Key Points
A prospectively isolatable and functionally homogeneous human megakaryocyte progenitor population resides in CMP fraction of adult bone marrow.
This newly identified unipotent megakaryocyte progenitor significantly contributes to normal and malignant megakaryopoiesis.
The developmental pathway for human megakaryocytes remains unclear, and the definition of pure unipotent megakaryocyte progenitor is still controversial. Using single-cell transcriptome analysis, we have identified a cluster of cells within immature hematopoietic stem- and progenitor-cell populations that specifically expresses genes related to the megakaryocyte lineage. We used CD41 as a positive marker to identify these cells within the CD34+CD38+IL-3RαdimCD45RA− common myeloid progenitor (CMP) population. These cells lacked erythroid and granulocyte-macrophage potential but exhibited robust differentiation into the megakaryocyte lineage at a high frequency, both in vivo and in vitro. The efficiency and expansion potential of these cells exceeded those of conventional bipotent megakaryocyte/erythrocyte progenitors. Accordingly, the CD41+ CMP was defined as a unipotent megakaryocyte progenitor (MegP) that is likely to represent the major pathway for human megakaryopoiesis, independent of canonical megakaryocyte-erythroid lineage bifurcation. In the bone marrow of patients with essential thrombocythemia, the MegP population was significantly expanded in the context of a high burden of Janus kinase 2 mutations. Thus, the prospectively isolatable and functionally homogeneous human MegP will be useful for the elucidation of the mechanisms underlying normal and malignant human hematopoiesis.
Introduction
During the process of differentiation into functional blood cells, hematopoietic stem cells (HSCs) lose their multipotency and differentiate into hematopoietic lineage–committed progenitor cells (HPCs). In normal hematopoiesis, megakaryocyte and erythroid lineage development are closely related and share certain stages. For example, both of these lineages use common lineage-deterministic transcription factors, such as GATA-1, FOG-1, and Gfi-1b,1,,,-5 and bipotent megakaryocyte/erythrocyte progenitor (MEP) can be isolated at the single-cell level in mouse and human using a fluorescence-activated cell sorter (FACS).6,7 However, the developmental pathways of megakaryocyte and erythroid lineages are still open to argument.8
The immunophenotypic definitions of lineage-committed progenitors have provided the methodology for purifying functionally homogeneous population, which is important not only to elucidate the concept of hematopoietic development but also to understand the underlying mechanism of lineage commitment. In murine hematopoiesis, a unipotent megakaryocyte progenitor (MegP) was identified downstream of a bipotent MEP after megakaryocyte and erythroid lineage bifurcation by using cell-surface markers such as CD9, CD150, CD41, and CD42b as positive markers.9,-11 Recently, several studies have demonstrated the existence of HSCs with platelet-biased differentiation potential at the apex of hematopoietic hierarchy in normal and stress hematopoiesis.12,,-15 In human hematopoiesis, MEPs with both megakaryocyte and erythroid potential have been identified within the CD34+CD38+ HPC fraction,7 and recent data from intensive single-cell profiling of human CD34+CD38+IL-3Rα−CD45RA− MEPs suggested that MegPs reside inside or downstream of MEPs.16 However, megakaryocyte-producing cells constitute only a minor proportion of MEPs,16 and the lineage potential of MEPs is largely skewed toward the erythroid lineage; ∼80% of single MEPs produce pure erythroid colonies.7 In addition, Mori et al17 identified CD71+CD105+ unipotent erythroid progenitors within MEPs, confirming that a majority of human MEPs (∼85%) are committed to the erythroid lineage. Notta et al18 analyzed lineage output of hematopoietic stem/progenitor cells (HSPCs) and predicted the existence of a megakaryocyte lineage–restricted progenitor during early hematopoiesis, such as at HSC and multipotent progenitor (MPP) stages. These data collectively indicate that commitment to the megakaryocyte lineage could occur at an earlier stage, bypassing erythroid versus megakaryocyte lineage bifurcation in MEPs. To date, however, the purification of such a unipotent MegP in adult human hematopoiesis has not been successful.8 By searching for a cell population expressing exclusively megakaryocyte lineage–affiliated genes at the single-cell level, we have been able to identify MegPs within the early CD34+CD38+ human HPC population: common myeloid progenitors (CMPs).7 Our data also suggest that human MegPs are involved in the pathogenic thrombopoiesis observed in patients with myeloproliferative neoplasms.
Methods
Clinical samples
Human adult bone marrow (BM) samples were obtained from healthy donors or purchased from AllCells. BM samples of adult hematological malignancies, diagnosed according to World Health Organization criteria, were enrolled in this study. Cord blood (CB) cells were collected during normal full-term deliveries after obtaining informed consent in accordance with the Declaration of Helsinki (provided by Japanese Red Cross Kyushu Cord Blood Bank, Fukuoka, Japan). CB samples were mainly used for in vivo experiments unless otherwise indicated. Informed consent was obtained from all patients and controls in accordance with the Helsinki Declaration of 1975 (revised in 1983). The Institutional Review Board of Kyushu University Hospital approved all research on human participants.
Mice
Single-cell gene-expression profiling
Single cells of target populations were directly sorted into 96-well polymerase chain reaction (PCR) plates and subjected to reverse transcription using a CellsDirect One-Step quantitative real-time PCR kit (Invitrogen, Carlsbad, CA). After PCR-based preamplification was performed (11 cycles), amplified complementary DNA (cDNA) samples were loaded onto a Fluidigm Dynamic Array Integrated Fluidic Chip and subjected to quantitative real-time PCR using the Biomark system (Fludigm, South San Francisco, CA). The primers and probes used in this assay were obtained from Applied Biosystems and are listed in supplemental Table 1 (available on the Blood Web site). Samples exhibiting no GAPDH signals were omitted from the analysis. As recommended by manufacturer, normalization by GAPDH level was not performed.
Antibodies, cell staining, and sorting
Sorting of HSCs and HPCs was accomplished by staining BM or CB cells with Pacific Blue–conjugated anti-CD41 (HIP8) monoclonal antibody, fluorescein isothiocyanate–conjugated anti-CD34 (561), phycoerythrin (PE)-conjugated anti-CD105 (43A3), Biotin-conjugated anti–interleukin-3Rα (IL-3Rα; 6H6) followed by streptavidin-PE/Cy7, allophycocyanin (APC)-conjugated anti-CD38 (HIT2), APC/Cy7-conjugated anti-CD45RA (HI100), and PerCP/Cy5.5-conjugated antibodies specific for the following lineage markers: anti-human CD3 (UCHT1), CD4 (SK3), CD8 (SK1), CD10 (HI10a), CD11b (ICRF44), CD14 (HCD14), CD19 (HIB19), CD20 (2H7), CD56 (HCD56), CD235ab (HIR2; BioLegend, San Diego, CA). HSCs were defined as lineage marker–negative (Lin−) CD34+ CD38− CD45RA−. HPCs were sorted as Lin− CD34+ CD38+ IL-3Rαdim CD45RA− (CMPs), Lin− CD34+ CD38+ IL-3Rαdim CD45RA+ (granulocyte-macrophage progenitors [GMPs]), and Lin− CD34+ CD38+ IL-3Rα− CD45RA− (MEPs) as described previously.7 All of these stem and progenitor cells were double sorted using the BD FACS Aria III cell-sorting system (BD Biosciences, San Jose, CA). For the analysis of megakaryocyte markers, BM HSPCs from healthy donors were analyzed by FACS using the following antibodies: PE-conjugated CD9 (HI9a), CD42b (HIP1), CD51 (NKI-M9), CD61 (VI-PL2), CD62P (AK4; BioLegend), and APC-conjugated CD41 (HIP8). For the analysis of xenograft models, we used the following antibodies: fluorescein isothiocyanate–conjugated anti-human CD235ab (HIR2), CD3 (HIT3a), PE-conjugated anti-mouse TER119, anti-human CD33 (WM53), PerCP/Cy5.5-conjugated anti-mouse CD45 (30-F11), APC-conjugated anti-human CD19 (HIB19), anti-mouse CD41 (MWReg30), APC/Cy7-conjugated anti-human CD45 (HI30), and Pacific Blue–conjugated anti-human CD41 (HIP8).
