Key Points
GPRASP family members negatively regulate murine HSC–in vivo hematopoietic repopulating activity.
GPRASP family members regulate CXCR4 stability and trafficking to affect HSC survival, quiescence, homing, and niche retention.
Abstract
Hematopoietic stem cell (HSC) transplantation (HSCT) is often exploited to treat hematologic disease. Donor HSCs must survive, proliferate, and differentiate in the damaged environment of the reconstituting niche. Illuminating molecular mechanisms regulating the activity of transplanted HSCs will inform efforts to improve HSCT. Here, we report that G-protein–coupled receptor–associated sorting proteins (GPRASPs) function as negative regulators of HSCT. Silencing of Gprasp1 or Gprasp2 increased the survival, quiescence, migration, niche retention, and hematopoietic repopulating activity of hematopoietic stem and progenitor cells (HSPCs) posttransplant. We further show that GPRASP1 and GPRASP2 promote the degradation of CXCR4, a master regulator of HSC function during transplantation. CXCR4 accumulates in Gprasp-deficient HSPCs, boosting their function posttransplant. Thus, GPRASPs negatively regulate CXCR4 stability in HSCs. Our work reveals GPRASP proteins as negative regulators of HSCT and CXCR4 activity. Disruption of GPRASP/CXCR4 interactions could be exploited in the future to enhance the efficiency of HSCT.
Introduction
Hematopoietic stem cell (HSC) transplantation (HSCT) represents the only curative treatment of many leukemias and blood disorders. Transplanted HSCs must reach and seed specific niches in recipient bone marrow (BM), a process termed “homing and engraftment.”1 Although recent advances in HSCT, such as milder conditioning regimens and haplotransplantation, have improved on recognized limitations of HSCT, including donor availability and conditioning-related morbidities,2-6 more could be done to improve the safety, efficacy, and accessibility of this critical therapy. For example, ex vivo manipulation of HSCs for gene therapy compromises their homing and engraftment,7-10 small cell numbers limit the utility of cord blood HSCs as a source for transplantation,11-16 and underrepresentation of minorities on national bone marrow registries contributes significantly to limited donor access for these groups.17-19 Moreover, our understanding of HSC/niche interactions is incomplete and especially limited when considering niches damaged by chronic hematologic disease (such as sickle cell anemia), conditioning regimens, or chemotherapy. Indeed, chronic BM niche damage likely contributes to the increased rates of graft failure seen in sickle cell patients who received transplants.20-22 Thus, a better understanding of the cellular and molecular mechanisms regulating HSC–in vivo repopulating activity and their niche interactions should illuminate new strategies to improve HSCT.
Multiple G-protein–coupled receptor (GPCR)-associated sorting protein (GPRASP) family members are expressed by long-term HSCs (LT-HSCs)23 (Figure 1). GPRASP proteins share a conserved C-terminal domain, and repeats of a highly conserved 15-aa sequence, known as a GPCR-associated sorting protein (GASP) domain, are present in multiple GPRASPs.24 The cellular function of most GPRASPs is currently unknown. However, several are linked to stemness, regeneration, and development.25-33 Molecularly, GPRASPs can bind and regulate the postendocytic trafficking of GPCRs via their C-terminal and GASP domains.34-42 A consensus GASP-binding motif is present in the C terminus of numerous GPCRs.43-45 GPCRs sense a variety of environmental inputs and signals and, therefore, are crucial regulators of cell signaling during embryonic development, homeostasis, and stress.46-50 Accordingly, GPRASPs likely function as crucial regulators of multiple cellular processes controlled by GPCRs. Although we recently reported Gprasp2 as a hit in a screen for novel HSC regulators, to date, nothing is known as to the cellular and molecular roles of the GPRASP protein family in HSCT.23
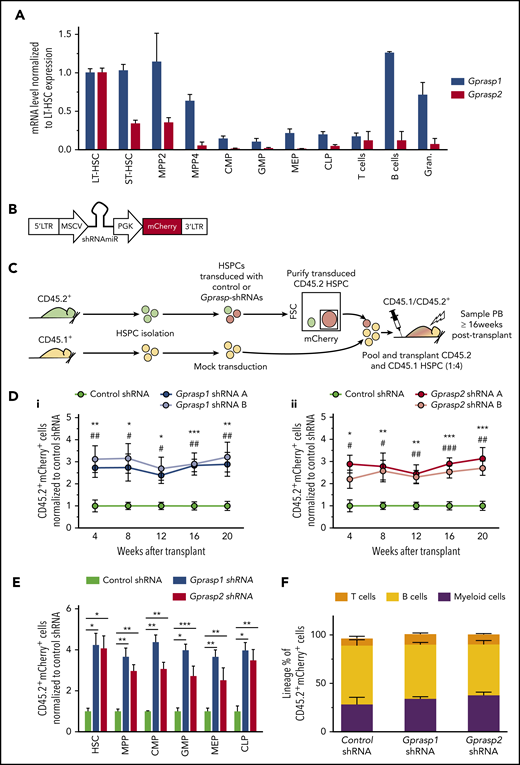
Silencing of Gprasp1 or Gprasp2 enhances HSPC–in vivo hematopoietic repopulating activity. (A) Messenger RNA (mRNA) expression analysis of Gprasp1 and Gprasp2 in the hematopoietic hierarchy. (B) Schematic of shRNA vector. (C) Experimental schematic. CD45.2+ “test” LSK cells were transduced with control or Gprasp shRNAs and then transplanted along with CD45.1+ “competitor” LSK cells into lethally irradiated recipients. Recipient peripheral blood (PB) or BM was analyzed for CD45.2+mCherry+ cells. (D) Enhanced PB-repopulating activity relative to HPSCs treated with control shRNA in (i) Gprasp1-deficient HSPCs and (ii) Gprasp2-deficient HSPCs. (E) Analysis of CD45.2+mCherry+ cells in recipient BM HSPC compartments 20 weeks posttransplant. (F) Distribution of mature lineages in CD45.2+mCherry+ PB in recipients of control or Gprasp-shRNA–treated HSPCs. Data in panels D, E, and F are from 3 independent transplants with 5 recipients per condition per transplant. Data represented as mean plus or minus standard error of the mean (SEM). */#, P < .05; **/##, P < .005; ***/###, P < .001 relative to recipients of control-shRNA HSPCs. In panel D, * refers to shRNA A; # refers to shRNA B. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; FSC, forward scatter; GMP, granulocyte/macrophage progenitor; Gran., granulocyte; LTR, long terminal repeat; MEP, megakaryocyte/erythrocyte progenitor; MPP, multipotent progenitor; MSCV, murine stem cell virus; PGK, phosphoglycerate kinase; shRNAmiR, microRNA-adapted short hairpin RNA; ST-HSC, short-term HSC.
