation depends on the HLA-B leader dimorphism and the mismatched HLA locus. The patient’s leader variant provides new information on the limits of HLA mismatching. The success of HLA-mismatched unrelated transplantation might be enhanced through the judicious selection of mismatched donors for a patient’s leader genotype.
Introduction
The HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-DPB1 genes comprise the hematopoietic cell transplantation (HCT) barrier.1-3 In HCT from unrelated donors, precise donor matching of the HLA peptide-binding region (PBR) is performed to achieve the degree of compatibility that is feasible between HLA-genotypically identical siblings.4-7 When HLA-matched donors are not available, mismatched donors with PBR differences that are better tolerated offer options for many patients.8,9
HLA matching of the PBR does not include consideration of other regions of the HLA gene that could harbor clinically relevant variation. HLA-B exon 1 is one such example wherein a sequence dimorphism at −21 gives rise to leader peptides with either methionine (M) or threonine (T) at the second position of the leader. M leaders promote higher HLA-E expression than T leaders, favoring robust T- and natural killer (NK) cell recognition of HLA-E, a mechanism that might control the progression of HIV infection.10-15 In HLA-B–mismatched unrelated HCT, the risk of acute graft-versus-host disease (GVHD) is higher when the patient has HLA-B M leaders and when the mismatched patient/donor HLA-B allotypes have different leaders.7 The data provide an approach for understanding permissive and nonpermissive HLA-B mismatches. However, whether HLA-B mismatching is necessary for the leader to have an effect on transplant outcome is unknown. The HLA-B leader is part and parcel of the extended HLA haplotype. If the leader in fact provides information on transplant outcome after unrelated donor transplantation in the setting of HLA-B matching but HLA-A, HLA-C, HLA-DRB1, or HLA-DQB1 mismatching, the leader might then provide an entirely novel approach for the selection of HLA-mismatched donors and inform risks that have historically been ascribed to the HLA mismatch itself.
Given that no genetic data are currently available to aid the prioritization of mismatched donors, a leader-based algorithm would fulfill an unmet need. Beyond the immediate clinical implications of the leader for donor selection, the information will advance understanding of the pathways involved in graft-versus-host allorecognition in transplantation.
Methods
Study population and design
We conducted a retrospective cohort analysis of 10 415 patients who received a transplant from an unrelated donor with one HLA-A, HLA-C, HLA-DRB1, or HLA-DQB1 mismatch between 1988 and 2016 and whose HLA and clinical data were contributed by members of the International Histocompatibility Working Group in Hematopoietic Cell Transplantation (IHWG) (supplemental Tables 1 and 2, available on the Blood Web site). There were no exclusion criteria. The transplants included 3399 HLA-A–, 3851 HLA-C–, 823 HLA-DRB1–, and 2342 HLA-DQB1–mismatched pairs. Their outcomes for GVHD, relapse, and mortality were compared vs a previous cohort of 1457 HLA-B mismatches.7
HLA
HLA-A, HLA-B, HLA-C, HLA-E, HLA-DRB1, HLA-DQB1, and HLA-DPB1 were typed as previously described.7 The leader genotype was determined from the exon 1 gene sequence for all HLA-B alleles in the study population with the exception of HLA-B*18:05 and B*39:20 present in 2 individuals. Average HLA-A and HLA-C expression, killer immunoglobulin-like receptor ligand, and donor-recipient HLA-DPB1 mismatching were determined as previously described.15-17
Protocols were approved by the institutional review boards of the National Institutes of Health, Office for Human Research Protections, and each participating IHWG center. Research was conducted in accordance with the Declaration of Helsinki.
