Key Points
EPCR is a reliable surface marker that detects human HSCs in culture.
EPCR is required for hematopoietic stem and progenitor cell function in vivo.
A small subset of human cord blood CD34+ cells express endothelial protein C receptor (EPCR/CD201/PROCR) when exposed to the hematopoietic stem cell (HSC) self-renewal agonist UM171. In this article, we show that EPCR-positive UM171-treated cells, as opposed to EPCR-negative cells, exhibit robust multilineage repopulation and serial reconstitution ability in immunocompromised mice. In contrast to other stem cell markers, such as CD38, EPCR expression is maintained when cells are introduced in culture, irrespective of UM171 treatment. Although engineered overexpression of EPCR fails to reproduce the effects of UM171 on HSC activity, its expression is required for the repopulating activity of human HSCs. Altogether, our results indicate that EPCR is a reliable and cell culture–compatible marker of UM171-expanded human cord blood HSCs.
Introduction
Human hematopoietic stem cells (HSCs) are defined by their ability to differentiate into all blood lineages and to self-renew to sustain hematopoiesis throughout adult life. The low HSC frequency in biological specimens as well as our inability to purify HSCs to homogeneity have challenged the molecular characterization of these cells. HSC frequencies of 1 in 10 to 20 can be achieved by depleting CD38- and CD45RA-expressing cells from uncultured human cord blood (CB) units and enriching for CD34-, CD90-, and CD49f-expressing cells,1,2 however, relying on these markers to evaluate HSC activity in vitro is not feasible because the expression of these markers changes dramatically when cells are introduced in culture3 (supplemental Figure 1, available on the Blood Web site). Dorell et al4 demonstrated that CD38 expression is lost in cultures initiated with CD34+CD38+ cells, which were reported to lack severe combined immunodeficiency–repopulating cell activity. CD90– cells were also shown to acquire the CD90 mark in vitro, producing cells with robust, long-term multilineage repopulating activity,2 highlighting the discrepancy between the HSC phenotype in vitro and repopulating activity in vivo. Therefore, to date, we are still lacking surrogate markers that define the hematopoietic stem and progenitor cell (HSPC) population ex vivo, and consequently, assessment of long-term HSC (LT-HSC) activity in expanded cultures still relies on time-consuming transplantation experiments in immunocompromised mice.5 Identifying surface molecules with expression levels that correlate with HSC repopulating activity is key to studying these cells and is expected to dramatically facilitate the development of high-throughput screening strategies aimed at identifying novel HSC agonists or genes involved in HSC expansion or differentiation.
Our group has previously shown that the pyrimidoindole derivative UM171 stimulates human HSC expansion in vitro. Transcriptome analysis of CD34+-enriched human CB populations revealed induction of endothelial protein C receptor (EPCR) gene expression on UM171 treatment.6 Originally known as a key constituent of endothelial barrier protection, EPCR is essential for the anti-inflammatory function of its binding partner, activated protein C (APC).7,,-10 EPCR has been previously shown to identify murine HSC populations.11,12 Recent studies have also demonstrated that the EPCR/PAR1 pathway regulates LT-HSC retention in the bone marrow (BM) by restricting nitric oxide production and Cdc42 activity, leading to increased integrin very late activation antigen 4 affinity.13 In line with this finding, mice that were genetically modified to express low levels of EPCR at the cell surface (Procrlow) showed defects in HSC BM homing and increased levels of circulating HSCs.13 Surprisingly, although EPCR is commonly used to sort murine HSC-enriched subsets, efforts to demonstrate its reliability for sorting human HSCs were unsuccessful.12
In this article, we identify EPCR as a highly reliable surface marker for expanded human HSCs, independently of the culture conditions or duration. We also demonstrate that EPCR defines a cell population with sustained short- and long-term repopulating activity and show that the transcriptional profile of expanded EPCR+ cells significantly matches that of freshly sorted HSCs. Finally, we provide evidence for a crucial role for EPCR in human HSC self-renewal.
Methods
Human CD34+ CB cell collection
Umbilical CB units were collected from consenting mothers according to an ethically approved protocol at CHU Sainte-Justine (Montreal, QC, Canada). Human CD34+ CB cells were isolated by using the EasySep positive selection kit (STEMCELL Technologies, catalog number 18056).
Flow cytometry and sorting
Fresh or cultured CB cells were labeled or sorted for different HSPC phenotypes by using the following antibodies. Mouse anti-human antibodies were used to detect CD34 (fluorescein isothiocyanate, BD Biosciences catalog number 555821 or BV421, BD Biosciences catalog number 562577), CD45RA (phycoerythrin [PE], BD Biosciences catalog number 555489 or allophycocyanin cyanine 7 (APC-CY7; BioLegend catalog number 304128), CD90 phycoerythrin-cyanine 7 (PE-CY7; BioLegend catalog number 328124), CD49f phycoerythrin-cyanine 5 (PE-CY5; BD Biosciences catalog number 551129 or BV241, BioLegend catalog number 313624), CD38 (peridinin chlorophyll [PerCP]-eFluor710, Myltenyi Biotec catalog number 46038842), CD133 (PE, Miltenyi Biotec catalog number 130-080-801) and c-Kit (APC-CY7, BioLegend catalog number 313228). EPCR-allophycocyanin-Vio770 (clone REA337) and its corresponding isotype (clone REA293) were purchased from Miltenyi Biotec. EPCR-APC (clone RCR-401) and its corresponding isotype (clone RTK2071) were purchased from BioLegend. Cells were analyzed on a BD FACSCanto flow cytometer (BD Biosciences), and cell sorting (under low-pressure conditions) was conducted on a BD FACSAria II cell sorter (BD Biosciences). Cell recovery after sorting and the purity of sorted populations were systematically evaluated after the sort for every experiment. Accordingly, cells were counted manually with a hemocytometer and assessed for viability using trypan blue. The recovery was generally 50% to 60% of cell numbers estimated with the FACSAria apparatus. The purity of the sorted populations was always monitored by rerunning a small fraction of the sorted populations into the FACSAria cell sorter; purity >95% had to be achieved to proceed with the samples. For every experiment, we cultured the same number of sorted cells (according to the manual count) in the presence or absence of UM171 and monitored the fold expansion resulting from UM171 treatment after 1, 7, or 12 days of culture.