In vitro culture
To test differentiation potential into whole-myeloid lineages, single cells were sorted directly onto Iscove’s Modified Dulbecco’s Medium–based methylcellulose medium (MethoCult H4034 Optimum; STEMCELL Technologies, Vancouver, Canada) in a 35-mm dish supplemented with recombinant human stem-cell factor (rhSCF; 20 ng/mL), rhIL-3 (20 ng/mL), rh–GM colony-stimulating factor (GM-CSF; 50 ng/mL), rh-thrombopoietin (TPO; 20 ng/mL), and rh-erythropoietin (EPO; 4 unit/mL; R&D Systems, Minneapolis, MN). For single-cell liquid culture assays, single cells were double sorted into wells of 96-well plates containing StemSpan SFEM II medium (Catalog No. 09655, STEMCELL Technologies) supplemented with rhSCF (20 ng/mL), rhIL-3 (20 ng/mL), rhIL-6 (20 ng/mL), rhIL-9 (20 ng/mL), rhGM-CSF (50 ng/mL), rhG-CSF (20 ng/mL), rhTPO (20 ng/mL), and rhEPO (4 unit/mL; R&D Systems) and cultured for 10 to 14 days. To test differentiation potential into a megakaryocyte lineage, MegaCult-C kit (Catalog No. 04973, STEMCELL Technologies) was used according to the manufacturer’s instructions. Cell characteristics of each culture were determined morphologically and cytochemically by May-Giemsa staining. To evaluate lineage relationships, 1500 HSPCs were cultured in 96-well plates with StemSpan SFEM II medium supplemented with Megakaryocyte Expansion supplement (Catalog No. 02696, STEMCELL Technologies) containing rhTPO, rhSCF, rhIL-6, rhIL-9, and 20% BIT 9500 Serum Substitute (containing bovine serum albumin, insulin, transferrin; STEMCELL Technologies). All cultures were incubated at 37°C in a humidified chamber under 5% carbon dioxide.
Cytospin preparation and morphological analysis
FACS-purified human HSPCs or cultured cells were centrifuged (at 600g for 1 minute) onto glass slides. Cytospin specimens were subjected to May-Giemsa staining.
In vivo assays
Mononuclear cells were concentrated by standard gradient centrifugation using LSM (Cosmo Bio Co., Ltd., Tokyo, Japan), and CD34-enriched CB cells were obtained using the EasySep Human CD34 Positive Selection Kit (STEMCELL Technologies). A total of 1 × 104 FACS-sorted Lin− CD34+ HSPC subpopulations were injected intrafemorally into 6- to 8-week-old BRGSKWv/Wv mice with irradiation preconditioning. BRGSKWv/Wv mice were irradiated at sublethal doses (150 cGy). Three weeks after transplantation, BM cells were harvested from femurs and subjected to FACS. Chimerism of human erythrocytes and platelets was determined based on the expression levels of human glycophorin A and TER119 (mouse erythroid) or human/mouse CD41 in the CD45− low forward-scatter fractions, respectively. Because lymphoid cells are barely detected in the BRGSKWv/Wv mice,20 engraftment of human myeloid and lymphoid cells was assessed by absolute counts. Animal experiments were conducted according to the guidelines of the Institutional Animal Committee of Kyushu University.
cDNA microarray analysis
Target populations of a minimum of 5000 cells were double sorted directly into Trizol reagent (Life Technologies). Total RNA was extracted according to the manufacturer’s instructions for Trizol. Cyanine3-labeled cRNA from each sample was prepared for messenger RNA (mRNA) amplification using the Low Input Quick Amp Labeling Kit (1 color; Agilent Technologies, Santa Clara, CA). cRNA (800 ng) was hybridized to the microarray chip (SurePrint G3 Human Gene Expression 8×60K v2; Agilent Technologies). Slides were scanned immediately after washing with Agilent SureScan Microarray Scanner (G4900DA) using the 1-color scan setting for 8×60k array slides (scan resolution of 3µm; dye channel was set to green, and green photomultiplier tube sensitivity was set to 100%). The scanned images were analyzed with Feature Extraction software (Agilent) using default parameters (protocol GE1_1105_Oct12 and grid: 039494_D_F_20120628) to obtain background subtracted and spatially detrended processed signal intensities. Raw expression data were imported and normalized using GeneSpring GX software (Agilent Technologies). The microarray data were deposited in Gene Expression Omnibus as GSE77439. Gene sets relevant to megakaryocyte lineage were obtained from the Molecular Signatures Database. Erythroid or GM lineage–specific genes were selected via extensive literature reviews.
Allele-specific PCR for JAK2 V617F detection
For the detection of Janus kinase 2 (JAK2) and calreticulin (CALR) mutations in patients with essential thrombocythemia (ET), patients’ genomic DNA samples were PCR amplified using the following primer sets: 5′-GCATCTTTATTATGGCAGAGAG-3′ and 5′-ACTGACACCTAGCTGTGATCC-3′ for JAK2 mutation; 5′-CCTGCAGGCAGCAGAGAAAC-3′ and 5′-ACAGAGACATTATTTGGCGCG-3′ for CALR mutation. The resultant PCR products were Sanger sequenced to identify mutations. Allele burden of JAK2 mutation was determined using the allele-specific probes (allele G probe: 5′-6-FAM-CGTCTCCACAGACAC-3′-BHQ1; allele T probe: 5′-HEX-CGTCTCCACAGAAAC-3′-BHQ1) as described.21
Statistical analysis
Data are presented as the mean plus or minus standard deviation. The significance of differences between groups was determined by Student t test for two groups and by 1-way analysis of variance followed by Tukey’s HSD test for multiple groups. Statistical analysis was performed with JMP (version 11.2) and GeneSpring GX software.
Results
Single-cell gene-expression profiling revealed distinct CMP subpopulation characterized by expression of megakaryocyte-specific genes
To identify the putative MegPs, we first tried to find a cluster of cells expressing megakaryocyte lineage–specific genes. To this end, we analyzed the expression profile of lineage-affiliated genes at the single-cell level in human HSPC populations predefined by Manz et al.7 Human megakaryocyte colony-forming unit (CFU) potential is concentrated in the CD45RA− fraction of CD34+ HSPCs, which consist of HSCs, CMPs, and MEPs; therefore, FACS-purified single cells from these populations were directly subjected to mRNA amplification and quantitative reverse transcription PCR analysis using the Fludigm Biomark system (Figure 1). Cell fate–determination transcription factors and cell surface markers specifically associated with each lineage3,22 were selected for evaluation (supplemental Table 1).