Statistical analysis
We examined the association of leader genotype and HLA mismatching with acute GVHD (grades II-IV and III-IV), chronic GVHD, relapse, death not preceded by relapse, disease-free survival, and overall mortality. The leader effect in HLA-B–mismatched transplantation was previously reported for GVHD risk, and the 1457 HLA-B–mismatched transplants were included for comparison with single HLA-A–, HLA-C–, HLA-DRB1–, and HLA-DQB1–mismatched transplants.7 Statistical tests for interaction were performed comparing the leader effect among single HLA-B mismatches vs the leader effect among single mismatches at HLA-A, HLA-C, HLA-DRB1, and HLA-DQB1. Tests for interaction were also performed comparing the leader effect among allele and antigen mismatches. Cox regression models were fit to compare the hazards of failure between appropriate groups for all end points other than acute GVHD, where logistic regression was used. Regression models were adjusted for patient age, donor age, source of cells, disease status, T-cell depletion, transplant type, use of total body irradiation, patient sex, donor sex, cytomegalovirus serologic status, patient race, donor race, HLA-DPB1 match status, average HLA-A and HLA-C expression, patient HLA-E genotype, donor killer immunoglobulin-like receptor, and year of transplantation (supplemental Table 2). To assess relapse, GVHD was modeled as a time-dependent covariate (ie, GVHD status is allowed to change after transplantation). Two-sided P values from regression models were obtained from the Wald test, and values <.05 were considered significant. Several comparisons were made, all focused on refinements of the concept of leader genotype and leader matching. The outcomes examined are highly correlated, minimizing the effect of multiple comparisons that result from the various outcomes. For this reason, no adjustments were made to the P values associated with the fitted regression models. All analyses were performed by using R version 3.4.1 and survival package in R.
Results
Patient leader genotype and GVHD
We hypothesized that acute GVHD risk correlates with patient M leaders after single HLA-A–, HLA-C–, HLA-DRB1–, or HLA-DQB1–mismatched HCT, an association that we observed after single HLA-B–mismatched HCT.7 The 10 415 HLA-B–matched pairs with one HLA-A, HLA-C, HLA-DRB1-, or HLA-DQB1 mismatch were therefore analyzed alongside the 1457 previously studied single HLA-B–mismatched pairs.7 The current study pairs are all HLA-B matched, and the patient and donor necessarily have the same leader genotype. Their leader frequencies (55.7% [5802 of 10 415], TT; 38.1% [3970 of 10 415], MT; and 6.2% [643 of 10 415], MM) were similar to those of HLA-B–mismatched patients and donors and of other populations.7,18 Also similar to HLA-B mismatches was the association of patient E*01:03 with lower disease-free survival and higher relapse (supplemental Table 3).7
In contrast to the association of patient leader genotype with grades III to IV acute GVHD among single HLA-B mismatches (odds ratio [OR], 2.26; 95% confidence interval [CI], 1.09-4.66; P = .03 for MM; OR, 1.41; 95% CI, 0.97-2.04; P = .07 for MT relative to TT), patient leader genotype did not affect the risk of grades III to IV acute GVHD among transplants with one HLA-A, HLA-C, HLA-DRB1, or HLA-DQB1 mismatch (OR, 1.08; 95% CI, 0.82-1.42; P = .59 for MM; OR, 1.05; 95% CI, 0.92-1.20; P = .43 for MT relative to TT). These results show that the effect of the patient’s leader genotype on acute GVHD risk is not the same among HLA-B–matched transplants as among HLA-B–mismatched transplants. A formal statistical test of interaction yielded P = .009, which indicates that the ORs of 2.26 and 1.08 (MM vs TT for HLA-B mismatches and mismatches other than HLA-B, respectively) and 1.41 and 1.05 (MT vs TT for HLA-B mismatches and mismatches other than HLA-B) are statistically different. The effect of the leader on acute GVHD among allele mismatches was not statistically different from the effect among antigen mismatches (interaction P = .33).