Silencing of Gprasp1 or Gprasp2 enhances HSPC–in vivo hematopoietic repopulating activity. (A) Messenger RNA (mRNA) expression analysis of Gprasp1 and Gprasp2 in the hematopoietic hierarchy. (B) Schematic of shRNA vector. (C) Experimental schematic. CD45.2+ “test” LSK cells were transduced with control or Gprasp shRNAs and then transplanted along with CD45.1+ “competitor” LSK cells into lethally irradiated recipients. Recipient peripheral blood (PB) or BM was analyzed for CD45.2+mCherry+ cells. (D) Enhanced PB-repopulating activity relative to HPSCs treated with control shRNA in (i) Gprasp1-deficient HSPCs and (ii) Gprasp2-deficient HSPCs. (E) Analysis of CD45.2+mCherry+ cells in recipient BM HSPC compartments 20 weeks posttransplant. (F) Distribution of mature lineages in CD45.2+mCherry+ PB in recipients of control or Gprasp-shRNA–treated HSPCs. Data in panels D, E, and F are from 3 independent transplants with 5 recipients per condition per transplant. Data represented as mean plus or minus standard error of the mean (SEM). */#, P < .05; **/##, P < .005; ***/###, P < .001 relative to recipients of control-shRNA HSPCs. In panel D, * refers to shRNA A; # refers to shRNA B. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; FSC, forward scatter; GMP, granulocyte/macrophage progenitor; Gran., granulocyte; LTR, long terminal repeat; MEP, megakaryocyte/erythrocyte progenitor; MPP, multipotent progenitor; MSCV, murine stem cell virus; PGK, phosphoglycerate kinase; shRNAmiR, microRNA-adapted short hairpin RNA; ST-HSC, short-term HSC.
Here, we examined the functional role of GPRASP proteins in HSCT. We report that individual silencing of multiple GPRASP family members in hematopoietic stem and progenitor cells (HSPCs) significantly enhances their repopulating activity posttransplant due to increased survival, quiescence, migration, and homing. We also report that both GPRASP1 and GPRASP2 promote CXCR4 degradation and that CXCR4 is required for the enhanced activity of Gprasp-deficient HSPCs. Thus, we describe for the first time the cellular and molecular mechanisms via which GPRASPs suppress HSCT and reveal these proteins as potential new targets for enhancing the efficiency of HSCT.
Methods
Material and method details regarding mice, cell lines, flow cytometry, gene-expression analysis, short hairpin RNAs (shRNAs), lentivirus production and transduction, microscopy, transplants, and statistics are provided in supplemental Methods (available on the Blood Web site).
Results
GPRASP1 and GPRASP2 negatively regulate HSPC–in vivo repopulating activity
Gprasp2 was a hit in our previous screen for HSC regulators.23 As the GPRASP family had never been previously linked to hematopoiesis and HSCs, we decided to interrogate GPRASP2 and related proteins for a role in HSCT. GPRASP1 is structurally similar to GPRASP2 and is the best-studied member of this protein family (supplemental Figure 1A). Hence, we interrogated murine BM HSPCs for Gprasp1 and Gprasp2 expression (Figure 1A). LT-HSCs (Lineage−Sca-1+cKit+ [LSK] Flt3−CD150+CD48−), short-term HSCs (ST-HSCs; LSK Flt3−CD150−CD48−), multipotent progenitor 2 (MPP2; LSK Flt3−CD150+CD48+), MPP4 (LSK Flt3+CD150−CD48+), common myeloid progenitors (CMPs; Lineage−cKit+Sca−CD34+CD16/32med), granulocyte/macrophage progenitors (GMP; Lineage−cKit+Sca−CD34+CD16/32high), megakaryocyte/erythrocyte progenitors (MEPs; Lineage−cKit+Sca−CD34−CD16/32−), and common lymphoid progenitors (CLPs; Lineage−cKitmedSca1medCD127+) were isolated by fluorescence-activated cell sorting (FACS) from C57Bl/6J mice and examined. As LT-HSCs highly expressed Gprasp1 and Gprasp2 relative to downstream populations, we tested them for a functional role in HSCT. Toward this, multiple shRNAs targeting Gprasp1 or Gprasp2 were designed and validated for robust knockdown efficiency in HSPCs (LSK cells) (supplemental Figure 1B). HSPCs isolated from CD45.2+ C57Bl/6J mice were then transduced with control, Gprasp1, or Gprasp2 shRNAs. Transduced cells also express an mCherry reporter (Figure 1B). Forty-eight hours posttransduction, 2000 CD45.2+mCherry+ cells were transplanted along with 8000 mock-transduced CD45.1+ HSPCs into lethally irradiated recipients (CD45.1+/CD45.2+) (Figure 1C). Recipients transplanted with HSPCs transduced with Gprasp1 or Gprasp2 shRNAs displayed significantly increased CD45.2+mCherry+ peripheral blood (PB) and BM repopulation (Figure 1D-E; supplemental Figure 1C) relative to recipients of control HSPCs. Enhanced repopulation persisted for >16 weeks posttransplant in all BM HSPC compartments and PB lineages examined (Figure 1E-F; supplemental Figure 1D).
To explore the role of Gprasp1 and Gprasp2 in HSPCs further, we next interrogated Gprasp1−/− and Gprasp2−/− HSPCs. Two thousand wild-type (WT), Gprasp1−/−, or Gprasp2−/− CD45.2+ HSPCs were transplanted along with 8000 competitor CD45.1+ HSPCs into lethally irradiated recipients (CD45.1+/CD45.2+). Surprisingly, Gprasp1−/−, Gprasp2−/−, and WT HSPCs displayed similar PB reconstitution (supplemental Figure 2A). To address these conflicting results, we examined the specificity of our Gprasp1 and Gprasp2 shRNAs. Gprasp1−/− or Gprasp2−/− HSPCs (CD45.2+) were transduced with control, Gprasp1, or Gprasp2 shRNAs. Forty-eight hours posttransduction, 2000 CD45.2+mCherry+ cells were transplanted along with 8000 mock-transduced CD45.1+ HSPCs into lethally irradiated recipients (CD45.1+/CD45.2+). Gprasp1 and Gprasp2 shRNAs had no effect on the repopulating activity of Gprasp1−/− and Gprasp2−/− HSPCs, respectively (supplemental Figure 1B). These data indicate that the enhanced repopulating activity of WT HSPCs treated with Gprasp1 or Gprasp2 shRNAs depends on the presence of Gprasp1 and Gprasp2, respectively (Figure 1D).
