TO THE EDITOR:
Anaplastic large cell lymphomas (ALCLs) represent a group of CD30-positive T-cell non-Hodgkin lymphomas with unifying morphological characteristics but variable clinical and genetic features. ALCLs are classified by their clinical presentation and the presence or absence of rearrangements of the anaplastic lymphoma kinase (ALK) gene. In addition to systemic ALK-positive ALCL, systemic ALK-negative ALCL, and primary cutaneous ALCL, the 2016 revision of the World Health Organization classification of lymphoid neoplasms recognizes breast implant–associated ALCL (BIA-ALCL) as a new provisional entity.1 BIA-ALCL arises in the capsule and/or effusion surrounding silicone or saline-filled textured breast implants an average of 9 years after placement.2,3 BIA-ALCL shares many features with other types of ALCL, including consistent expression of CD30; cases reported to date have been ALK negative.1,2,4 The prognosis of patients with BIA-ALCL is favorable if the tumor is confined to the capsule and effusion and complete resection is performed; invasion through the capsule with the presence of an associated mass and lymph node involvement are adverse prognostic features.2,5,6
Our group has identified 2 additional recurrent rearrangements in ALCLs that to date have been mutually exclusive with ALK rearrangements. Rearrangements involving the DUSP22-IRF4 locus on 6p25.3 are associated with loss of expression of DUSP22, a putative tumor-suppressor gene encoding dual-specificity phosphatase 22.7 Rearrangements of the TP53 homolog TP63 on 3q28 lead to the formation of fusion genes, most commonly with the partner TBL1XR1 on 3q26.8 ALCLs lacking all 3 rearrangements (ie, ALK, DUSP22, and TP63) have been referred to as “triple-negative” ALCLs.9 Laurent et al examined 9 BIA-ALCLs for DUSP22 rearrangements, and all cases were negative.4 However, a comprehensive study of the genetic subtype of BIA-ALCL cases has not been reported.
Under an institutional review board–approved protocol, we evaluated the genetic subtype and other pathologic features in the capsules from 36 patients with BIA-ALCL. Detailed methods are given in supplemental Materials (available on the Blood Web site). All patients were women, with a mean age of 56 years (range, 33-76 years). Notably, all cases had a triple-negative genetic subtype (Table 1; supplemental Figure 1). This result is in contrast to the substantial genetic heterogeneity observed in other types of ALCL (Figure 1A). Among systemic ALCLs, ∼55% of cases carry ALK rearrangements and overexpress ALK fusion proteins.10 In systemic ALK-negative ALCLs, ∼30% have DUSP22 rearrangements and 8% have TP63 rearrangements.9 DUSP22 rearrangements have been associated with excellent outcomes, whereas patients with TP63 rearrangements have very poor outcomes.9,11 Although we did not find TP63 rearrangements in the present study, it is possible a larger study might identify such cases, given that only a minority of BIA-ALCLs are clinically aggressive (Table 1; previously reported series2,4,12 ). Primary cutaneous ALCLs are characteristically ALK negative, and DUSP22 and TP63 rearrangements occur at about the same frequency as in systemic ALK-negative ALCL.8,13 The findings we report in BIA-ALCL tissue specimens are consistent with the previously reported lack of characteristic lymphoma-associated translocations in karyotypic studies of BIA-ALCL–derived cell lines.14,15
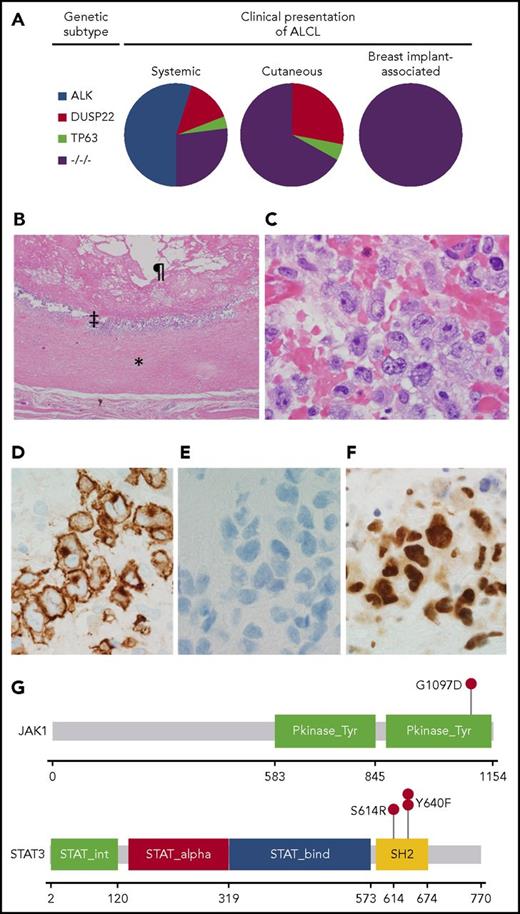