These results suggest that the clinical significance of the HLA-B leader may depend on the mismatched HLA locus. We therefore examined the impact of a patient’s leader genotype on other outcomes among single mismatches at loci other than HLA-B. Relative to TT patients, MM patients had modest increases in mortality, failure (relapse or death) for disease-free survival, relapse, and chronic GVHD (Table 1). Disease-free survival decreased with increasing numbers of M leaders, suggesting a biological step-up of risk from TT to MT to MM. In summary, the MM genotype is associated with higher risks of certain outcomes after HLA-A–, HLA-C–, HLA-DRB1–, or HLA-DQB1–mismatched transplantation compared with TT. Whereas the leader affects primarily GVHD in HLA-B–mismatched HCT, the leader may influence mortality and relapse after HLA-A–, HLA-C–, HLA-DRB1–, or HLA-DQB1–mismatched HCT despite little influence on grades III to IV GVHD.
Leader genotype and the mismatched HLA locus
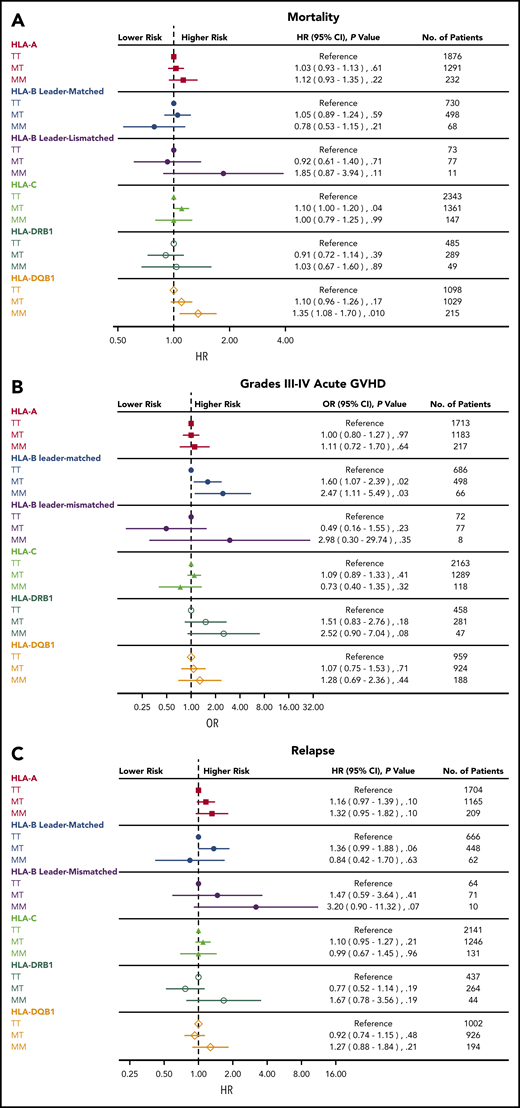
The results described here assume that the impact of the patient’s leader genotype on outcome is the same across all loci outside of HLA-B. For example, the risk of acute GVHD for HLA-A–mismatched MM patients relative to HLA-A–mismatched TT patients is assumed to be identical to the risk of acute GVHD for HLA-C–mismatched MM patients relative to HLA-C–mismatched TT patients. We formally tested whether the association of the patient’s leader genotype with outcome is similar across the different mismatched loci (statistical test of interaction). In fact, they were not the same for mortality, grades III to IV acute GVHD, or relapse (P = .06, .06 and .10, respectively). This result warrants analyses of the leader for each mismatched locus (Figure 1).
Association of patient leader genotype with clinical outcome. Risks of mortality (A), grades III to IV acute GVHD (B), and relapse (C) associated with patient leader genotypes according to mismatched HLA locus.
Association of patient leader genotype with clinical outcome. Risks of mortality (A), grades III to IV acute GVHD (B), and relapse (C) associated with patient leader genotypes according to mismatched HLA locus.