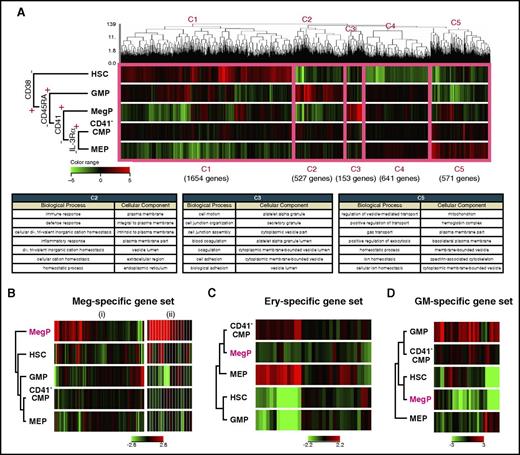
Single-cell gene expression profiling revealed a distinct CMP subpopulation characterized by expression of megakaryocyte-specific genes. Heat map visualization of single-cell gene expression profiling of BM-derived HSPCs. Gene expression levels of individual single-cells from (A) HSC, (B) MEP, and (C) CMP fractions are shown. Genes related to erythroid (Ery), megakaryocyte (Meg), or GM lineage are listed on the left side of the heat map. Annotation for each gene is presented in supplemental Table 1. Representative heat maps from 3 independent samples are shown. (B) Most MEPs expressed erythroid-specific genes (red rectangle) but not megakaryocyte-specific genes. (C) A cluster of cells that exclusively expressed megakaryocyte-specific genes at a high level was observed (green rectangle).
Single-cell gene expression profiling revealed a distinct CMP subpopulation characterized by expression of megakaryocyte-specific genes. Heat map visualization of single-cell gene expression profiling of BM-derived HSPCs. Gene expression levels of individual single-cells from (A) HSC, (B) MEP, and (C) CMP fractions are shown. Genes related to erythroid (Ery), megakaryocyte (Meg), or GM lineage are listed on the left side of the heat map. Annotation for each gene is presented in supplemental Table 1. Representative heat maps from 3 independent samples are shown. (B) Most MEPs expressed erythroid-specific genes (red rectangle) but not megakaryocyte-specific genes. (C) A cluster of cells that exclusively expressed megakaryocyte-specific genes at a high level was observed (green rectangle).
The majority of single CD34+CD38−CD45RA− HSCs did not express lineage-affiliated genes at a detectable level in this assay (Figure 1A). In contrast, 85% of single human CD34+CD38+CD45RA− IL-3Rα− MEPs7 expressed detectable levels of erythroid genes, including hemoglobin α1 (HBA1), EPO receptor (EPOR)23 and transferrin receptor 2 (TFR2),24 and erythroid lineage–determination transcription factors such as KLF-1 and Gfi-1b,3,22 forming a cluster (Figure 1B). Approximately 40% of this cluster expressed endoglin (CD105, represented as ENG), a marker for an erythroid lineage–committed progenitor residing within the MEP.17 The vast majority of MEPs expressed genes commonly associated with both megakaryocytic and erythroid differentiation, such as Gfi-1b and NF-E2, whereas expression of genes exclusively associated with megakaryocyte lineage was not detected (Figure 1B). The skewed expression of erythroid-related transcripts in MEPs is compatible with the biased differentiation potential of human MEPs into erythroid cells as demonstrated in previous studies.7,17
In contrast, only ∼50% of predefined single CD34+CD38+CD45RA− IL-3Rαdim CMPs expressed detectable levels of genes affiliated with erythroid, megakaryocyte, and/or myeloid lineages. Interestingly, a fraction (8%) of CMPs expressed megakaryocyte lineage–specific transcription factors and platelet-specific genes but not myeloid or erythroid genes and seemed to form a distinct cluster (Figure 1C). These genes included cell-surface molecules of mature platelets such as P-selectin/CD62P (SELP), thrombopoietin receptor (MPL),25 von Willebrand factor (VWF),26 and CD41 (ITGA2B),27 as well as megakaryocyte lineage–specific transcription factor FLI-1. Therefore, we hypothesized that this cluster could represent the unipotent MegP.
Immunophenotypic and morphological characteristics suggested that CD41+ CMPs were megakaryocyte lineage–committed cells
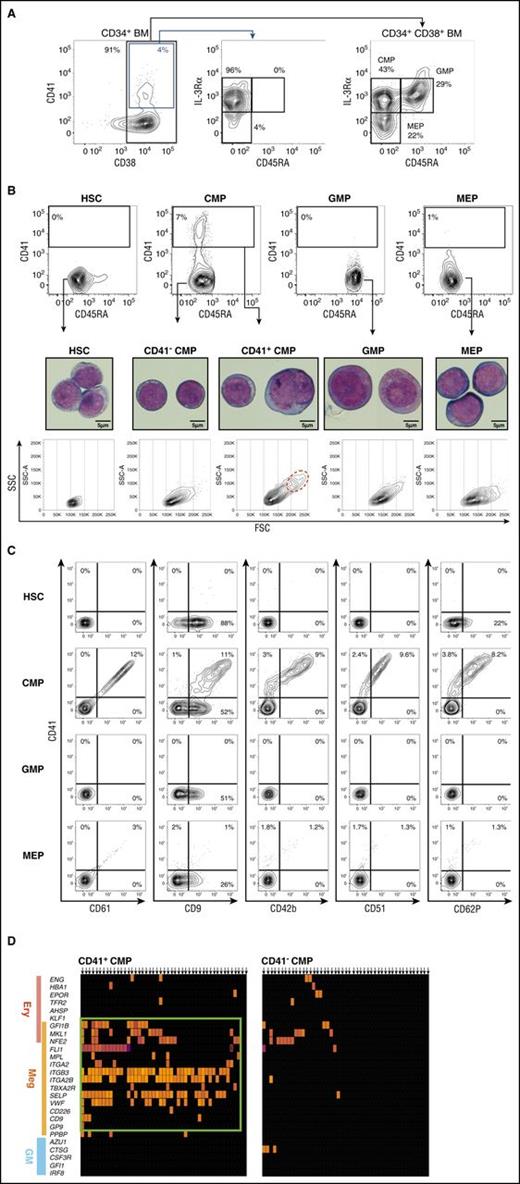
To isolate candidate MegPs, we searched for surface markers expressed exclusively in this population from a panel of megakaryocyte-related genes and found that CD41 is a useful positive marker. We then analyzed the distribution of surface CD41 among CD34+ HSPCs from human adult BM by FACS. CD41-positive cells were found exclusively in the CD45RA− IL-3Rαdim HSPC fraction (Figure 2A). Nearly 8% (7.9% ± 3.2%; n = 10) of predefined CMPs7 expressed CD41 (CD41+ CMPs), whereas other HSPC populations such as HSCs, GMPs, and MEPs did not express significant levels of CD41 (Figure 2B). CD41+ CMPs were also positive for other megakaryocyte markers, including CD9, CD42b, CD51, CD61, and CD62P (Figure 2C). CD61 (ie, integrin β3) levels were proportional to those of CD41 (integrin α2b), in agreement with the fact that these molecules form a heterodimeric complex. CD9, which was reported as the positive marker for the murine megakaryocyte progenitor,9 was found in all HSPC populations, including GMPs. Of note, CD105, a marker for MEPs,17 was negative in the CD41+ CMP fraction (supplemental Figure 1A).