We next considered genetic compensation for the absence of Gprasp1 or Gprasp2 by other GPRASPs. We first explored whether Gprasp1 might compensate for the absence of Gprasp2 and vice versa. However, although transduction of Gprasp2−/− HSPC with Gprasp1-shRNAs significantly enhanced HSPC repopulating activity, this effect was similar in magnitude to that seen when Gprasp1 was knocked-down in WT HSPC (supplemental Figure 2C; Figure 1D). Similar results were observed when Gprasp2 was knocked-down in Gprasp1−/− HSPC (supplemental Figure 2C; Figure 1D). We next examined WT, Gprasp1−/−, and Gprasp2−/− LT-HSCs by reverse transcription–quantitative polymerase chain reaction for the expression of all known Gprasp genes and discovered upregulation of Bhlhb9 in both Gprasp1−/− and Gprasp2−/− LT-HSCs (supplemental Figure 3A). In WT mice, Bhlhb9 expression is detectable across the hematopoietic hierarchy (supplemental Figure 3B). However, its expression increases significantly in Gprasp1−/− and Gprasp2−/− LT-HSCs and MPP2s. Therefore, multiple shRNAs targeting Bhlhb9 were tested for robust knockdown efficiency in HSPCs (supplemental Figure 3C). WT, Gprasp1−/−, or Gprasp2−/− HSPCs were next transduced with control or Bhlhb9 shRNAs. Forty-eight hours posttransduction, 5000 CD45.2+mCherry+ HSPCs were transplanted along with 5000 mock-transduced HSPCs (CD45.1+) into lethally irradiated recipients (CD45.1/CD45.2+). Bhlhb9 silencing did not perturb the activity of WT HSPCs (supplemental Figure 3D). However, Bhlhb9 silencing significantly enhanced the repopulating activity of Gprasp1−/− and Gprasp2−/− HSPCs at 4 weeks posttransplant (supplemental Figure 3E-F). Surprisingly, Bhlhb9-deficient Gprasp1−/− and Gprasp2−/− HSPCs displayed diminished repopulating activity beginning 12 weeks posttransplant (supplemental Figure 3E-F). Interestingly, this loss of PB repopulation did not correlate with a loss of CD45.2+mCherry+ cells in recipient BM HSPCs (supplemental Figure 3G-H). Indeed, CD45.2+mCherry+ LT-HSCs accumulated in the BM of recipients of Bhlhb9-shRNA–treated Gprasp1−/− HSPCs at 20 weeks posttransplant (supplemental Figure 3G). A differentiation block or compromised cell survival may exist downstream of Bhlhb9-deficient Gprasp1−/− or Gprasp2−/− HSPCs; further work will be required to definitely determine this precise mechanism. In sum, these data support a model in which Bhlhb9 can compensate for genetic loss of Gprasp1 and Gprasp2 in HSCs. BHLHB9 thus represents a third GPRASP family member capable of regulating HSPC function posttransplant under certain conditions.
GPRASPs regulate CXCR4 dynamics and stability
We next sought to understand how GPRASPs function as negative regulators of HSPCs. GPRASPs regulate GPCR trafficking by directing targets to the lysosome for degradation after internalization.34-41,43,45 Therefore, we hypothesized that loss of GPRASPs might stabilize a GPCR (or GPCRs) that facilitates HSPC transplantation. An in silico search for functionally relevant GPCRs with putative C-terminal GASP-binding motifs revealed that CXCR4, a regulator of HSC function,51-60 contains a close match in its C terminus (Figure 2A; supplemental Figure 4A). Cell-surface CXCR4 is activated by binding to its ligand, stromal cell–derived factor 1 (SDF-1),61-64 triggering its internalization. Because GPRASPs are implicated in the trafficking of internalized GPCRs, we examined the effect of Gprasp1 and Gprasp2 silencing on CXCR4 cellular localization. C57Bl/6J HSPCs were transduced with control, Gprasp1, or Gprasp2 shRNAs. Forty, 48, and 72 hours later, transduced cells were treated with 100 ng/mL SDF-1 to activate CXCR4 internalization. Cells were then examined for cytoplasmic and cell-surface CXCR4 by flow cytometry either immediately or after being washed and cultured with fresh media for 3 hours to allow for CXCR4 recycling to the cell surface. The ratio between intracellular and extracellular CXCR4 was then calculated (Figure 2B-C).
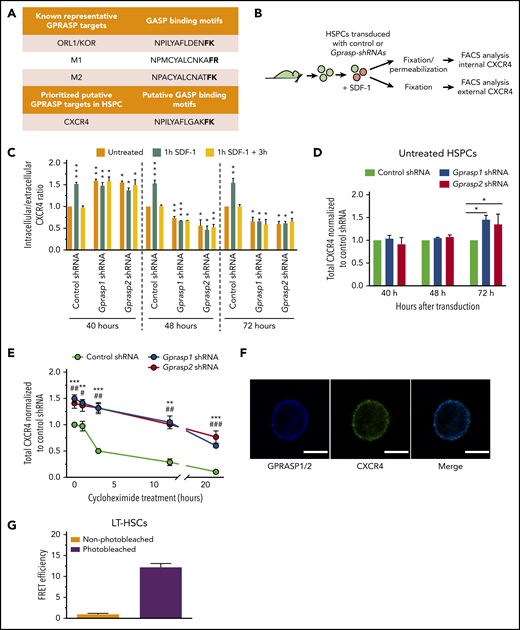
Silencing of Gprasp1 or Gprasp2 perturbs CXCR4 localization. (A) GASP-binding motifs in the C terminus of known and putative GPRASP targets. (B) Experimental schematic. LSK cells were treated with Gprasp1, Gprasp2, or control shRNAs, cultured for 72 hours and then treated for 1 hour with stromal cell–derived factor 1 (SDF-1) to induce CXCR4 internalization. Permeabilized and nonpermeabilized cell were then examined for CXCR4 expression and intracellular-to-extracellular ratio calculated. (C) CXCR4 intracellular-to-extracellular ratio. (D) Total CXCR4 40 to 72 hours posttransduction in Gprasp1, Gprasp2, or control-shRNA–treated HSPCs assessed by flow cytometry. (E) Total CXCR4 72 hours posttransduction in Gprasp1, Gprasp2, or control-shRNA–treated HSPCs treated with 5 μg/mL cycloheximide assessed by flow cytometry. (F) LT-HSCs were examined for GPRASP1/2 and CXCR4 colocalization via confocal microscopy. GPRASP1/2-405 and CXCR4-488 stain; scale bar, 5 μm. (G) High Förster resonance energy transfer (FRET) efficiency between GPRASP1/2-405 and CXCR4-488 in LT-HSCs indicates a physical proximity of <10 nm. Data in panels C, D, E, and G from at least 3 independent experiments. Data represented as mean plus or minus SEM. */#, P < .05; **/##, P < .005; ***/###, P < .001 relative to control. In panel E, * refers to Gprasp1 shRNA; # refers to Gprasp2 shRNA. FACS, fluorescence-activated cell sorting; M1, muscarinic receptor 1; M2, muscarinic receptor 2; ORL1/KOR, opioid receptor-like 1/κ opioid receptor.