Overall mortality was adversely affected by MM relative to the TT genotype primarily when the mismatch was at HLA-DQB1 (hazard ratio [HR], 1.35 [95% CI, 1.08-1.70; P = .01] for MM patients compared with TT patients). In addition, MT patients undergoing HLA-C–mismatched transplantation had a modest increase in mortality compared with HLA-C–mismatched TT patients (HR, 1.10; 95% CI, 1.00-1.21; P = .04). The risk of relapse was generally consistently higher for MM patients than for TT patients (but not statistically significant) when the mismatch was at HLA-A (HR, 1.32; P = .10), HLA-DRB1 (HR, 1.67; P = .19), or HLA-DQB1 (HR, 1.27; P = .21). The risk of grades III to IV GVHD was higher for MM patients compared with TT patients after HLA-B–mismatched leader-matched transplantation (OR, 2.47; 95% CI, 1.11-5.49; P = .03) as shown previously,7 but this detrimental effect of MM genotype on acute GVHD was not observed with other mismatched loci with the possible exception of mismatches at HLA-DRB1 (OR, 2.52; 95% CI, 0.90-7.04; P = .08). These results suggest that the risks of mortality, GVHD, and relapse associated with M leaders vary with the specific mismatched HLA locus.
Impact of HLA mismatching on outcome
In current clinical practice, mismatched donors are selected according to the mismatched locus4,5,7 ; however, the results described here show that risks of specific locus mismatches vary according to the patient’s leader genotype. Among all TT patients, mismatching at HLA-A, HLA-B, HLA-C, and HLA-DRB1 each increased mortality and grades III to IV GVHD relative to HLA-DQB1 mismatches (Figure 2). These results show that among TT patients, HLA-DQB1 mismatches are associated with lower risks than mismatches at other loci. Among all MT patients, the risks of mortality, acute GVHD, and relapse were each increased with mismatching at all loci relative to HLA-DQB1; one possible exception was mismatches at HLA-DRB1, where mortality and relapse seem to be similar to those for HLA-DQB1 mismatches. In contrast to TT and MT patients, there was little suggestion that MM patients had an increased risk of mortality for any mismatch relative to HLA-DQB1 mismatches, with the possible exception of mismatching at HLA-A (HR, 1.25) and HLA-B leader mismatching (HR, 1.94) (Figure 2). Conversely, grades III to IV GVHD were increased in MM patients at all mismatched loci relative to HLA-DQB1 with the possible exception of HLA-C.
Association of HLA mismatching with clinical outcome. Risks of mortality (A), grades III to IV acute GVHD (B), and relapse (C) associated with one HLA-A, HLA-B, HLA-C, or HLA-DRB1 mismatch relative to one HLA-DQB1 mismatch among patients with TT, MT, and MM leader genotypes.
Association of HLA mismatching with clinical outcome. Risks of mortality (A), grades III to IV acute GVHD (B), and relapse (C) associated with one HLA-A, HLA-B, HLA-C, or HLA-DRB1 mismatch relative to one HLA-DQB1 mismatch among patients with TT, MT, and MM leader genotypes.
In summary, the risks of mortality, GVHD, and relapse associated with HLA mismatching vary with the HLA-B leader genotype of the patient. For a given patient HLA-B leader genotype, there is a hierarchy of donors with preferred and nonpreferred HLA mismatches (Figure 2). Preference for HLA-DQB1–mismatched donors and avoidance of HLA-B–mismatched leader-mismatched donors may lower the risk of acute GVHD and improve survival without increasing relapse.
Acute GVHD and relapse according to leader genotype and mismatched HLA locus
Patients who develop clinical acute GVHD may have lower risk of relapse compared with patients without GVHD.19,20 Given the differential effect of the HLA-B leader on relapse and acute GVHD according to the mismatched HLA locus, we assessed the impact of acute GVHD on the risk of relapse and whether risk varies according to the leader or the mismatched HLA locus. Modeling GVHD as a time-dependent covariate, the occurrence of grades II to IV GVHD did very little to lower the risk of relapse (HR, 0.96; 95% CI, 0.87-1.07; P = .47). Moreover, there was little suggestion that this effect varied across different mismatched loci (interaction P = .17) and even less suggestion that the effect differed according to the patient’s leader genotype (interaction P = .74). According to the specific mismatched locus, the hazards of relapse with grades II to IV GVHD vs without grades II to IV GVHD were 0.90 (95% CI, 0.74-1.10; P = .31), 1.01 (95% CI, 0.74-1.38; P = .94), 1.04 (95% CI, 0.87-1.25; P = .67), 1.58 (95% CI, 0.98-2.55; P = .06), and 0.87 (95% CI, 0.68-1.12; P = .28) for mismatches at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1, respectively. For the leader genotypes, the hazards of relapse were 0.98 (95% CI, 0.85-1.14; P = .80), 0.96 (95% CI, 0.81-1.13; P = .61), and 0.85 (95% CI, 0.56-1.28; P = .43) for the TT, MT, and MM genotypes. In summary, MM patients have increased risk of both relapse and GVHD compared with TT patients. These results suggest a role for HLA-B M leaders in the pathways involved in relapse and GVHD after transplantation.