CD41 is expressed exclusively in a fraction of CMPs. (A) Gating strategy for BM HPCs (CMP, GMP, MEP) is shown.7 CD41 expression was concentrated in the CD38+ IL-3RαdimCD45RA− population, which is identical to the CMP immunophenotype. (B) CD41 expression patterns in human BM HSPC fractions (top). CD41 was solely and exclusively expressed in CMPs (7.9% ± 3.2%) but not in GMPs (0.0% ± 0.2%) or MEPs (0.3% ± 0.4%). Representative FACS plots are shown. Morphological characteristics of HSPCs were examined via May-Grünwald-Giemsa staining (scale bars, 5 μm). CD41+ CMPs showed immature morphological features resembling those of HSCs and CD41− CMPs. A fraction of CD41+ CMPs (1.2% ± 0.8%; n = 3) consisted of polyploid cells, indicating initiation of the endomitotic process for megakaryocyte development (middle). Side-scatter/forward-scatter (SSC/FSC) dot plots of each HSPC fraction (bottom). CD41+ CMPs were heterogeneous in size compared with other populations. Polyploid cells were highly enriched in FSChigh fraction (red dotted circle). (C) CD41+ CMPs expressed various megakaryocyte lineage markers. To avoid the effects of spectral overlaps, combinations of PE- and APC-conjugated antibodies were used. (D) Single-cell gene expression profiles of CMPs. Most CD41+CMPs, but not CD41−CMPs, expressed megakaryocyte-specific genes at high levels.
CD41 is expressed exclusively in a fraction of CMPs. (A) Gating strategy for BM HPCs (CMP, GMP, MEP) is shown.7 CD41 expression was concentrated in the CD38+ IL-3RαdimCD45RA− population, which is identical to the CMP immunophenotype. (B) CD41 expression patterns in human BM HSPC fractions (top). CD41 was solely and exclusively expressed in CMPs (7.9% ± 3.2%) but not in GMPs (0.0% ± 0.2%) or MEPs (0.3% ± 0.4%). Representative FACS plots are shown. Morphological characteristics of HSPCs were examined via May-Grünwald-Giemsa staining (scale bars, 5 μm). CD41+ CMPs showed immature morphological features resembling those of HSCs and CD41− CMPs. A fraction of CD41+ CMPs (1.2% ± 0.8%; n = 3) consisted of polyploid cells, indicating initiation of the endomitotic process for megakaryocyte development (middle). Side-scatter/forward-scatter (SSC/FSC) dot plots of each HSPC fraction (bottom). CD41+ CMPs were heterogeneous in size compared with other populations. Polyploid cells were highly enriched in FSChigh fraction (red dotted circle). (C) CD41+ CMPs expressed various megakaryocyte lineage markers. To avoid the effects of spectral overlaps, combinations of PE- and APC-conjugated antibodies were used. (D) Single-cell gene expression profiles of CMPs. Most CD41+CMPs, but not CD41−CMPs, expressed megakaryocyte-specific genes at high levels.
The morphology of purified HSPCs was examined by May-Grünwald-Giemsa staining (Figure 2B). The CD41+ CMPs exhibited immature myeloblast-like characteristics resembling those of HSCs and CD41− CMPs, and nearly 20% of them were larger than GMPs or MEPs (Figure 2B; supplemental Figure 1B). They did not have azurophilic granules or basophilic cytoplasm, which were abundant in GMPs and MEPs, respectively (Figure 2B). Interestingly, ∼1% of the CD41+ CMPs but not other CD34+ HSPCs seemed to possess 4N-nuclei, suggesting that the initiation of endomitosis and polyploidization has taken place in this population.28 These 4N-nuclei cells resided in large-cell fractions of the CD41+ CMPs (red dotted circle in Figure 2B).
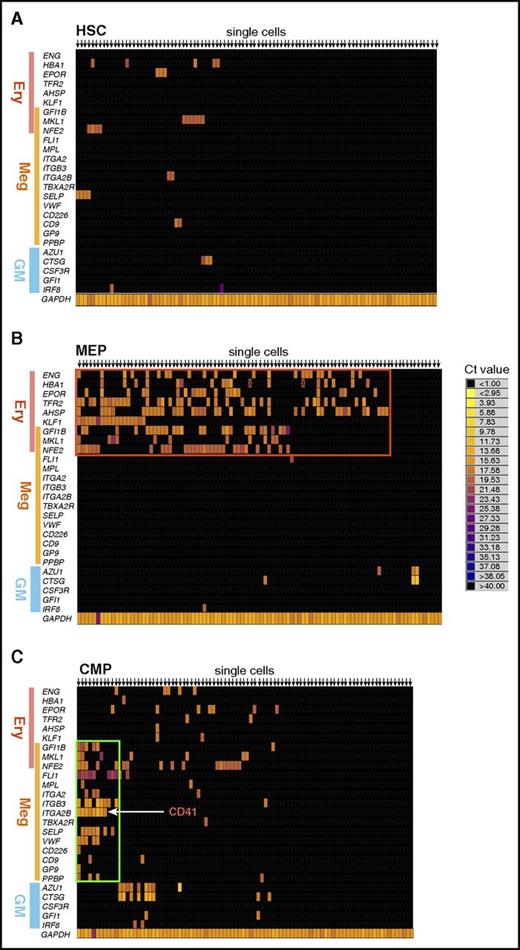
We then reanalyzed the gene expression profile of individual CD41+ CMP cells at the single-cell level (Figure 2D). The majority of single CD41+ CMP cells expressed megakaryocyte lineage–specific genes, whereas they lacked the expression of erythroid- and myeloid-specific genes, suggesting that CD41+ CMPs are a homogeneous and megakaryocyte lineage–biased population. In contrast, most CD41− CMP cells did not express hematopoietic lineage–affiliated genes, presumably representing their immaturity. There were no clusters having megakaryocyte lineage–only profiles within any HSPC population other than predefined CMPs.
CD41+ CMP is human MegP that possesses robust megakaryocyte-specific lineage potential
Next, we tested differentiation potential of CD41+ CMPs by evaluating CFU activity. CFU assays were performed under myeloerythroid culture conditions using methylcellulose supplemented with fetal bovine serum and pertinent cytokines (Figure 3A-B). This culture system is efficient for the growth of myeloerythroid CFUs but is not appropriate for CFU-megakaryocyte (CFU-Meg) evaluation. CD41− CMPs produced a variety of colonies, including CFU-mixed, whereas GMPs and MEPs produced only CFU-GM and erythroid burst-forming units or erythroid CFUs, respectively, with high plating efficiency. CD41+ CMPs, however, did not give rise to any of these myeloerythroid CFUs (Figure 3B, left). As expected, CD41− CMPs, GMPs, and MEPs gave rise to lineage-restricted colonies in single-cell liquid culture assays; however, CD41+ CMPs failed to form erythroid/myeloid colonies under the same condition (Figure 3B, right). We merely observed small clusters consisting of few large cells, some of which presumably were megakaryoctic lineage cells (data not shown). Following these observations, we next tested megakaryocytic potential on the collagen-based semisolid medium Megacult-C, which is optimized for CFU-Megs (Figure 3C-D). Strikingly, >70% of single CD41+ CMPs gave rise to CFU-Megs, and one-third of CFU-Meg colonies contained >50 megakaryocytes. In contrast, ∼10% of CD41− CMPs produced mixed colonies, and only a few percent of MEPs produced small CFU-Megs (Figure 3D). Thus, CD41+ CMPs produced almost 30 times as many colonies as MEPs on a per-cell basis (Figure 3D). In addition, CD41+ CMPs gave rise to progenies with megakaryocyte features, such as bleb formation and endomitosis (some of them with 8N nuclei), as early as culture day 7 (Figure 3E).28 These data indicated that the CD41+ CMPs are fully committed to the megakaryocyte lineage, whereas predefined MEPs are largely erythroid lineage committed.