Silencing of Gprasp1 or Gprasp2 perturbs CXCR4 localization. (A) GASP-binding motifs in the C terminus of known and putative GPRASP targets. (B) Experimental schematic. LSK cells were treated with Gprasp1, Gprasp2, or control shRNAs, cultured for 72 hours and then treated for 1 hour with stromal cell–derived factor 1 (SDF-1) to induce CXCR4 internalization. Permeabilized and nonpermeabilized cell were then examined for CXCR4 expression and intracellular-to-extracellular ratio calculated. (C) CXCR4 intracellular-to-extracellular ratio. (D) Total CXCR4 40 to 72 hours posttransduction in Gprasp1, Gprasp2, or control-shRNA–treated HSPCs assessed by flow cytometry. (E) Total CXCR4 72 hours posttransduction in Gprasp1, Gprasp2, or control-shRNA–treated HSPCs treated with 5 μg/mL cycloheximide assessed by flow cytometry. (F) LT-HSCs were examined for GPRASP1/2 and CXCR4 colocalization via confocal microscopy. GPRASP1/2-405 and CXCR4-488 stain; scale bar, 5 μm. (G) High Förster resonance energy transfer (FRET) efficiency between GPRASP1/2-405 and CXCR4-488 in LT-HSCs indicates a physical proximity of <10 nm. Data in panels C, D, E, and G from at least 3 independent experiments. Data represented as mean plus or minus SEM. */#, P < .05; **/##, P < .005; ***/###, P < .001 relative to control. In panel E, * refers to Gprasp1 shRNA; # refers to Gprasp2 shRNA. FACS, fluorescence-activated cell sorting; M1, muscarinic receptor 1; M2, muscarinic receptor 2; ORL1/KOR, opioid receptor-like 1/κ opioid receptor.
As expected, HSPCs transduced with control shRNA increased their CXCR4 intracellular-to-extracellular ratio in response to SDF-1 compared with untreated cells, which returned to baseline after washout of SDF-1 (Figure 2C; supplemental Figure 4Bi-ii). For untreated HSPCs transduced with Gprasp1 or Gprasp2 shRNAs, the CXCR4 intracellular-to-extracellular ratio 40 hours posttransduction was higher than untreated controls (ie, mimicking the effect of SDF-1 on controls) (Figure 2C). SDF-1 treatment did not further perturb this ratio, suggesting an accumulation of cytoplasmic CXCR4 in Gprasp-deficient HSPCs at this time point (Figure 2C; supplemental Figure 4Biii-iv). However, Gprasp-deficient HSPCs analyzed 48 and 72 hours posttransduction displayed a significantly lower CXCR4 intracellular-to-extracellular ratio than untreated control HSPCs (again regardless of SDF-1 treatment), reflecting increased CXCR4 at the cell membrane (Figure 2C; supplemental Figure 4B). Furthermore, total CXCR4 increased in Gprasp-deficient HSPCs relative to control HSPCs by 72 hours posttransduction (Figure 2D). To assess whether increased CXCR4 levels resulted from a block in protein degradation, HSPCs transduced with control, Gprasp1, or Gprasp2 shRNAs were cultured for 1, 3, 12, or 24 hours with 5 μg/mL cycloheximide, an inhibitor of translation.65 Here, we observed delayed CXCR4 degradation in Gprasp-deficient HSPCs relative to control (Figure 2E). Altogether, these data indicate that silencing of either Gprasp1 or Gprasp2 in HSPCs results in an accumulation of CXCR4 first in the cytoplasm and then at the cell surface. The increasing levels of total CXCR4 in Gprasp-deficient HSPCs further suggests that Gprasp silencing hinders CXCR4 degradation.
CXCR4 contains a putative GASP-binding motif (Figure 2A). This suggests that GPRASPs may regulate CXCR4 via a physical interaction. To asses this, LT-HSCs were examined by confocal microscopy for GPRASP1/GPRASP2 and CXCR4 colocalization. GPRASP1/GPRASP2 and CXCR4 appeared to colocalize in the cytoplasm of LT-HSCs, suggesting a physical distance of <200 nm (Figure 2F).66 To enhance resolution, we performed Förster resonance energy transfer (FRET) acceptor photobleaching on LT-HSCs stained for GPRASP1/GPRASP2 and CXCR4. Here, we observed an average FRET efficiency > 12% (Figure 2G), which strongly suggests that GPRASP1/GPRASP2 and CXCR4 exist within a distance of 10 nm and thus physically interact in LT-HSCs.67 We repeated these experiments using Neuro2a and human neuroblastoma cells (SH-SY5Y), which express high levels of GPRASP1, GPRASP2, and CXCR4 and have larger cytoplasms than LT-HSCs (supplemental Figure 4C). Neuro2A and SH-SY5Y cells displayed average FRET efficiencies of 25.56% and 19.17%, respectively (supplemental Figure 4D), suggesting that CXCR4 physically interacts with GPRASP1 and/or GPRASP2 in these cells. These results strongly suggest that CXCR4 is a direct molecular target of GPRASP1 and GPRASP2.
Gprasp1 or Gprasp2 silencing improves HSPC survival, quiescence, migration, homing, and niche retention during transplant
CXCR4 can promote HSC migration, niche retention, survival, and quiescence.51-60 Thus, we assessed whether silencing of Gprasp1 or Gprasp2 perturbed these functions in HSPCs. We first tested Gprasp1- and Gprasp2-deficient HSPCs for apoptosis, cell-cycle status, and proliferation ex vivo. C57Bl/6J HSPCs were transduced with control, Gprasp1, or Gprasp2 shRNAs and then cultured ex vivo for 5 days, at which point mCherry+ cells were examined by flow cytometry. Gprasp1- and Gprasp2-shRNA–treated HSPCs showed reduced apoptosis, increased G0, and decreased S/G2/M relative to controls (Figure 3A-B; supplemental Figure 5A-B). We also tracked the ex vivo growth of Gprasp1- and Gprasp2-deficient HSPCs by interrogating carboxyfluorescein succinimidyl ester (CFSE) retention and cell viability (Figure 3C; supplemental Figure 5C-F). For the first 10 days of culture, Gprasp1- and Gprasp2-deficient cells displayed higher viability and CFSE retention than control cells, suggesting increased survival and reduced proliferation (Figure 3C; supplemental Figure 5C-F). Not surprisingly, overall cell growth was the same for Gprasp-deficient and control cells during this early culture period, as increased survival combined with reduced proliferation resulted in a zero-sum gain in overall cell numbers. However, longer ex vivo culture (10-21 days) yielded accumulating Gprasp1- and Gprasp2-deficient cells relative to controls (Figure 3C).