Discussion
Historically, 40% of unrelated donor transplantations performed worldwide have used HLA-matched unrelated donors as the stem cell source, 38% of whom were unrelated donors with one mismatch, and the remainder of patients received a transplant from donors with more than one mismatch.7 An unmet need in unrelated HCT is a better understanding of the features of HLA mismatches associated with increased transplant-related complications and lower survival. If the rules that govern (non)-permissive HLA mismatches were known, this information could be used to judiciously select donors with acceptable mismatches, thereby broadening the availability of transplantation as a curative therapy.
Most approaches for understanding permissive HLA mismatches have focused on sequence polymorphisms that reside within the PBR of HLA molecules because of its role in antigen presentation and T-cell recognition. HLA-A and HLA-B mismatches are associated with some of the highest risks.7 The polymorphic HLA-B leader peptide that binds and stabilizes HLA-E, primarily when methionine is present at position 2 of the peptide, affects in vitro recognition of HLA-E by NK and T cells12,13 but is not currently assessed in patients and candidate unrelated donors for transplantation. The fact that the patient’s HLA-B leader genotype can be used to identify permissive HLA-B mismatches7 provided strong rationale to conduct the current study, directed at the clinical impact of the leader genotype in the context of mismatches at other HLA class I and II loci.
The current study was designed to test hypotheses concerning the clinical significance of the HLA-B leader dimorphism in HLA-B–matched but HLA-A–, HLA-C–, HLA-DRB1–, or HLA-DQB1–mismatched unrelated donor transplantation. The hypotheses were developed to address whether HLA-B mismatching is required for there to be important consequences of the leader dimorphism on transplant outcome. Inherent in this hypothesis is the concept that should the leader, in fact, inform transplant outcome, the information could have major implications on our understanding of the risks associated with HLA-A, HLA-C, HLA-DRB1, and HLA-DQB1 mismatching, the key criteria for the selection of unrelated donors. We tested these hypotheses in a large cohort of unrelated donor transplants with one HLA-A, HLA-C, HLA-DRB1, or HLA-DQB1 mismatch performed by the international community and uncovered a new relationship between the HLA-B leader dimorphism and relapse and survival. In contrast to the association of the leader genotype with GVHD after HLA-B–mismatched transplantation, we found that the leader genotype may primarily affect survival and relapse after HLA-A–, HLA-C–, HLA-DRB1–, or HLA-DQB1–mismatched transplantation. Knowledge of the leader’s effects on outcome clarified the significance of mismatching for the classic HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 genes. Of the five loci currently assessed in donors before transplantation, HLA-DQB1 mismatches were the best tolerated, particularly among TT and MT patients. The relative (dis)advantages associated with HLA-A, HLA-C, HLA-B, HLA-DRB1, or HLA-DQB1 mismatching are less evident for MM patients who benefit from complete HLA matching. Thus, a new paradigm for the selection of HLA-mismatched unrelated donors for future patients may include consideration of the patient’s leader genotype.
