Key Points
Eleven pedigrees were identified with biallelic pathogenic variants in RASGPR2, which encodes platelet CalDAG-GEFI.
CalDAG-GEFI deficiency is a severe, recessive, nonsyndromic platelet function disorder with defective aggregation to multiple agonists.
Abstract
Heritable platelet function disorders (PFDs) are genetically heterogeneous and poorly characterized. Pathogenic variants in RASGRP2, which encodes calcium and diacylglycerol-regulated guanine exchange factor I (CalDAG-GEFI), have been reported previously in 3 pedigrees with bleeding and reduced platelet aggregation responses. To better define the phenotype associated with pathogenic RASGRP2 variants, we compared high-throughput sequencing and phenotype data from 2042 cases in pedigrees with unexplained bleeding or platelet disorders to data from 5422 controls. Eleven cases harbored 11 different, previously unreported RASGRP2 variants that were biallelic and likely pathogenic. The variants included 5 high-impact variants predicted to prevent CalDAG-GEFI expression and 6 missense variants affecting the CalDAG-GEFI CDC25 domain, which mediates Rap1 activation during platelet inside-out αIIbβ3 signaling. Cases with biallelic RASGRP2 variants had abnormal mucocutaneous, surgical, and dental bleeding from childhood, requiring ≥1 blood or platelet transfusion in 78% of cases. Platelets displayed reduced aggregation in response to adenosine 5′-diphosphate and epinephrine, but variable aggregation defects with other agonists. There were no other consistent clinical or laboratory features. These data enable definition of human CalDAG-GEFI deficiency as a nonsyndromic, recessive PFD associated with a moderate or severe bleeding phenotype and complex defects in platelet aggregation.
Introduction
Heritable platelet function disorders (PFDs) are genetically heterogeneous rare diseases characterized by mucocutaneous, surgical, and traumatic bleeding1,2 and reduced platelet responses to activating agonists.3,4 Human PFDs have been associated with causal variants in >30 genes, often associated with distinctive clinical or laboratory phenotypes that inform selection of candidate genes for diagnosis.5 Several new PFDs with less distinctive phenotypes have recently been identified by using high-throughput sequencing.6-8 However, to date, these have only been reported in small numbers of pedigrees and are incompletely characterized.
One such example is the PFD associated with loss-of-function variants in RASGRP2, which encodes calcium and diacylglycerol-regulated guanine exchange factor I (CalDAG-GEFI). This PFD was first identified in a consanguineous pedigree with a homozygous p.G248W variant associated with bleeding and reduced platelet light transmission aggregation (LTA) responses to multiple agonists.9 CalDAG-GEFI deficiency has been reported subsequently in only 3 pedigrees worldwide.10,11
To better define the phenotype and mutational spectrum of CalDAG-GEFI deficiency, we analyzed data from a collection of 2042 cases with unexplained bleeding or platelet disorders (BPDs) to identify 11 new affected pedigrees.
Study design
RASGRP2 was investigated in 2 study collections in which all participants were enrolled in the National Institute for Health Research BioResource - Rare Diseases between 2012 and 2016 after providing informed written consent (UK Research Ethics Committee approval 13/EE/0325). The first collection comprised 1472 cases or pedigree members with unexplained BPDs and 5422 cases with other rare inherited disorders who underwent whole-exome or genome sequencing.12 The second collection comprised 570 other BPD cases analyzed by using the ThromboGenomics high-throughput platform,13 which captures variants in 81 tier 1 genes for BPD, designated by the ISTH Genomics Scientific and Standardization Subcommittee.