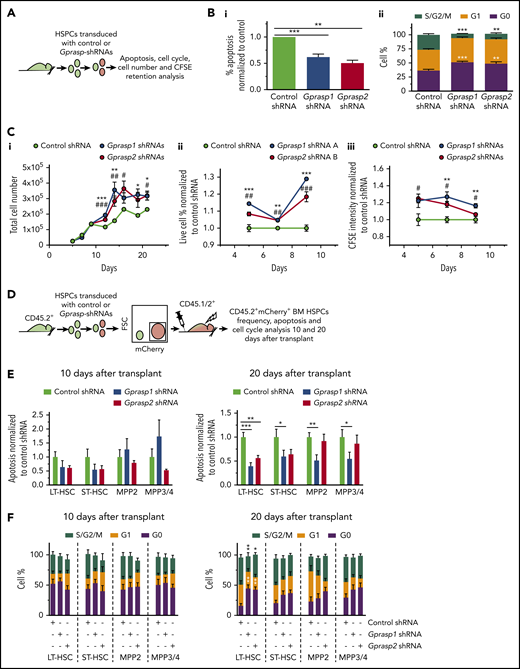
Silencing of Gprasp1 or Gprasp2 enhances HSPC survival and quiescence. (A) Experimental schematic. (B-C) LSK cells transduced with Gprasp1, Gprasp2, or control shRNAs were assayed by flow cytometry: (B) for (i) apoptosis or (ii) cell cycle status; (C) for (i) expansion, (ii) viability, or (iii) carboxyfluorescein succinimidyl ester (CFSE) retention. (D) Experimental schematic. CD45.2+ LSK cells transduced with Gprasp1, Gprasp2, or control shRNAs were transplanted into lethally irradiated CD45.1/CD45.2+ mice. Recipient CD45.2+mCherry+ HSPCs were analyzed for apoptosis (E) and cell cycle (F) 10 and 20 days posttransplant by flow cytometry. Data in panels E and F from 4 independent transplants with 5 recipients per condition per transplant. Data represented as mean plus or minus SEM. */#, P < .05; **/##, P < .005; ***/###, P < .001 relative to control. In panel C, * refers to Gprasp1 shRNA, # refers to Gprasp2 shRNA.
Silencing of Gprasp1 or Gprasp2 enhances HSPC survival and quiescence. (A) Experimental schematic. (B-C) LSK cells transduced with Gprasp1, Gprasp2, or control shRNAs were assayed by flow cytometry: (B) for (i) apoptosis or (ii) cell cycle status; (C) for (i) expansion, (ii) viability, or (iii) carboxyfluorescein succinimidyl ester (CFSE) retention. (D) Experimental schematic. CD45.2+ LSK cells transduced with Gprasp1, Gprasp2, or control shRNAs were transplanted into lethally irradiated CD45.1/CD45.2+ mice. Recipient CD45.2+mCherry+ HSPCs were analyzed for apoptosis (E) and cell cycle (F) 10 and 20 days posttransplant by flow cytometry. Data in panels E and F from 4 independent transplants with 5 recipients per condition per transplant. Data represented as mean plus or minus SEM. */#, P < .05; **/##, P < .005; ***/###, P < .001 relative to control. In panel C, * refers to Gprasp1 shRNA, # refers to Gprasp2 shRNA.
We next examined apoptosis and cell-cycle status in Gprasp-silenced BM HSPCs acutely posttransplant. Toward this, CD45.2+ C57Bl/6J HSPCs were transduced with control, Gprasp1 or Gprasp2 shRNAs. Forty-eight hours later, 8000 CD45.2+mCherry+ cells were transplanted into lethally irradiated recipients (CD45.1/CD45.2+). Recipient CD45.2+mCherry+ BM HSPCs were examined 10 and 20 days posttransplant (Figure 3D). By 20 days posttransplant, CD45.2+mCherry+ HSPCs displayed reduced apoptosis, increased G0, and decreased S/G2/M in recipients of Gprasp-deficient HSPCs relative to control recipients (Figure 3E-F; supplemental Figure 5G-H). Reduced apoptosis was observed in reconstituted (ie, CD45.2+mCherry+ recipient BM) Gprasp1-deficient LT-HSCs, ST-HSCs, MPP2, MPP3, and MPP4. In contrast, reduced apoptosis was only observed in reconstituted Gprasp2-deficient LT-HSC and ST-HSC compartments (Figure 3E; supplemental Figure 5G). Interestingly, increased G0 and decreased S/G2/M appeared only in reconstituted Gprasp1- and Gprasp2-deficient LT-HSCs (Figure 3F; supplemental Figure 5H).
The increased survival and quiescence of Gprasp-deficient LT-HSCs was maintained even up to 16 weeks postsecondary transplantation (supplemental Figure 6A-B). Gprasp-deficient LT-HSCs also maintained elevated levels of total CXCR4 at this time point (supplemental Figure 6C). Interestingly, Gprasp-deficient HSPCs displayed only modestly enhanced repopulating activity in secondary recipients relative to controls (supplemental Figure 6D-E), perhaps reflecting their increased quiescence during secondary transplantation. In sum, Gprasp1- and Gprasp2-deficient cells display increased survival and quiescence both ex vivo and by 20 days posttransplant, which likely contributes to their long-term competitive repopulating advantage in primary transplants.
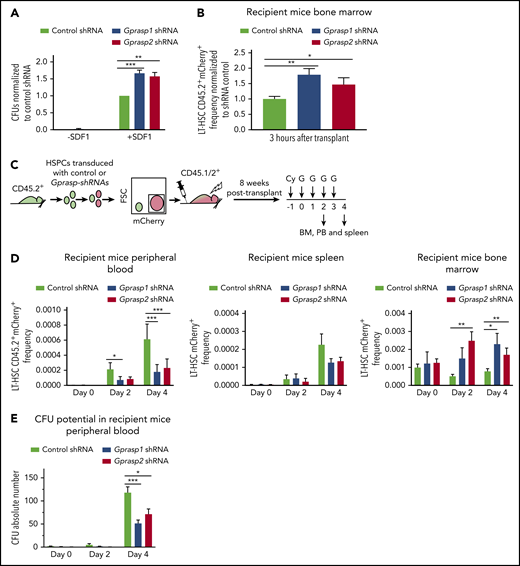
We next assessed whether Gprasp1 or Gprasp2 silencing perturbs HSPC migration in response to SDF-1 and BM homing. C57Bl/6J HSPCs were transduced with control, Gprasp1, or Gprasp2 shRNAs. Forty-eight hours later, mCherry+ HSPCs were assayed for SDF-1–induced migration in trans-well assays. Significantly more Gprasp1- and Gprasp2-deficient HSPCs migrated in response to SDF-1 relative to controls (Figure 4A), suggesting increased sensitivity to SDF-1. Few cells migrated in the absence of SDF-1 (Figure 4A). Total colony-forming unit (CFU) input was the same for all conditions (supplemental Figure 7A). To determine whether Gprasp1- and Gprasp2-deficient HSPCs displayed increased homing in vivo, C57Bl/6J HSPCs were transduced with control, Gprasp1, or Gprasp2 shRNAs. Forty-eight hours later, CD45.2+mCherry+ cells were collected and transplanted into irradiated recipients. Recipient BM was examined for CD45.2+mCherry+ LT-HSCs 3 hours posttransplant. More CD45.2+mCherry+ LT-HSCs were detected in the BM of recipients of Gprasp1- or Gprasp2-shRNA–treated HSPCs relative to recipients of control cells (Figure 4B).