Pathological and genetic findings in BIA-ALCL. (A) Distribution of genetic subtypes in systemic, cutaneous, and breast implant–associated ALCL. −/−/−, triple-negative. (B) Low-power hematoxylin and eosin–stained image of a capsulectomy specimen showing the fibrous capsule (*), an inner layer of lymphoma cells (‡), and the original location of the implant and surrounding effusion (¶). Image was taken using an Olympus DP71 camera, Olympus BX51 microscope, and Olympus cellSens image acquisition software (original magnification ×40). (C) High-power hematoxylin and eosin–stained image of the capsule shows a cluster of large pleomorphic cells (original magnification ×1000). (D) Immunohistochemistry for CD30 shows strong and uniform staining in the neoplastic cells (original magnification ×1000). (E) Immunohistochemistry for ALK shows absence of staining in the neoplastic cells (original magnification ×1000). (F) Immunohistochemistry for pSTAT3Y705 shows strong nuclear staining in the neoplastic cells (original magnification ×1000). (G) Mutations in JAK1 and STAT3 in BIA-ALCL. Pkinase_Tyr, protein tyrosine kinase domain; SH2, Src homology 2 domain; STAT_alpha, all-α domain; STAT_bind, DNA binding domain; STAT_int, protein interaction domain.
Pathological and genetic findings in BIA-ALCL. (A) Distribution of genetic subtypes in systemic, cutaneous, and breast implant–associated ALCL. −/−/−, triple-negative. (B) Low-power hematoxylin and eosin–stained image of a capsulectomy specimen showing the fibrous capsule (*), an inner layer of lymphoma cells (‡), and the original location of the implant and surrounding effusion (¶). Image was taken using an Olympus DP71 camera, Olympus BX51 microscope, and Olympus cellSens image acquisition software (original magnification ×40). (C) High-power hematoxylin and eosin–stained image of the capsule shows a cluster of large pleomorphic cells (original magnification ×1000). (D) Immunohistochemistry for CD30 shows strong and uniform staining in the neoplastic cells (original magnification ×1000). (E) Immunohistochemistry for ALK shows absence of staining in the neoplastic cells (original magnification ×1000). (F) Immunohistochemistry for pSTAT3Y705 shows strong nuclear staining in the neoplastic cells (original magnification ×1000). (G) Mutations in JAK1 and STAT3 in BIA-ALCL. Pkinase_Tyr, protein tyrosine kinase domain; SH2, Src homology 2 domain; STAT_alpha, all-α domain; STAT_bind, DNA binding domain; STAT_int, protein interaction domain.
The morphological features in all BIA-ALCLs were similar. All cases were characterized by large cells with pleomorphic, multilobated, or wreath-like nuclei and abundant cytoplasm, often localized along the inner surface of the fibrous capsule surrounding the implant (Figure 1B-C). This predominance of large, pleomorphic cells is consistent with the triple-negative genetic subtype observed in BIA-ALCLs, as large, pleomorphic cells are less common in ALCLs with DUSP22 or TP63 rearrangements based on our published and unpublished observations.8,16 The frequencies of expression of T-cell antigens, cytotoxic markers, and T-cell receptor proteins in BIA-ALCL (supplemental Table 1) were similar to those previously reported in this disease4 as well as in systemic ALK-negative ALCL with triple-negative genetics.9 All cases showed strong and uniform reactivity for CD30 (Figure 1D) and were negative for ALK (Figure 1E). Similar to other forms of ALCL,17 a subset of cases expressed p63 protein but lacked TP63 rearrangements. Immunohistochemistry for pSTAT3 was positive in all 27 cases tested (100%), with staining in 30% to 100% of tumor cell nuclei (Figure 1F). In situ hybridization for Epstein-Barr virus was negative in all cases tested.
The findings in the present study demonstrate remarkable consistency of BIA-ALCLs in genetic subtype, morphology, and presence of activated STAT3 that contrasts with other types of ALCL. While ALK fusion proteins consistently drive STAT3 activation in ALK-positive ALCL,18 only 38% to 47% of ALK-negative ALCLs are positive for pSTAT3 by immunohistochemistry.19-21 STAT3 activation in ALK-negative ALCLs is mediated by somatic JAK1 and/or STAT3 mutations, a variety of non-ALK kinase gene fusions, and possibly other events.20,22 Blombery et al studied 2 BIA-ALCLs and identified JAK1 G1079V in 1 case and STAT3 S614R in the other,23 while Di Napoli et al identified STAT3 S614R in 1 of 5 informative cases.24 To understand the spectrum of JAK-STAT gene mutations further, we evaluated 15 BIA-ALCLs with adequate DNA for mutations of JAK1, JAK3, STAT3, STAT5A, and STAT5B. All mutations identified were orthogonally validated.