Collection of clinical and laboratory phenotypes and coding with Human Phenotype Ontology (HPO ) terms were performed as previously described.12 Variants from high-throughput sequencing were obtained by using Isaac (Illumina, Inc., San Diego, CA) or as described previously.12 Variants were not considered to be potentially pathogenic if the Variant Effect Predictor14 identified them as synonymous relative to canonical transcript ENST00000354024, or if the allele frequency was >1 per 1000 in reference databases.12,15 Index cases in the National Institute for Health Research (NIHR) BioResource were classified as having an indicator phenotype of CalDAG-GEFI deficiency if they had HPO terms indicating abnormal bleeding and reduced LTA responses to ≥3 agonists. Associations between rare and nonsynonymous RASGRP2 variants in the NIHR BioResource and the presence or absence of the indicator phenotype were then assessed by using the BeviMed method.16
Results and discussion
The CalDAG-GEFI deficiency indicator phenotype of bleeding and reduced LTA responses to ≥3 agonists were present in 119 (9.7%) of 1229 BPD index cases from the NIHR BioResource. Analysis of all the rare, nonsynonymous variants in the BPD and non-BPD index cases revealed a strong statistical association between the indicator phenotype and the presence of ≥2 RASGRP2 alleles, obtaining a posterior probability of association with recessive inheritance of 1 by inference using the BeviMed method (Figure 1A). The association was driven by 8 different variants observed in 8 BPD index cases as homozygous or compound heterozygous alleles (Figures 1B and 2A, pedigrees I-K). There were an additional 35 RASGRP2 variants, which were not associated with the indicator phenotype or which were present as a single allele (Figure 1A). In the ThromboGenomics collection, there were 4 rare, nonsynonymous RASGRP2 variants that were biallelic (Figure 1B) in 3 index cases with bleeding and reduced LTA responses to ≥3 agonists (Figure 2A, pedigrees I-K). Since this analysis, pedigree G has also been reported elsewhere.18
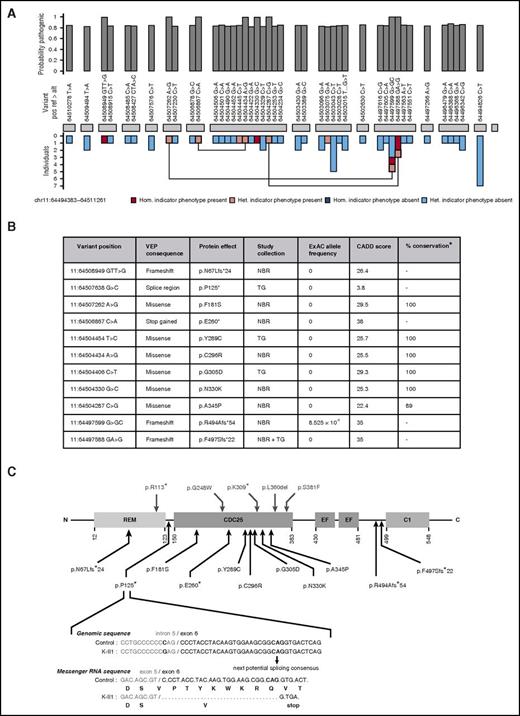
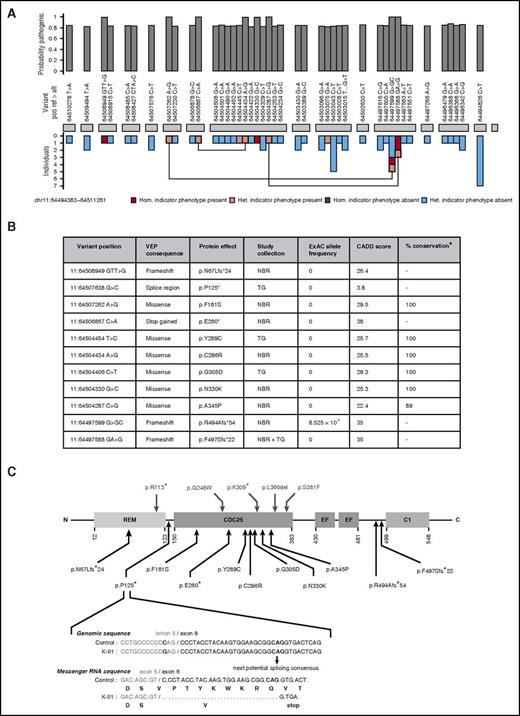
Identification and characteristics of pathogenic variants in RASGRP2. (A) BeviMed inference analysis applied to rare, nonsynonymous RASGRP2 variants observed in all index cases from the NIHR BioResource. Cases were designated as having the indicator phenotype of CalDAG-GEFI deficiency if they had HPO terms indicating bleeding and reduced light transmission aggregation responses to ≥3 activating agonists. The indicator phenotype was present in 119 index cases and was absent in 5982 index cases. The posterior probability of the association model was 1 (prior was 0.1), and the posterior probability of recessive inheritance was 1 (prior was 0.5). The RASGRP2 exons are represented by gray blocks. The bar chart above shows the marginal posterior probabilities of pathogenicity for individual variants observed in all NIHR BioResource index cases, conditional on an association under a recessive mode of inheritance. The bar chart beneath indicates whether the variant was observed in an index case with (pink) or without (blue) the indicator phenotype and whether it was present as a heterozygous (het.) or homozygous (hom.) allele. Variants observed in index cases as compound heterozygous alleles are linked. (B) The characteristics of the 11 likely pathogenic RASGRP2 variants across the NIHR BioResource (NBR) and ThromboGenomics (TG) collections. Variants were annotated against the canonical transcript ENST00000354024 by using the Variant Effect Predictor (VEP).14 