CD41+CMP possesses robust megakaryocyte-specific lineage potential in vitro. (A-B) Colony assays with conventional semisolid culture system (MethoCult supplemented with cytokines; n = 6). (A) Representative overall view of colonies derived from single BM-derived CD41+ CMPs and MEPs after 14 days on 35-mm dishes. CD41+ CMPs did not give rise to colonies under this culture condition (left). Images were created by using the image-stitching function of BZ-X700 (Keyence, Osaka, Japan) at low magnification (×4). (B) Single-cell colony assays. CFUs derived from 100 single HSPCs in Methocult (left) or in serum-free liquid culture (right) were enumerated (means ± standard deviations [SDs]; n = 6). CD41+ CMPs barely produced colonies in both assays. (C-D) Megakaryocyte lineage potential was evaluated using the specific culture system with collagen-based medium and cytokines, which is highly optimized for CFU-Meg (Megacult-C; n = 6). (C) Representative overall views of colonies derived from CD41+ CMPs and MEPs after 14 days using 4× (upper; scale bars, 250 µm) and 20× (lower; scale bars, 50 µm) objectives (BZ-X700). CFU-Megs derived from MEPs (pink arrow) were found far less frequently, and the size of each CFU-Meg was much smaller compared with those from CD41+ CMPs. Staining was performed according to the manufacturer's instructions. (D) Bar graphs represent numbers of burst-forming unit (BFU)/CFU-Meg colonies derived from 1000 cells. CD41+ CMPs gave rise exclusively to megakaryocyte colonies without any nonmegakaryocyte colonies. Both the number and size of megakaryocyte colonies produced from CD41+ CMPs far exceeded those of MEPs (means ± SDs; n = 6; megakaryocyte). (E) Cytospin preparation after 7-day culture of CD41+ CMPs in liquid medium (May-Giemsa staining). Progenies of CD41+ CMPs possessed distinct morphological features of megakaryocytes, such as polyploidy and bleb formation. The composite image from 3 representative views were shown. Images were captured by a BH2 microscope (Olympus, Tokyo, Japan) and Digital Sight DS-5M (Nikon, Tokyo, Japan) with 60× objective lens. E, erythroid; GEMM, mixed.
CD41+CMP possesses robust megakaryocyte-specific lineage potential in vitro. (A-B) Colony assays with conventional semisolid culture system (MethoCult supplemented with cytokines; n = 6). (A) Representative overall view of colonies derived from single BM-derived CD41+ CMPs and MEPs after 14 days on 35-mm dishes. CD41+ CMPs did not give rise to colonies under this culture condition (left). Images were created by using the image-stitching function of BZ-X700 (Keyence, Osaka, Japan) at low magnification (×4). (B) Single-cell colony assays. CFUs derived from 100 single HSPCs in Methocult (left) or in serum-free liquid culture (right) were enumerated (means ± standard deviations [SDs]; n = 6). CD41+ CMPs barely produced colonies in both assays. (C-D) Megakaryocyte lineage potential was evaluated using the specific culture system with collagen-based medium and cytokines, which is highly optimized for CFU-Meg (Megacult-C; n = 6). (C) Representative overall views of colonies derived from CD41+ CMPs and MEPs after 14 days using 4× (upper; scale bars, 250 µm) and 20× (lower; scale bars, 50 µm) objectives (BZ-X700). CFU-Megs derived from MEPs (pink arrow) were found far less frequently, and the size of each CFU-Meg was much smaller compared with those from CD41+ CMPs. Staining was performed according to the manufacturer's instructions. (D) Bar graphs represent numbers of burst-forming unit (BFU)/CFU-Meg colonies derived from 1000 cells. CD41+ CMPs gave rise exclusively to megakaryocyte colonies without any nonmegakaryocyte colonies. Both the number and size of megakaryocyte colonies produced from CD41+ CMPs far exceeded those of MEPs (means ± SDs; n = 6; megakaryocyte). (E) Cytospin preparation after 7-day culture of CD41+ CMPs in liquid medium (May-Giemsa staining). Progenies of CD41+ CMPs possessed distinct morphological features of megakaryocytes, such as polyploidy and bleb formation. The composite image from 3 representative views were shown. Images were captured by a BH2 microscope (Olympus, Tokyo, Japan) and Digital Sight DS-5M (Nikon, Tokyo, Japan) with 60× objective lens. E, erythroid; GEMM, mixed.
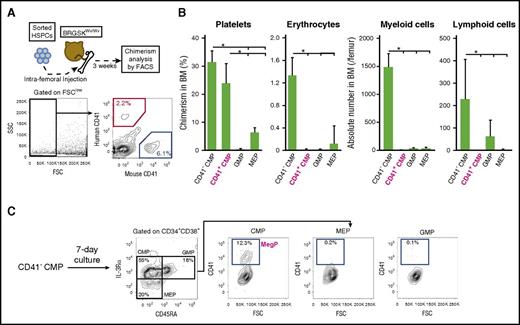
Differentiation potential of CD41+ CMPs was further tested in vivo via a xenotransplantation assay using a novel immunodeficient mouse strain, BRGSKWv/Wv, which allows reconstitution of human megakaryocyte and erythroid lineage hematopoiesis with high efficiency.20 Because CB-derived HSPCs reportedly engraft more efficiently than those of BM,29,30 we transplanted CB CD41+ CMPs or other progenitors into sublethally irradiated BRGSKWv/Wv mice and analyzed their potential to generate progenies of each hematopoietic lineage 3 weeks after transplantation (Figure 4A). Of note, CB-derived CD41+ CMPs exhibited unipotent megakaryocytic differentiation in vitro, as observed in BM-derived CD41+ CMPs (supplemental Figure 2A-C). As expected, CD41+ CMPs developed CD41+ platelets with chimerism levels similar to those of CD41− CMPs but significantly exceededing those of MEPs. In contrast, they did not give rise to CD33+ myeloid cells, CD19+ lymphoid cells, or GPA+ erythrocytes (Figure 4B; supplemental Figure 3A). Human platelet engraftment was not evident 2 weeks after transplantation, although we could detect those of human myeloid, B-lymphoid, and erythroid cells (data not shown). Collectively, the CD41+ CMP population has strong unipotent megakaryocyte-specific lineage potential both in vitro and in vivo. We thus defined the CD41+ CMP as the human MegP.
CD41+CMPs show strong platelet-producing ability in a xenotransplantation model. (A) Experimental schema of xenotransplantation. CB-derived HSPCs were injected into BRGSKWv/Wv mice via femur. After 3 weeks, chimerism of progenies in each lineage (CD33+ myeloid cells, CD19+ B cells, CD3+ T cells, GPA+ erythrocytes, CD41+ platelets) was analyzed using FACS. For the evaluation of erythrocytes or platelets, the forward-scatter (FSC) detector voltage was magnified, and FSClow cells (black rectangle) were analyzed. (B) Chimerism of human platelets, erythrocytes, and myeloid and lymphoid cells was determined after transplantation of each progenitor fraction. Engraftment of human myeloid and lymphoid cells was assessed by absolute counts. Bars indicate means ± standard deviations. *P < .05 (1-way analysis of variance followed by Tukey’s HSD test; n = 3). (C) Immunophenotypic changes of BM-derived CD41− CMPs after a 7-day liquid culture. CD41− CMPs gave rise to CD41+ CMPs (CD34+ CD38+ IL-3Rαdim CD45RA− CD41+). Representative FACS plots from 3 independent experiments are shown. SSC, side scatter.