Loss of Gprasp1 or Gprasp2 effects HSPCs migration, homing, and niche retention. (A) Control, Gprasp1, or Gprasp2-shRNA-treated HSPCs were assayed for SDF-1 responsiveness in trans-well migration assays. Each trans-well bottom chamber was tested for colony-forming units (CFUs) as a readout for migrating cells. (B) LT-HSCs CD45.2+mCherry+ frequency in the BM of recipients of control, Gprasp1- or Gprasp2-shRNA–treated LSK cells 3 hours posttransplant. (C) Experimental schematic. CD45.2+ LSK cells transduced with Gprasp1, Gprasp2, or control shRNAs were transplanted into lethally irradiated CD45.1/CD45.2+ mice. After stable engraftment, recipient BM was mobilized by cytoxan (Cy)/G-CSF (G) treatment. (D) LT-HSC CD45.2+mCherry+ frequency in PB, spleen, and BM at 0, 2, and 4 days postmobilization. (E) CFUs in the PB of mobilized recipients described in panel C. Data in panels B through E from ≥5 independent transplants. N ≥ 3 recipients per condition per transplant. Data represented as mean plus or minus SEM. *P < .05; ** P < .005; ***P < .001 significantly different to control.
Loss of Gprasp1 or Gprasp2 effects HSPCs migration, homing, and niche retention. (A) Control, Gprasp1, or Gprasp2-shRNA-treated HSPCs were assayed for SDF-1 responsiveness in trans-well migration assays. Each trans-well bottom chamber was tested for colony-forming units (CFUs) as a readout for migrating cells. (B) LT-HSCs CD45.2+mCherry+ frequency in the BM of recipients of control, Gprasp1- or Gprasp2-shRNA–treated LSK cells 3 hours posttransplant. (C) Experimental schematic. CD45.2+ LSK cells transduced with Gprasp1, Gprasp2, or control shRNAs were transplanted into lethally irradiated CD45.1/CD45.2+ mice. After stable engraftment, recipient BM was mobilized by cytoxan (Cy)/G-CSF (G) treatment. (D) LT-HSC CD45.2+mCherry+ frequency in PB, spleen, and BM at 0, 2, and 4 days postmobilization. (E) CFUs in the PB of mobilized recipients described in panel C. Data in panels B through E from ≥5 independent transplants. N ≥ 3 recipients per condition per transplant. Data represented as mean plus or minus SEM. *P < .05; ** P < .005; ***P < .001 significantly different to control.
CXCR4 surface levels increase on BM HSCs but decrease on circulating HSCs after mobilization with cytoxan/granulocyte–colony-stimulating factor (G-CSF; Cy/G).68 Thus, daily G-CSF treatment overcomes CXCR4-mediated HSC BM retention and only the highest CXCR4-expressing HSCs remain in the BM. To interrogate a role for Gprasp1 and Gprasp2 in HSC niche retention, CD45.2+ C57Bl/6J HSPCs were transduced with control, Gprasp1, or Gprasp2 shRNAs. Forty-eight hours later, 8000 CD45.2+mCherry+ HSPCs were transplanted into lethally irradiated recipients (CD45.1/CD45.2+). Eight weeks posttransplant, recipient mice were treated with Cy/G to induce HSC mobilization. CD45.2+mCherry+ LT-HSC frequency was analyzed by flow cytometry in recipient PB, BM, and spleen at multiple time points posttreatment (Figure 4C; supplemental Figure 7B). Although increasing numbers of both control shRNA and Gprasp-shRNA–transduced LT-HSCs were seen in the PB and spleen of recipients 2 and 4 days posttreatment relative to day 0, significantly more of these cells were detected in the PB of recipients of controls relative to recipients of Gprasp-deficient HSPCs (Figure 4D; supplemental Figure 7B). Consistently, far fewer CFUs were detected in the PB of recipients of Gprasp-deficient HSPCs relative to recipients of control cells 4 days posttreatment (Figure 4E). Concomitantly, more CD45.2+mCherry+ LT-HSCs were detected in the BM of recipients of Gprasp-deficient HSPCs relative to controls, indicating stronger niche retention (Figure 4D; supplemental Figure 7B).
In sum, these data reveal that Gprasp1 and Gprasp2 negatively regulate HSPC survival, quiescence, migration, homing, and niche retention posttransplantation, which is highly consistent with a role in facilitating CXCR4 turnover.
CXCR4 is necessary for enhanced survival, quiescence, and repopulating activity of Gprasp1- and Gprasp2-deficient HSPCs
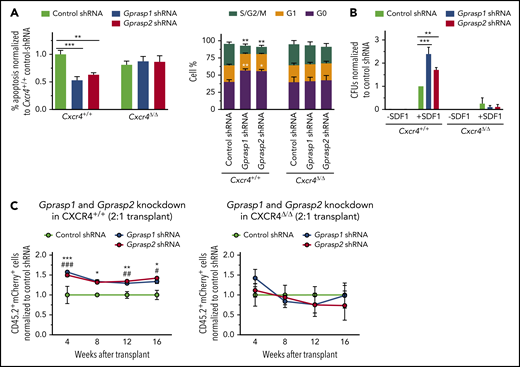
To definitively establish CXCR4 as central to GPRASP regulation of HSPC function, we next tested whether the enhanced survival, quiescence, and migration of Gprasp-deficient HSPCs depends on the presence of CXCR4. Toward this, 8- to 10-week old Cxcr4fl/flRosa26+/+ or Cxcr4fl/flRosa26ERT2-Cre/+ mice were treated daily with tamoxifen (TAM) for 5 days to induce genetic deletion of Cxcr4. Cxcr4+/+ or Cxcr4Δ/Δ HSPCs were then isolated from the BM of TAM-treated Cxcr4fl/flRosa26+/+ or Cxcr4fl/flRosa26ERT2-Cre/+ mice, respectively, and transduced with control, Gprasp1, or Gprasp2 shRNAs. After 5 days in culture, apoptosis and cell-cycle status were analyzed by flow cytometry. As expected, Gprasp1- and Gprasp2-shRNA–treated Cxcr4+/+ HSPCs showed reduced apoptosis, increased quiescence (G0 cells), and reduced proliferation (S/M/G2 cells) relative to Cxcr4+/+ control HSPCs (Figure 5A). In contrast, Cxcr4Δ/Δ HSPCs transduced with Gprasp1 or Gprasp2 shRNAs displayed no differences in apoptosis or cell-cycle status when compared with Cxcr4Δ/Δ control HSPCs (Figure 5A). Similar results were seen when CXCR4 activity was blocked by treating cells with an anti–SDF-1 antibody (supplemental Figure 8A-B). Augmented migration in response to SDF-1 was also no longer seen after Cxcr4 deletion when Gprasp genes were silenced (Figure 5B; supplemental Figure 8C). These results establish CXCR4 as necessary for the reduced apoptosis, increased quiescence, and enhanced migration of Gprasp-deficient HSPCs ex vivo.