Population allelic frequencies are derived from the Exome Aggregation Consortium (ExAC).15 The likely pathogenicity of the variants is expressed as the Combined Annotation Dependent Depletion (CADD) score,17 and the percentage conservation as the proportion of CalDAG-GEFI orthologs in 9 species that have the same amino acid as human CalDAG-GEFI. (C) Localization and predicted consequence of the likely pathogenic RASGRP2 variants identified in the NIHR BioResource and ThromboGenomics collections (below the protein diagram) and the previously reported variants9-11 (above the diagram). The protein domains indicated are the Ras exchange motif (REM), catalytic domain (CDC25), calcium-binding EF hands (EF) and diacylglycerol-binding domain (C1). The exploded view shows the predicted consequence of the 11:64507638 G>C variant identified as a homozygous allele in ThromboGenomics case K II.1. Bioinformatic analysis of the variant sequence predicts preferential use of an alternative exonic splice acceptor, resulting in codon deletion, frameshift, and a stop gain at codon 125.
Identification and characteristics of pathogenic variants in RASGRP2. (A) BeviMed inference analysis applied to rare, nonsynonymous RASGRP2 variants observed in all index cases from the NIHR BioResource. Cases were designated as having the indicator phenotype of CalDAG-GEFI deficiency if they had HPO terms indicating bleeding and reduced light transmission aggregation responses to ≥3 activating agonists. The indicator phenotype was present in 119 index cases and was absent in 5982 index cases. The posterior probability of the association model was 1 (prior was 0.1), and the posterior probability of recessive inheritance was 1 (prior was 0.5). The RASGRP2 exons are represented by gray blocks. The bar chart above shows the marginal posterior probabilities of pathogenicity for individual variants observed in all NIHR BioResource index cases, conditional on an association under a recessive mode of inheritance. The bar chart beneath indicates whether the variant was observed in an index case with (pink) or without (blue) the indicator phenotype and whether it was present as a heterozygous (het.) or homozygous (hom.) allele. Variants observed in index cases as compound heterozygous alleles are linked. (B) The characteristics of the 11 likely pathogenic RASGRP2 variants across the NIHR BioResource (NBR) and ThromboGenomics (TG) collections. Variants were annotated against the canonical transcript ENST00000354024 by using the Variant Effect Predictor (VEP).14 Population allelic frequencies are derived from the Exome Aggregation Consortium (ExAC).15 The likely pathogenicity of the variants is expressed as the Combined Annotation Dependent Depletion (CADD) score,17 and the percentage conservation as the proportion of CalDAG-GEFI orthologs in 9 species that have the same amino acid as human CalDAG-GEFI. (C) Localization and predicted consequence of the likely pathogenic RASGRP2 variants identified in the NIHR BioResource and ThromboGenomics collections (below the protein diagram) and the previously reported variants9-11 (above the diagram). The protein domains indicated are the Ras exchange motif (REM), catalytic domain (CDC25), calcium-binding EF hands (EF) and diacylglycerol-binding domain (C1). The exploded view shows the predicted consequence of the 11:64507638 G>C variant identified as a homozygous allele in ThromboGenomics case K II.1. Bioinformatic analysis of the variant sequence predicts preferential use of an alternative exonic splice acceptor, resulting in codon deletion, frameshift, and a stop gain at codon 125.