CD41+CMPs show strong platelet-producing ability in a xenotransplantation model. (A) Experimental schema of xenotransplantation. CB-derived HSPCs were injected into BRGSKWv/Wv mice via femur. After 3 weeks, chimerism of progenies in each lineage (CD33+ myeloid cells, CD19+ B cells, CD3+ T cells, GPA+ erythrocytes, CD41+ platelets) was analyzed using FACS. For the evaluation of erythrocytes or platelets, the forward-scatter (FSC) detector voltage was magnified, and FSClow cells (black rectangle) were analyzed. (B) Chimerism of human platelets, erythrocytes, and myeloid and lymphoid cells was determined after transplantation of each progenitor fraction. Engraftment of human myeloid and lymphoid cells was assessed by absolute counts. Bars indicate means ± standard deviations. *P < .05 (1-way analysis of variance followed by Tukey’s HSD test; n = 3). (C) Immunophenotypic changes of BM-derived CD41− CMPs after a 7-day liquid culture. CD41− CMPs gave rise to CD41+ CMPs (CD34+ CD38+ IL-3Rαdim CD45RA− CD41+). Representative FACS plots from 3 independent experiments are shown. SSC, side scatter.
Human MegPs are derived from CD41− primitive CMPs
CD41− CMPs are capable of producing abundant platelets; therefore, it is possible that CD41− CMPs can give rise to MegPs. We thus tested differentiation potential of CD41− CMPs using a serum-free liquid culture system. After culture for 7 days, CD41− CMPs differentiated into secondary CMPs, MEPs, and GMPs, as reported previously7 (Figure 4C). In these secondary progenitor populations, CD41+ cells were found exclusively in CMPs but not in MEPs or GMPs (Figure 4C). Importantly, secondary CD41+ CMPs (MegPs) efficiently gave rise to CFU-Meg colonies in Megacult-C assay (supplemental Figure 3B). Given that MEPs did not give rise to secondary CD41+ CMPs under the same culture condition (data not shown), we propose that MegPs are progenies of CD41− CMPs.
Genome-wide gene expression analysis shows MegPs and MEPs to be distinct HPC populations
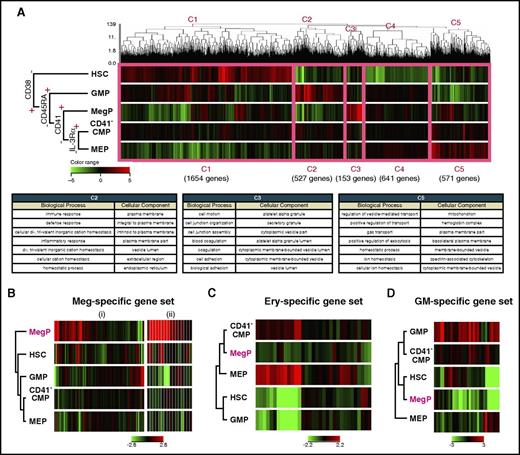
We next performed global gene-expression profiling of BM-derived CD34+CD38− HSCs and CD34+CD38+ HPC populations, including the newly identified MegPs. Figure 5A shows results of unsupervised hierarchical clustering analysis of cDNA microarray data. The dendrogram covering all of these populations indicated that the expression of CD38 demarcated HSCs from the other HPC populations. Five major gene clusters were formed by the dendrogram (C1-C5 in Figure 5A). Clusters 1, 2, 3, and 5 were composed of genes that were mainly upregulated in HSCs, GMPs, MegPs, and MEPs, respectively. Cluster 4 represented the group of genes without significant differences in expression among HPC populations. Gene ontology terms related to cluster 2, biological process and cellular component, included immune response, defense response, inflammatory response, response to wounding, and leukocyte activation, indicating strong association of GMPs with the GM lineage. Likewise, gene ontology terms related to cluster 3 (specifically upregulated in MegPs) were platelet-associated terms, such as coagulation, hemostasis, and platelet alpha granule. Cluster 5 genes were related to erythroid-associated terms such as gas transport, ion homeostasis, and hemoglobin complex (Figure 5A). The unsupervised clustering clearly clustered the analyzed genes into groups of functionally important genes for each HPC population, suggesting that global gene expression profiles reflect individual functions and cell fates.
MegP transcriptome was distinct from those of other HSPCs. (A) Gene expression profiling of BM-derived HSPCs analyzed by microarray. Unsupervised hierarchical clustering analysis on differentially expressed genes among analyzed populations is shown in the dendrogram together with a heat map representing gene expression level by color (top). The thick pink line demarcates 5 distinct clusters (C1-C5). C2, C3, and C5 genes were upregulated in lineage-committed HPCs (GMPs, MegPs, and MEPs, respectively). The biological meanings of C1-C5 cluster genes were investigated using the Database for Annotation, Visualization and Integrated Discovery tool, and results are listed in the table (bottom). (B-D) Gene expression patterns of hematopoietic lineage-affiliated gene sets are shown in heat map displays. MegPs predominantly expressed megakaryocyte-specific genes. The following two gene sets were obtained from the Molecular Signatures Database: (i) REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION and (ii) REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN. Ery, erythroid.
MegP transcriptome was distinct from those of other HSPCs. (A) Gene expression profiling of BM-derived HSPCs analyzed by microarray. Unsupervised hierarchical clustering analysis on differentially expressed genes among analyzed populations is shown in the dendrogram together with a heat map representing gene expression level by color (top). The thick pink line demarcates 5 distinct clusters (C1-C5). C2, C3, and C5 genes were upregulated in lineage-committed HPCs (GMPs, MegPs, and MEPs, respectively). The biological meanings of C1-C5 cluster genes were investigated using the Database for Annotation, Visualization and Integrated Discovery tool, and results are listed in the table (bottom). (B-D) Gene expression patterns of hematopoietic lineage-affiliated gene sets are shown in heat map displays. MegPs predominantly expressed megakaryocyte-specific genes. The following two gene sets were obtained from the Molecular Signatures Database: (i) REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION and (ii) REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN. Ery, erythroid.
Next, we investigated the expression levels of canonical lineage–affiliated genes (supplemental Table 2) in each progenitor fraction. Strikingly, genes related to megakaryocytes and platelets were significantly enriched in MegPs but not in MEPs (Figure 5B). In contrast, genes related to the erythroid (eg, erythroid lineage–specific transcription factors and hemoglobin synthesis–related genes) and GM lineages (eg, myeloid lineage–specific surface markers, cytokine receptors and enzymes) were downregulated in MegPs (Figure 5C-D). Interestingly, CD41− CMPs exhibited none of these lineage-specific signatures, suggesting their immaturity compared with other HPCs.
Transcription factors, including GATA-1, FOG-1, Gfi-1b, and NF-E2,4,5 which play critical roles in commitment to both megakaryocyte and erythroid lineages, were upregulated in both MegP and MEP populations (supplemental Figure 4). FLI1 and KLF1 are key transcription factors essential for megakaryocyte and erythroid lineage development, respectively.31 FLI1 was highly upregulated in MegPs but was downregulated in MEPs, whereas KLF1 expression showed a reciprocal pattern (supplemental Figure 4). Thus, transcriptome profiling provided definitive evidence that MegPs and MEPs are largely committed toward megakaryocyte and erythroid lineages, respectively.