Loss of CXCR4 abolishes the effects of Gprasp1 or Gprasp2 silencing on HSPCs.Cxcr4+/+ or Cxcr4Δ/Δ HSPCs transduced with Gprasp1, Gprasp2, or control shRNAs were assayed by flow cytometry for apoptosis, cell-cycle status (A), or migration assays toward an SDF-1 gradient in trans-well assays (B). (C) Cxcr4+/+ or Cxcr4Δ/Δ CD45.2+ LSK cells were transduced with control or Gprasp shRNAs and transplanted with CD45.1+ LSK cells into irradiated recipients at a 2:1 ratio. Recipients’ PB was analyzed for CD45.2+mCherry+ cells. Data in panel C from ≥2 independent transplants with 5 recipients per condition per transplant. Data represented as mean plus or minus SEM. */#, P < .05; **/##, P < .005; ***/###, P < .001 significantly different to control. In panel C, * refers to Gprasp1 shRNA; # refers to Gprasp2 shRNA.
Loss of CXCR4 abolishes the effects of Gprasp1 or Gprasp2 silencing on HSPCs.Cxcr4+/+ or Cxcr4Δ/Δ HSPCs transduced with Gprasp1, Gprasp2, or control shRNAs were assayed by flow cytometry for apoptosis, cell-cycle status (A), or migration assays toward an SDF-1 gradient in trans-well assays (B). (C) Cxcr4+/+ or Cxcr4Δ/Δ CD45.2+ LSK cells were transduced with control or Gprasp shRNAs and transplanted with CD45.1+ LSK cells into irradiated recipients at a 2:1 ratio. Recipients’ PB was analyzed for CD45.2+mCherry+ cells. Data in panel C from ≥2 independent transplants with 5 recipients per condition per transplant. Data represented as mean plus or minus SEM. */#, P < .05; **/##, P < .005; ***/###, P < .001 significantly different to control. In panel C, * refers to Gprasp1 shRNA; # refers to Gprasp2 shRNA.
We next tested whether CXCR4 is critical for the enhanced in vivo repopulating activity of Gprasp-deficient HSPCs. Cxcr4+/+ or Cxcr4Δ/Δ HSPCs (CD45.2+) were transduced with control, Gprasp1, or Gprasp2 shRNAs. Cxcr4−/− HSPCs display poor in vivo repopulating activity due to compromised survival and homing into the BM niche.53,54,56,58,60 However, stable Cxcr4Δ/Δ HSPC engraftment occurs when large cell numbers are transplanted.57 Thus, we performed these experiments by transplanting shRNA-treated Cxcr4+/+ and Cxcr4Δ/Δ HSPCs (CD45.2+) at a 2:1 ratio with competitor HSPCs (CD45.1+). Stable Cxcr4Δ/Δ-derived PB reconstitution was observed under these conditions (Figure 5C; supplemental Figure 8D). As expected, Gprasp silencing enhanced the repopulating activity of Cxcr4+/+ cells (Figure 5C; supplemental Figure 7D). In contrast, Gprasp silencing failed to enhance the repopulating activity of Cxcr4Δ/Δ HSPCs (Figure 5C; supplemental Figure 8D), strongly suggesting a Cxcr4 requirement for the enhanced in vivo repopulating activity of Gprasp-deficient HSPCs. Cxcr4 deletion efficiency was examined for every experiment involving TAM treatment of Cxcr4fl/flRosa26ERT2-Cre/+ mice (supplemental Figure 8E).
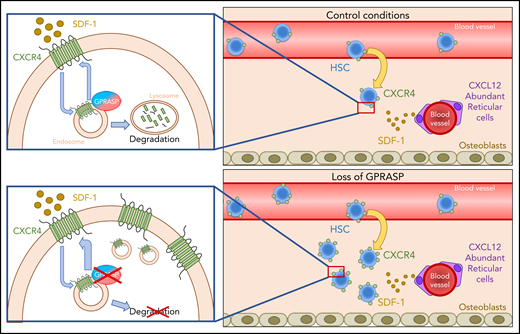
In summary, we have shown that silencing of Gprasp1 or Gprasp2 enhances the in vivo repopulating activity of HSPCs by increasing CXCR4 cell-surface levels. This acutely results in increased sensitivity to SDF-1, more efficient homing and engraftment in the BM niche, and, ultimately, increased survival and quiescence posttransplant. We further demonstrate that GPRASP1 and GPRASP2 act as novel regulators of CXCR4 stability by promoting the degradation of this critical GPCR (Figure 6).
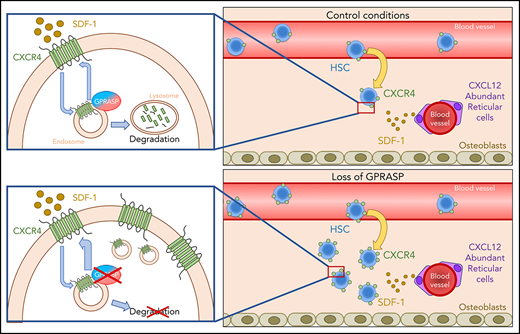
GPRASPs regulate HSC migration, survival, and repopulation after transplant by promoting CXCR4 degradation. Under control conditions, both GPRASP1 and GPRASP2 are necessary for the proper degradation of CXCR4. Upon Gprasp1 or Gprasp2 knockdown, CXCR4 degradation is blocked. This provokes an accumulation of this protein, allowing a higher recycling rate to the cell surface and makes the cells more sensitive to the SDF-1–chemoattractive effect, enhancing their migrating, survival, and repopulation potential after transplant.
GPRASPs regulate HSC migration, survival, and repopulation after transplant by promoting CXCR4 degradation. Under control conditions, both GPRASP1 and GPRASP2 are necessary for the proper degradation of CXCR4. Upon Gprasp1 or Gprasp2 knockdown, CXCR4 degradation is blocked. This provokes an accumulation of this protein, allowing a higher recycling rate to the cell surface and makes the cells more sensitive to the SDF-1–chemoattractive effect, enhancing their migrating, survival, and repopulation potential after transplant.
Discussion
Here, we report that multiple GPRASP proteins (ie, GPRASP1, GPRASP2, and BHLHB9) can negatively regulate HSPC transplantation and function. Our work is the first to implicate this protein family in hematopoiesis and HSC biology. We show GPRASPs interacting directly with CXCR4 to regulate its cellular trafficking and degradation in HSCs. Loss of GPRASPs stabilizes CXCR4 pools, resulting in more CXCR4 recycling to the cell surface, which subsequently enhances HSC homing and engraftment in the BM and survival during transplantation.