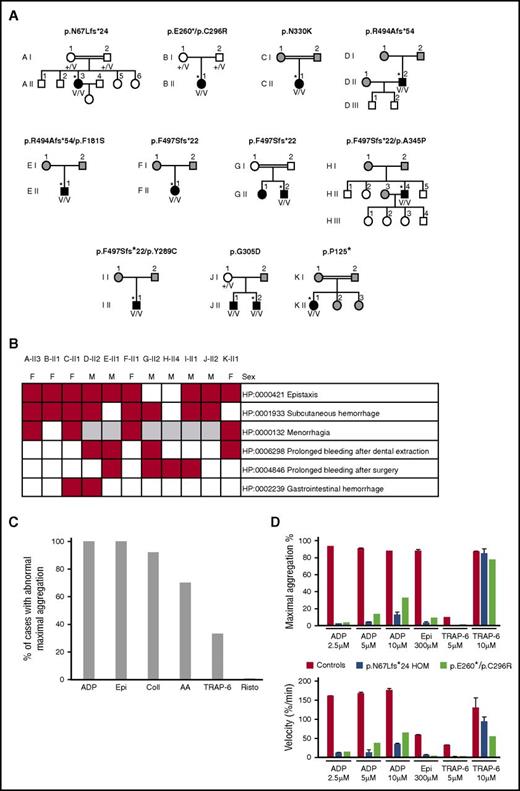
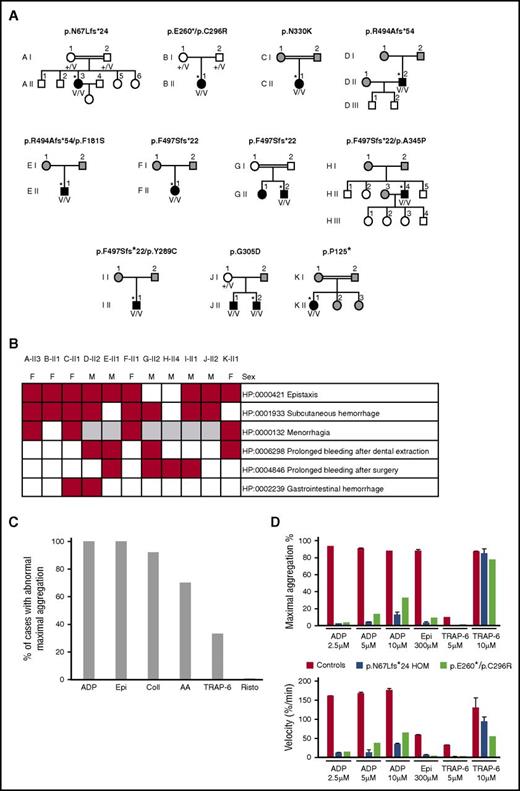
Characteristics of the 11 index cases with likely pathogenic biallelic RASGRP2 variants. (A) Pedigrees of the index cases (*) with likely pathogenic RASGRP2 variants indicating the genotype of the index cases. The black symbols indicate cases with abnormal bleeding and reduced platelet aggregation responses. The white symbols indicate pedigree members without bleeding symptoms, and the gray symbols indicate pedigree members unavailable for evaluation. +/V, heterozygous; V/V, compound heterozygous or homozygous for the variant allele. (B) Annotation of 11 index cases with HPO terms for bleeding symptoms. (C) Summary light transmission aggregation for all 11 index cases. Data are expressed as the proportion of index cases with any reported defect in aggregation responses to ADP, epinephrine (Epi), collagen (Coll), arachidonic acid (AA), TRAP-6, and ristocetin (Risto). (D) Detailed light transmission aggregation response in cases AII-3 and BII-1 in response to the stated agonist concentrations. Data are presented as the maximum aggregation and initial velocity of the aggregation responses.