Alloreactive donor T cells cause GVHD, and through the course of mounting graft-versus-host responses, they are cytotoxic against residual patient cancer cells, leading to lowered relapse (“graft-versus-leukemia”).19,20 Interestingly, MM patients had higher risk of relapse compared with TT patients, suggesting a potential role for NK cells in relapse in these patients. Of note, compared with MM patients without GVHD, MM patients with GVHD had a hazard of 0.84 of relapse. Although not statistically significant, the extent to which this might reflect classic graft-versus-leukemia and whether a deleterious MM-associated effect could “neutralize” a beneficial T-cell–associated effect, remain to be determined in the future when a larger transplant experience is available.
The foundation of donor selection for HCT rests on precise matching of exons 2 and 3 of class I and exon 2 of class II HLA genes to minimize patient/donor differences within the PBR.21 The current study confirms that HLA-B exon 1 harbors clinically relevant variation, regardless of matching or mismatching with donor, highlighting the importance of variation outside of regions that are traditionally tested, which may affect transplant outcome in a manner distinct from mismatching within the PBR.22 Dissection of the effects associated with the leader genotype from those associated with HLA mismatching of the mature protein provides new information on the biology of GVHD. The novel association between the leader dimorphism in survivorship after transplantation suggests that prospective consideration of the patient’s HLA-B leader may enhance the selection of mismatched donors for transplantation and provide important information for assessment of risks pretransplant based on the patient’s leader genotype.
Publication-related data are available on reasonable request by e-mail from the corresponding author.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mike Haagenson, Dawn Miller, and Mark Gatterman for outstanding support.
This work was funded by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (AI069197, E.W.P., T.G., P.S., M.M., M.H., C.M., and S.R.S.), National Cancer Institute (CA100019, E.W.P., T.G., P.S., M.M., and C.M.; CA18029, E.W.P., T.G., and M.M.; CA72978, E.W.P.; CA015704, T.G. and P.S; and 5U24CA076518, M.H. and S.R.S), and the National Heart, Lung, and Blood Institute (HL069294, M.H. and S.R.S); the US Office of Naval Research (N00014-17-1-2388 and N00014-17-1-2850, M.H. and S.R.S); Swiss FNRS No. 310030_173237/1; and the Philanthropy Settlement Foundation (J.V.). This project was funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research (Contract No. HHSN261200800001E).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The views expressed in this article do not necessarily reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, or any other agency of the US government.
Authorship
Contribution: E.W.P. designed the study; P.S. and T.G. performed the statistical analysis; and E.W.P. drafted the manuscript. All authors assembled the data, critically reviewed and edited the manuscript, and approved the final version.
Conflict-of-interest disclosure: E.W.P., T.G., M.H., M.M., P.S., C.M., and S.R.S. report grants from the National Institutes of Health. M.H. and S.R.S. report grants from the US Office of Naval Research. M.H. reports grants from Actinium Pharmaceuticals, Amgen, Inc., Amneal Biosciences, Angiocrine Bioscience, Inc, Anthem, Inc., Bluebird Bio, Inc., Bristol-Myers Squibb Company, Chimerix, CSL Behring, Cyto-Sen Therapeutics, Daiichi Sankyo, Gamida Cell, GlaxoSmithKline, Incyte Corporation, Janssen, Jazz Pharmaceuticals, Kite Pharma, Mesoblast, Miltenyi Biotech, Neovii Biotech, OncoImmune, Pfizer, Pharmacyclics LLC, Regeneron, Sanofi, Seattle Genetics, Shire, Takeda, and Medac; and grants and consulting fees from Magenta. M.C. reports support from the Frederick National Laboratory for Cancer Research and the Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard University. J.V. reports grants from the Swiss FNRS and the Philanthropy Settlement Foundation; and personal fees from Astellas and One Lambda. The remaining authors declare no competing financial interests.
A complete list of the members of the International Histocompatibility Working Group in Hematopoietic Cell Transplantation appears in supplemental Table 1.
Correspondence: Effie W. Petersdorf, University of Washington, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, D4-115, Seattle, WA 98109; e-mail: epetersd@fredhutch.org.