GPCRs are one of the most abundant protein families encoded by the human genome. Approximately 800 sequence-verified human GPCRs have been identified, 369 of which respond to known ligands. These ligands are diverse and include amino acids, Ca2+ ions, peptides, lipid-like substances, and large glycoprotein hormones, making GPCRs a common target for pharmaceutical applications.69 Consequently, to further illuminate therapeutic agents targeting GPCRs, a deep understanding of their interacting partners is needed. GPRASPs regulate the intracellular trafficking and degradation of GPCRs.34-41,43,45 Here, we identified a GASP-binding motif in the C terminus of CXCR4, a GPCR that is also one of the better characterized actors in HSC migration, niche retention, survival, and quiescence.51-60 This discovery implicated CXCR4 as a putative GPRASP target. Indeed, loss of multiple GPRASP proteins stabilizes CXCR4 in HSPCs.
Induced deletion of CXCR4 in adult mice results in significant loss of BM HSCs and increased sensitivity to myelotoxic injury, indicating CXCR4/SDF-1 as crucial for preserving BM HSCs.70 Cxcr4−/− HSCs do not transplant efficiently.53,54,56,58,60 Conversely, ectopic Cxcr4 enhances HSCT,71 similarly to loss of Gprasp1 or Gprasp2. Cxcr4 also promotes HSC quiescence and suppresses apoptosis of hematopoietic progenitors.51-54,57,59 We found that Gprasp1- and Gprasp2-deficient HSPCs display increased G0, decreased G2SM, and reduced apoptosis relative to controls ex vivo and posttransplant, phenocopying the known effects of Cxcr4.51-54,57,59
SDF-1, also known as CXCL12 and a potent chemoattractant of HSPCs, elicits its effects by binding to CXCR4 on HSPCs.72,73 Indeed, the CXCR4/SDF-1 axis is a critical regulatory pathway affecting HSCT.56,60 CXCR4 expression and cell localization drive HSC sensitivity to SDF-1, which is crucial for efficient HSCT because SDF-1 promotes HSC migration, homing, engraftment, and BM niche retention.46 Here, we show GPRASP1 and GPRASP2 physically interact with CXCR4 and that Gprasp1 and Gprasp2 loss blocks CXCR4 degradation, which results in accumulating CXCR4 and its accrual at the cell membrane. This accrual then promotes the survival, quiescence, migration, homing, and niche retention of Gprasp-silenced cells, thereby enhancing their acute repopulating activity. These phenotypes are dependent on CXCR4 and appear restricted to LT-HSCs, which correlates with the enriched expression of Gprasp1 and Gprasp2 by HSCs in the hematopoietic hierarchy.
As mentioned, Gprasp silencing ultimately promotes HSC quiescence, both ex vivo and in vivo. How can we reconcile increased quiescence with the enhanced repopulating activity of Gprasp-deficient HSPCs? Increased quiescence and survival of Gprasp-deficient HSCs manifests between 10 and 20 days posttransplant. In contrast, Gprasp-deficient HSPCs display enhanced BM homing within hours of transplantation. Thus, a simple model suggests that Gprasp-deficient HSPCs gain an immediate advantage posttransplant due to enhanced homing, niche retention, and sensitivity to SDF-1, which then promotes their survival and quiescence over the next several weeks. Consistent with these data, Gprasp-deficient HSPCs display only modestly enhanced repopulating activity in secondary transplants, likely due to their deeper quiescent state. In other words, as HSCs reconstitute recipient hematopoiesis and reassert homeostasis, they return to quiescence, which ultimately protects them from the risks inherent in excessive proliferation.74
Our work suggests that GPRASP1 and GPRASP2 each play a key role in regulating CXCR4 in HSCs. This multilayered regulation reflects the importance of CXCR4 activity to HSC function. The inability of GPRASP1 and GPRASP2 to compensate for each other in knockdown studies suggests that these proteins may participate in distinct steps of CXCR4 processing following internationalization. Remarkably, our discovery that a third GPRASP (BHLHB9) can partially compensate for loss of GPRASP1 or GPRASP2 reinforces the significance of GPRASPs as novel HSC regulators. Further work is necessary to delineate the precise biochemical role of each of these proteins in CXCR4 processing.
Although Cxcr4 is expressed by many hematopoietic cell types, Gprasp1 and Gprasp2 expression is generally restricted to LT-HSCs. Our functional data demonstrate highly LT-HSC–specific functions for Gprasp1 and Gprasp2. Bhlhb9, which is broadly expressed across hematopoietic populations, appears to both compensate for genetic loss of Gprasp1 and Gprasp2 in HSCs and possibly also play a role in the differentiation or survival of maturing hematopoietic cells posttransplant. Bhlhb9 antiapoptotic and differentiation roles in neuronal cells75,76 and during chondrogenesis77 are consistent with our model. Further exploration of Bhlhb9 deficiency should clarify its precise role in hematopoiesis.
In sum, here we discovered GPRASP proteins as new regulators of HSC survival, quiescence, migration, homing, and niche retention posttransplantation by controlling CXCR4 degradation. GPRASP proteins, and their molecular targets, represent windows into better understanding the molecular pathways influencing HSCT. Indeed, both GPRASP1 and GPRASP2 are expressed by human BM CD34+ cells (data not shown). Our data implicate GPRASPs as potential targets for inducing CXCR4 stabilization, which could be clinically valuable. Better understanding these pathways should illuminate novel strategies to boost the efficiency of HSCT.
For original data, please e-mail the corresponding author.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the McKinney-Freeman and Clements laboratories for critical discussions and reading of the manuscript; D. Ashmun, D. Langfitt, L. He, S. Schwemberger, and S. Wollard for FACS support; V. Frohlich and S. King for microscopy support; and A. Reap, C. Davis-Goodrum, and K. Millican for help with mouse injections. Gprasp1−/− and Gprasp2−/− mice were generously provided by Jennifer Whistler (University of California San Francisco [UCSF]) and Frédéric Simonin (Centre National de la Recherche Scientifique [CNRS] Unité Mixte de Recherche [UMR] 7242 Biotechnologie et Signalisation Cellulaire [BSC]), respectively. GPRASP1/2 antibody was generously provided by Frédéric Simonin (CNRS UMR7242 BSC).
This work was supported by the American Lebanese Syrian Associated Charities (ALSAC) (S.M.-F.) and the National Institute for Diabetes and Digestive and Kidney Diseases at the National Institutes of Health (R01 DK104028 [S.M.-F.], R01 DK116835 [S.M.-F.]).
S.M.-F. is a Scholar of the Leukemia & Lymphoma Society.
Authorship
Contribution: A.M.-H. and S.M.-F. wrote, reviewed, and revised the manuscript and designed the experiments; M.F. participated in the experimental design; A.M.-H., A.C., C.C., and C.B. performed the experiments; and X.Z. and G.K. performed the statistical analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shannon Mckinney-Freeman, St. Jude Children's Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105-3678; e-mail: shannon.mckinney-freeman@stjude.org.