Characteristics of the 11 index cases with likely pathogenic biallelic RASGRP2 variants. (A) Pedigrees of the index cases (*) with likely pathogenic RASGRP2 variants indicating the genotype of the index cases. The black symbols indicate cases with abnormal bleeding and reduced platelet aggregation responses. The white symbols indicate pedigree members without bleeding symptoms, and the gray symbols indicate pedigree members unavailable for evaluation. +/V, heterozygous; V/V, compound heterozygous or homozygous for the variant allele. (B) Annotation of 11 index cases with HPO terms for bleeding symptoms. (C) Summary light transmission aggregation for all 11 index cases. Data are expressed as the proportion of index cases with any reported defect in aggregation responses to ADP, epinephrine (Epi), collagen (Coll), arachidonic acid (AA), TRAP-6, and ristocetin (Risto). (D) Detailed light transmission aggregation response in cases AII-3 and BII-1 in response to the stated agonist concentrations. Data are presented as the maximum aggregation and initial velocity of the aggregation responses.
Of the 11 likely pathogenic RASGRP2 variants, 4 resulted in frameshift or a stop codon when annotated against the RASGRP2 canonical transcript (Figure 1B-C). One additional variant (11:64507638 G>C: pedigree K) was at the −3 position in the intron 5 splice region, adjacent to the splice acceptor site. The Alamut Visual splicing module (Interactive Biosoftware, Rouen, France) predicted this to result in loss of the native splice acceptor site and co-option of a cryptic acceptor site in exon 6, leading to the deletion of 9 codons and a stop gain at codon 125 (Figure 1C). Thus, a total of 5 variants were predicted to prevent full-length CalDAG-GEFI expression.
All 6 missense variants predicted substitutions of amino acids that were conserved in CalDAG-GEFI orthologs in 9 distantly related species, except for human residue A345, which is a T residue in the Xenopus ortholog (Figure 1B; supplemental Table 1, available on the Blood Web site). All had high CADD pathogenicity scores (>20; within the top 1% of deleterious variants genome-wide)17 and were either absent or at a very low allele frequency in the Exome Aggregation Consortium population database (Figure 1B).15 All the predicted substitutions were in the CDC25 domain of CalDAG-GEFI, which is essential for guanine nucleotide exchange activity and directly interacts with Rap1, the major platelet target for CalDAG-GEFI in the inside-out αIIbβ3 signaling pathway.19 This suggests that the missense variants reduce platelet function by altering CalDAG-GEFI function, although we cannot exclude reduced protein expression, as observed previously for the RASGRP2 p.Ser381Phe variant.11 There were no consistent phenotype differences between the cases with missense and high-impact variants.
The 11 likely pathogenic RASGRP2 variants occurred in a total of 11 unrelated cases (7 males) as homozygous (7 cases) or compound heterozygous (4 cases) alleles, indicating autosomal recessive inheritance. One pedigree (J) contained a sibling with biallelic RASGRP2 variants (Figure 2A). All of the index cases had a first diagnosis of a bleeding disorder during childhood (age 1-14 years; supplemental Table 2). Abnormal bleeding was predominantly mucocutaneous or followed surgery or dental extraction. Gastrointestinal bleeding occurred in 2 cases (Figure 2B), but there was no intracranial bleeding. Of the 9 index cases with available data, 7 had required red cell or platelet transfusion on ≥1 occasion, suggesting severe bleeding. There were no other consistently reported phenotypes. Platelet numbers and sizes were normal (supplemental Table 2).