MegPs are pathophysiologically important population in myeloproliferative neoplasms
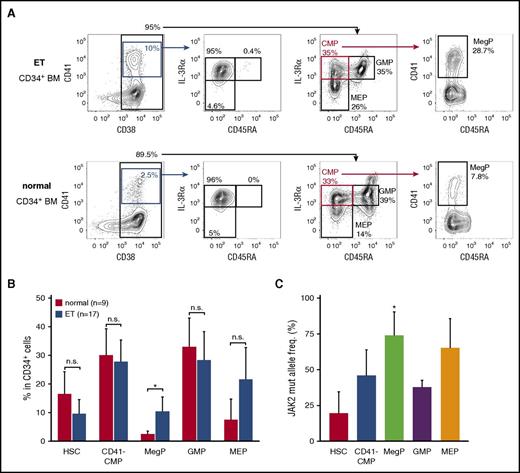
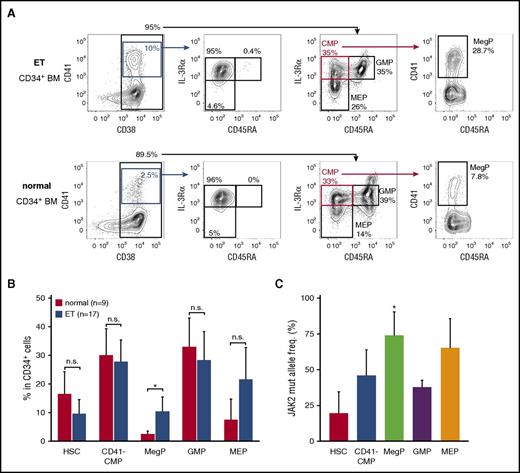
To address the significance of MegPs in human pathophysiology, we analyzed frequencies of the MegP population in the BM of patients with ET, a myeloproliferative neoplasm characterized by sustained and clonal proliferation of megakaryocytes and platelets. Strikingly, MegP fractions were significantly expanded in the BM of patients with ET (Figure 6A-B). MEPs were also expanded in the majority of patient cases of ET, although statistical significance was not observed (Figure 6B).
MegP is a pathophysiologically important population. (A) We analyzed MegP fractions in BM of 9 healthy donors and 17 patients with ET. Representative FACS profiles of control (bottom) and ET (top) samples are shown. MegPs were significantly expanded in ET BM. (B) Proportions of each HSPC fraction in CD34+ BM cells are shown. Bars indicate means of 9 healthy donors or 17 ET patients ± standard deviations (SDs). *P < .05 (Student t test). (C) The allele frequency of JAK2 V617F mutations in each HSPC population was evaluated. JAK2 V617F allele burden was significantly increased in the MegP fraction. Bars indicate means of 5 patient cases of ET ± SDs. *P < .05 (Tukey’s HSD test).
MegP is a pathophysiologically important population. (A) We analyzed MegP fractions in BM of 9 healthy donors and 17 patients with ET. Representative FACS profiles of control (bottom) and ET (top) samples are shown. MegPs were significantly expanded in ET BM. (B) Proportions of each HSPC fraction in CD34+ BM cells are shown. Bars indicate means of 9 healthy donors or 17 ET patients ± standard deviations (SDs). *P < .05 (Student t test). (C) The allele frequency of JAK2 V617F mutations in each HSPC population was evaluated. JAK2 V617F allele burden was significantly increased in the MegP fraction. Bars indicate means of 5 patient cases of ET ± SDs. *P < .05 (Tukey’s HSD test).
In our cohort, 10 of 17 patients with ET tested positive for the JAK2 V617F mutation, a major driver mutation found in 50% to 60% of those with ET,32 whereas 7 patients were positive for the CALR mutation (supplemental Table 3).33,34 We therefore tested whether the expansion of MegPs in patients with ET was associated with the JAK2 V617F allele burden. The JAK2 V617F allele burden was the lowest in HSCs and the highest in MegPs and MEPs (Figure 6C), suggesting that the mutation promotes expansion of MegPs and MEPs during ET pathogenesis, although it does not enhance HSC self-renewal capacity sufficient to outcompete normal HSCs. In fact, MegPs from patients with ET gave rise to slightly bigger colonies when analyzed by colony assay (supplemental Figure 5), although no statistical significance was observed because of small sample size. MEPs of patients with ET did not produce megakaryocyte colonies under the same condition (data not shown), indicating that MEPs of patients with ET did not acquire the capacity to differentiate toward megakaryocytes.
Discussion
In the present study, we successfully identified the unipotent MegP of adult human hematopoiesis. Using CD41 as a positive marker, we could prospectively isolate human MegPs, which exclusively express megakaryocyte-related genes, from the conventional CMP population.7 The purified CD34+CD38+CD45RA− IL-3RαdimCD41+ MegP was strictly committed to the megakaryocyte lineage, and its potential to produce megakaryocytes and platelets far exceeded that of the MEP. We propose that the unipotent progenitor population endowed with robust megakaryocyte producing potential (MegPs) resides in the CMP fraction.
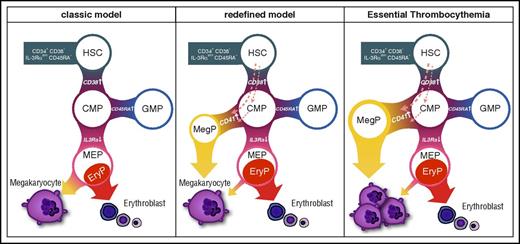
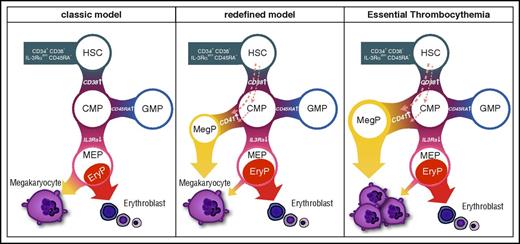
Our data also suggest that progenitors in CD41− CMP or more primitive fractions, but not MEPs, give rise to MegPs (Figure 4C). Considering recent reports on platelet-biased HSCs12,,-15 and the fact that HSCs express megakaryocyte-associated genes (Figure 5B), it is possible that MegPs are progenies of the putative megakaryocyte lineage–committed HSCs18 or those of the more recently reported MEP population residing in the CD34+CD38dim fraction.35 Furthermore, MegPs might still be heterogeneous populations, because expression levels of some megakaryocyte/erythroid transcription factors (eg, GFI1B, NFE2, FLI1) varied in the MegP fraction (Figure 2D). Methodological advances for prospective isolation of HSCs/HSPCs are necessary to clarify these issues. Our proposed model of the MegP pathway is shown in Figure 7.
Revised model of human megakaryocyte development. Unipotent MegP defined by CD41 expression in the CMP fraction branches off from the early stage of hematopoietic hierarchy. EryP, unipotent erythrocyte progenitor.
Revised model of human megakaryocyte development. Unipotent MegP defined by CD41 expression in the CMP fraction branches off from the early stage of hematopoietic hierarchy. EryP, unipotent erythrocyte progenitor.
