Myelodysplastic syndrome (MDS) is a heterogenous myeloid lineage malignancy characterized by blood cell morphological dysplasia, ineffective clonal hematopoiesis, and risk of secondary transformation to acute myeloid leukemia (sAML). Genomic sequencing of large MDS cohorts has led to the identification of recurrent genetic abnormalities that carry independent prognostic significance and overlap with mutational changes in sAML. However, no set of mutations is sufficient to predict the transformation of MDS raising the question of how an identical genotype produces MDS in one patient and sAML in another? We hypothesize there are therapeutically targetable cellular processes altered by the initiating genetic changes in MDS that predict transformation to sAML.
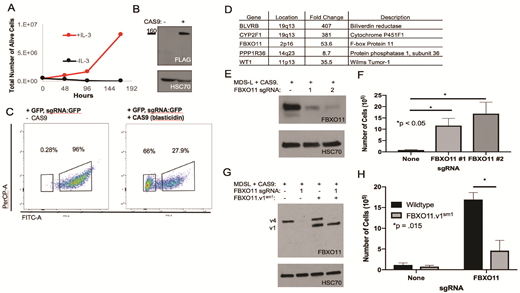
To uncover novel cellular pathways involved in MDS transformation, we performed an unbiased genome-wide CRISPR/Cas9 in the human MDS-L cell line. MDS-L was established from bone marrow mononuclear cells in a 52-year-old male patient and requires IL3-containing media for growth in vitro (Figure 1A). GFP expressing MDS-L cells were transduced with Cas9 and a sgRNA against GFP to confirm functional Cas9 expression in MDS-L (Figures 1B and C). In Cas9 expressing MDS-L cells, we then transduced the Brunello sgRNA CRISPR library and subjected the cells to IL-3 starvation for 4 weeks. Cells surviving IL-3 starvation were then expanded and harvested for genomic DNA. High throughput sequencing of the barcoded DNA produced raw reads that were analyzed using the PinAPL-Py web-based software. sgRNAs appearing in duplicate with absolute read counts over 1000 or in triplicate over 100 were considered significant.
We identified 5 genes that conferred resistance to IL-3 starvation, which included FBXO11 (Figure 1D). The Fbox protein FBXO11 is a component of the SCF ubiquitin ligase complex and regulates its substrates via ubiquitination and proteasomal degradation. FBXO11 is mutated in up to 20% of diffuse large B-cell lymphomas and its loss in breast cancer models leads to increased metastases. Therefore, we hypothesized FBXO11 may also function as a tumor suppressor in the transformation of MDS to AML.
We confirmed in the Bloodspot gene expression database that there are decreased levels of FBXO11 in a variety of AML samples, including complex karyotype, compared to normal HSCs. To validate the results of the screen, we synthesized two sgRNAs targeting FBXO11, transduced these into MDS-L cells, and detected reduced FBXO11 expression (Figure 1E). Loss of FBXO11 expression promoted survival in IL-3 free media, confirming the selection readout of the screen (Figure 1F). We then designed a silent mutation in the shorter isoform of FBXO11 (FBXO11v1sm1) that rendered resistance to CRISPR/Cas9 (Figure 1G) and observed that overexpression of FBXO11v1sm1 re-sensitized cells to cytokine starvation (Figure 1H). Whether there are different functions between FBXO11 variant 1 and 4 are currently being explored. We are actively performing RNA sequencing and ubiquitin proteomics in FBXO11 knockout cells to identify its downstream targets and assaying for reduced expression of FBXO11 in primary patient MDS and AML samples. Based on our studies, we predict that SCF ubiquitin ligase component FBXO11 is a tumor suppressor regulating the transformation of MDS to secondary AML.
Crispino:Sierra Oncology: Consultancy; MPN Research Foundation: Membership on an entity's Board of Directors or advisory committees; Forma Therapeutics: Research Funding; Scholar Rock: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.