All 11 index cases had reduced platelet LTA responses with adenosine 5′-diphosphate (ADP) and epinephrine and, in some cases, also with collagen, arachidonic acid, and TRAP-6 (Figure 2C-D). Detailed analysis of A-II3 and B-II1 showed additionally that the velocity of aggregation to all tested doses of ADP, epinephrine, and TRAP-6 was reduced compared with controls (Figure 2D). The normal platelet ultrastructure, nucleotide content, and release of α (P-selectin exposure) and dense (CD63 exposure) granules in response to high-dose TRAP-14 indicated no defect in platelet granule biosynthesis. However, dense granule release with ADP was diminished in most tested cases, mirroring the functional defects observed with LTA (supplemental Table 2), similar to previous reports.9-11
This report of 11 new pedigrees significantly expands the reported cases with human CalDAG-GEFI deficiency and enables description of this disorder as an autosomal recessive, nonsyndromic PFD that is associated with moderate or severe bleeding, similar to other disorders of αIIbβ3 integrin signaling20,21 and some types of Glanzmann thrombasthenia in which there is reduced expression of functional αIIbβ3 integrin.22 In addition, we show that there is a consistent laboratory phenotype of reduced aggregation responses to ADP and epinephrine in all reported cases, but no defect in dense granule secretion, distinguishing this disorder from δ-storage pool disease. This distinctive phenotype is likely to assist genetic diagnosis of further pedigrees.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jean-Christophe Gris (Nîmes, France), Ségolène Claeyssens (Toulouse, France), and Yeon Ahn (Miami, FL) for providing the genomic DNA for index cases A, B, and G and Alan Nurden (Bordeaux, France) for contributing to the initial characterization of the platelet function abnormality of the index cases in pedigrees A, B, G, and K.
The NIHR BioResource–Rare Diseases is funded by the National Institute for Health Research of England (www.nihr.ac.uk; award RG65966). S.K.W. is supported by a Medical Research Council Clinical Research Training Fellowship (MR/K023489/1). C.M.M. and M.A.L. are supported by the NIHR Imperial College Biomedical Research Centre. K.F. and C.V.G. are supported by the Fund for Scientific Research-Flanders (Vlaanderen, Belgium; grant G.0B17.13N) and the Research Council of the University of Leuven (BOF KU Leuven, Belgium, grant OT/14/098). W.H.O. is supported by the British Heart Foundation, the European Commission, the Medical Research Council, the NIHR, the Wellcome Trust, and the National Health Service Blood and Transplant. A.D.M. is supported by the NIHR Biomedical Research Centre at the University Hospitals Bristol National Health Service Foundation Trust and the University of Bristol.
The views expressed in this publication are those of the authors and not necessarily those of the National Health Service, the NIHR, or the UK Department of Health.
Authorship
Contribution: S.K.W. and M.C. wrote the paper with assistance from M.-C.A., E.T., and A.D.M.; M.C., D.G., E.T., and S.K.W. analyzed data; E.B., K.H., M.P.L., C.M.M., P.N., S.G.O., S.R.-V., and C.V.G. provided samples and clinical data; K.D. managed the ThromboGenomics Program; S.P. coordinated the NIHR BioResource–Rare Diseases BPD project, including ethics and governance; S.T. and C.W. provided sample logistics, quality control, and whole genome sequencing oversight; and K.F., M.A.L., and W.H.O. contributed to the study design.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the NIHR BioResource–Rare Diseases Consortium appears in the online appendix.
Correspondence: Andrew D. Mumford, School of Clinical Sciences, University of Bristol, Research Floor Level 7, Bristol Royal Infirmary, Upper Maudlin St, Bristol BS2 8HW, United Kingdom; e-mail a.mumford@bristol.ac.uk.
References
Author notes
S.K.W. and M.C. contributed equally to this study.
E.T. and A.D.M. contributed equally to this study.