CD41 is integrin α2b, a functional molecule of platelets that serves as a receptor for platelet fibrinogen and von Willebrand factor. Murine CD41 is transiently upregulated in the recently revised CMPs that reside within the Lin−Sca-1+c-Kit+CD34+ MPP population in young mice,36 whereas it is upregulated in long-term HSCs in old mice, marking a myeloid-biased HSC population.37 In human hematopoiesis, we show that CD41 is not expressed in HSCs at mRNA and protein levels but is upregulated in concert with commitment into the megakaryocyte lineage (Figures 1 and 2). Furthermore, combinatorial expression of megakaryocyte genes was only evident in MegPs (Figures 1 and 2D), suggesting that MegPs are the earliest and only megakaryocyte lineage–committed population that can be prospectively isolated by FACS.
It is important to note that our analysis (Figure 3B) and the original report7 show that conventional human MEPs, defined as the CD34+CD38+CD45RA− IL-3Rα− population, exhibit unipotent activity to produce erythroid cells.17 In agreement with these observations, the human MegP was not generated through the MEP stage (Figure 4C), and MegPs and MEPs exhibit distinct transcriptional profiles (Figure 5; supplemental Figure 4). These data collectively suggest that MegPs and MEPs are developmentally distinct populations. Furthermore, the expression of positive markers for MegPs (CD41) and erythroid progenitors (CD105)17 were confined to IL-3Rαdim and negative fractions, respectively, showing a mutually exclusive expression pattern (supplemental Figure 1A). Thus, the main developmental pathway for platelets is not through a megakaryocyte/erythroid bifurcation stage within the MEP fraction but instead through MegPs within the CMP fraction in human hematopoiesis. CD41 is reportedly expressed in a small fraction of MEPs, and the CD41+ MEPs exhibit megakaryocyte lineage potential.16 Because CD41 expression peaks in IL-3Rαdim populations (Figure 2B; supplemental Figure 1A), it might be technically difficult to completely segregate MegPs from MEPs.
The physiological importance of human MegPs was also studied using BM samples of patients with ET. MegPs, but not HSCs or GMPs, were significantly expanded in patients with ET, and JAK2 V617F allele burden was highest at the MegP stage (Figure 6C). Given that the JAK2 V617F allele burden is associated with clinical consequences for patients with ET,38,39 JAK2 V617F–driven MegP expansion might be the basis for the pathological thrombocytosis observed in patients with ET. In fact, MegPs obtained from BM of patients with ET tended to give rise to bigger colonies than those from controls (supplemental Figure 5). Because mutations of JAK2, CALR, and MPL are the main drivers of ET pathogenesis, it remains to be seen how each mutation affects MegP functions and whether the pathological MegPs should be targeted for ET therapy.
In summary, we have identified the unipotent MegP in human adult BM. The isolatable MegP population will be useful for understanding physiological and pathological human megakaryocyte development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all laboratory members for valuable discussions and Arisa Matsuyama and Chihiro Matsuo for animal care, as well as the Japanese Red Cross Kyushu Cord Blood Bank for providing cord blood samples.
This work was supported in part by a Grant-in-Aid for Scientific Research (C) (25461424) (H.I.), a Grant-in-Aid for Scientific Research (C) (26461424) (K.T.), Grants-in-Aid for Scientific Research on Innovative Areas (22130001 and 22130002) (K.A.), Grants-in-Aid for Challenging Exploratory Research (24659463 and 15K15365) (K.A.), Grants-in-Aid for Scientific Research (A) (25253069 and 16H02662) (K.A.), a Grant-in-Aid for Challenging Exploratory Research (15K15364) (Y. Kunisaki), a Grant-in-Aid for Scientific Research (B) (15H04859) (Y. Kunisaki), Grants-in-Aid for Young Scientists (A) (26713034 and 16H06250) (Y. Kikushige), a Grant-in-Aid for Scientific Research (B) (23390254) (T. Miyamoto), Grants-in-Aid for Scientific Research (C) (25461453 and 16K09875) (K.K.), and Grants-in-Aid for Scientific Research on Innovative Areas (25115002 and 16H05340) (T. Miyamoto). This work was also supported in part by the Social Medical Corporation and ChiyuKai Foundation.
Authorship
Contribution: K.M., H.I., T. Maeda, and K.A. coordinated the project, designed and performed the experiments, analyzed the data, and wrote the manuscript; T.J., H.K., A.Y., T.S., Y.U., J.O., S.D., and H.T. performed the experiments; Y. Kunisaki, Y. Kikushige, Y.M., T.I., Y.A., K.K., K.T., and T. Miyamoto reviewed the data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Koichi Akashi, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan; e-mail: akashi@med.kyushu-u.ac.jp.

![Figure 3. CD41+ CMP possesses robust megakaryocyte-specific lineage potential in vitro. (A-B) Colony assays with conventional semisolid culture system (MethoCult supplemented with cytokines; n = 6). (A) Representative overall view of colonies derived from single BM-derived CD41+ CMPs and MEPs after 14 days on 35-mm dishes. CD41+ CMPs did not give rise to colonies under this culture condition (left). Images were created by using the image-stitching function of BZ-X700 (Keyence, Osaka, Japan) at low magnification (×4). (B) Single-cell colony assays. CFUs derived from 100 single HSPCs in Methocult (left) or in serum-free liquid culture (right) were enumerated (means ± standard deviations [SDs]; n = 6). CD41+ CMPs barely produced colonies in both assays. (C-D) Megakaryocyte lineage potential was evaluated using the specific culture system with collagen-based medium and cytokines, which is highly optimized for CFU-Meg (Megacult-C; n = 6). (C) Representative overall views of colonies derived from CD41+ CMPs and MEPs after 14 days using 4× (upper; scale bars, 250 µm) and 20× (lower; scale bars, 50 µm) objectives (BZ-X700). CFU-Megs derived from MEPs (pink arrow) were found far less frequently, and the size of each CFU-Meg was much smaller compared with those from CD41+ CMPs. Staining was performed according to the manufacturer's instructions. (D) Bar graphs represent numbers of burst-forming unit (BFU)/CFU-Meg colonies derived from 1000 cells. CD41+ CMPs gave rise exclusively to megakaryocyte colonies without any nonmegakaryocyte colonies. Both the number and size of megakaryocyte colonies produced from CD41+ CMPs far exceeded those of MEPs (means ± SDs; n = 6; megakaryocyte). (E) Cytospin preparation after 7-day culture of CD41+ CMPs in liquid medium (May-Giemsa staining). Progenies of CD41+ CMPs possessed distinct morphological features of megakaryocytes, such as polyploidy and bleb formation. The composite image from 3 representative views were shown. Images were captured by a BH2 microscope (Olympus, Tokyo, Japan) and Digital Sight DS-5M (Nikon, Tokyo, Japan) with 60× objective lens. E, erythroid; GEMM, mixed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/25/10.1182_blood-2016-09-741611/6/m_blood741611f3.jpeg?Expires=1769779484&Signature=DzCfFx8ws3Xg5PjmUYhAtvWwuX9aaVzBrNU3pGCcKbPsxuY3oRIXSR8MB65uj-Vg81Xpcj~1JoXHs7xnNm0EDjv7LqL3kpURhC9FvN1HAPfdrus6hPjVeD59YZT7v69YhXMYNZ~PHDJwa-~qufWRgEa9BhtPcEOtuycZWECb328ZX2Q7OHSLMffgQntKFRQI0aDQmrUR0doCiPUSY0QYJaT73TYZRtjobX2Xi-t8ih~T6wlFStVNYbeXbE0ipcbq5yYsaGozmuwUOMrgpp2VruqIiFPjE0PFmtuOMXB2lQJ4QPZyyxGupoeWwZXoC4CFpWUbat~AGFsUL9T6Haw7iQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)