Four BIA-ALCLs (27%) had mutations known or predicted to be activating in one of the JAK-STAT genes (Table 1; supplemental Table 2; Figure 1G). One case had JAK1 G1097D, which has been shown to constitutively activate STAT320 and involves the same codon as the JAK1 mutation reported by Blombery et al.23 Another case had STAT3 S614R, found by both Blombery et al23 and Di Napoli et al24 and also present in the TLBR1 BIA-ALCL cell line.25 Two cases had the known gain-of-function mutation STAT3 Y640F.20,26 One case had a previously unreported STAT5B mutation, T479M, which was in the DNA-binding domain rather than the SH2 domain hotspot and was not predicted to be activating. No missense mutations were identified in JAK3 or STAT5A. Collectively, our sequencing data and the uniform expression of pSTAT3 extend earlier findings4 that suggest that BIA-ALCL is characterized by oncogenic JAK-STAT3 activation. Importantly, pharmacological inhibition of the JAK-STAT3 pathway has efficacy in preclinical ALCL models, suggesting a potential role for targeted therapies.15,18,20,25 Of note, only 1 patient was found to have both an activating JAK-STAT gene mutation and relatively advanced clinical disease (tumor stage T3 or T4 and/or nodal disease5 ). This lack of correlation might be due to the limited statistical power of the current series, but it also may suggest that molecular factors other than JAK-STAT3 pathway activation are important determinants of clinical aggressiveness. Since pSTAT3Y705 was expressed in all BIA-ALCLs regardless of mutational status, it is likely that more comprehensive genetic analysis will reveal alternative mechanisms of STAT3 activation, as shown in other forms of ALK-negative ALCL.20,22
Taken together, these observations indicate that BIA-ALCLs not only have unique clinical manifestations but also are consistently of triple-negative genetic subtype and show activation of the JAK-STAT3 signaling pathway. Similar to other ALCLs, this activation is associated with mutations of JAK1 or STAT3 in some cases. The absence of ALK, DUSP22, and TP63 rearrangements suggest less heterogeneity than in other ALCL subtypes. Future studies should evaluate whether additional rearrangements are present, such as those involving VAV1, FRK, ROS1, and TYK2.20,22,27 The relative molecular homogeneity of BIA-ALCL in the setting of its unique clinical presentation provides support for its inclusion as a distinct entity in the World Health Organization classification and suggests that these cases share a unifying pathogenetic mechanism, likely related to the inflammatory milieu involving the particulate textured surface of breast implants in which they arise.
Acknowledgments
This work was supported by grants from the National Cancer Institute, National Institutes of Health (R01 CA177734 [A.L.F.]) (P30 CA15083 [Mayo Clinic Cancer Center] and P50 CA97274 [University of Iowa/Mayo Clinic Lymphoma SPORE]), the National Center for Advancing Translational Science (Clinical and Translational Science Award UL1 TR000135), the Damon Runyon Cancer Research Foundation (CI-48-09) (A.L.F.), the Plastic Surgery Foundation, the American Society of Plastic Surgeons (MD Anderson Cancer Center), and the Department of Laboratory Medicine and Pathology and the Center for Individualized Medicine, Mayo Clinic.
Authorship
Contribution: N.O., R.N.M., and A.L.F. designed the study; G.S.B., C.A.H., R.L.B., E.D.M., N.N.B., K.S., M.W.C., L.J.M., R.N.M., and A.L.F. provided samples and interpreted data; N.O. and A.L.F. performed pathology review and interpretation; R.P.K. and C.A.S. scored and interpreted fluorescence in situ hybridization testing; D.S.V., R.H., and M.M. designed and performed next-generation sequencing studies; H.K.B. performed nucleic acid extraction and analysis; S.D. performed bioinformatic analysis; A.L.F. drafted the manuscript; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: A.L.F. and R.P.K. are inventors of technology discussed in this manuscript for which Mayo Clinic holds unlicensed patents. The remaining authors declare no competing financial interests.
Correspondence: Andrew L. Feldman, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN 55905; e-mail: feldman.andrew@mayo.edu; and Roberto N. Miranda, Department of Hematopathology, M.D. Anderson Cancer Center, Houston, TX 77030; e-mail: roberto.miranda@mdanderson.org.
References
Author notes
The online version of this article contains a data supplement.
R.N.M. and A.L.F. contributed equally to this study.