HSPC cell culture
Human CB-derived CD34+ or CD34+CD45RA– cells were cultured in HSC expansion media consisting of StemSpan Serum-Free Expansion Medium (STEMCELL Technologies catalog number 09650) supplemented with 100 ng/mL stem cell factor (R&D Systems catalog number 255-SC), 100 ng/mL FMS-like tyrosine kinase 3 ligand (R&D Systems catalog number 308-FK), 50 ng/mL thrombopoietin (R&D Systems catalog number 288-TP), and 10 μg/mL low-density lipoproteins (STEMCELL Technologies catalog number 02698). Cells were seeded at a density of 1 × 103 cells/mL, and fresh HSC expansion media supplemented with UM171 (STEMCELL Technologies catalog number 72914) (38 nM), StemRegenin 1 (SR1) (Alichem catalog number 41864) (750 nM), or vehicle (0.1% dimethyl sulfoxide [DMSO]) was added periodically to the culture to keep the cell density around 3 to 6 × 105 cells/mL. For transplantation experiments, the fed-batch culture system was used as previously described.14 A total of 1 × 105 CD34+ or CD34+CD45RA– CB cells/mL were injected into 25-mL bags (American Fluoroseal Corporation catalog number 2 PF-0025) connected to a syringe-loaded pumping system and maintained on an orbital shaker at 37°C with 5% CO2. The pump was set to continuously deliver HSC expansion media supplemented with vehicle (0.1% DMSO), UM171 (38 nM), or a combination of UM171 (38 nM), and SR1 (750 nM) at a flow rate of 0.7 μl/min.
Colony-forming assay
Frequencies of colony-forming cells were estimated by plating 250 to 1000 EPCR–, EPCRlow, or EPCR+ cells sorted form uncultured or 7- or 12-day UM171-expanded CD34+CD45RA– populations. Cells were cultured in 2% methylcellulose Iscove's Modified Dulbecco's Medium– based (Gibco catalog number 12440053) media supplemented with 20% heat-inactivated fetal bovine serum (Wisent Bioproducts catalog number 115681), 1% bovine serum albumin (Wisent Bioproducts catalog number 800-195-LG), 2 mM l-glutamine, 100 ng/mL stem cell factor, 10 ng/mL interleukin-3, 10 ng/mL interleukin-6, 3 U/mL erythropoietin, 200 µg/mL holo-transferrin (Sigma catalog number T4132), 10 ng/mL GM-CSF (Shenandoah Biotechnology catalog number 100-08), 50 ng/mL thrombopoietin (Shenandoah Biotechnology catalog number 100-05) and 10−4 M 2-mercaptoethanol. After 14 days of culture, plates were scored for colony-forming unit-granulocyte, erythrocyte, monocyte/macrophage, megakaryocyte (CFU-GEMM).
Culture of EPCR-expressing progeny
Human CB-derived CD34+ cells were expanded for 3 days with UM171 (38 nM) before they were stained with mouse anti-human EPCR antibody (APC, BioLegend catalog number 351906). Total (unpurified), EPCR–/low, and EPCRHi cells were sorted and put in culture for 7 days in media supplemented with UM171 (38 nM) before HSC phenotype staining and transplantation assays were performed.
Transplantation assays
All experiments with animals were conducted using protocols approved by University of Montreal Animal Care Committee. EPCR cell subsets purified from uncultured or expanded CD34+CD45RA– CB cells were transplanted by tail vein injection into sublethally irradiated (250 cGy, <24 hours before transplantation) 8- to 16-week-old female NOD-Scid IL-2Rɣ null (NSG, The Jackson Laboratory) mice. The animal technicians performed the transplantation experiments blindly.
The engraftment of human CB cells in the BM of NSG mice was monitored by flow cytometry at 12 and 24 weeks posttransplantation. BM cells were collected by femoral aspiration (at week 12) or by flushing the 2 femurs, tibias, and hips when animals were sacrificed at week 24. The criteria used for the successful engraftment and evaluation of HSC activity were the same as previously reported by our group6 . We used an engraftment criterion of >0.1% human CD45+ cells in the BM as assessed by flow cytometry to establish a biologically significant cutoff. For limiting dilution analysis, cells were transplanted at 3 to 5 different cell doses with 5 to 8 mice per condition. The results from these experiments were analyzed using the ELDA software from the Walter and Eliza Hall Institute of Medical Research.15 Differences in HSC frequencies were analyzed using the χ2 test. P values < 0.05 were considered significant (Mann-Whitney U test).
For secondary transplantations, 80% of total BM cells from primary NSG recipients (24 weeks posttransplantation) were injected into secondary sublethally irradiated NSG mice. BM cells of the recipient mice were harvested and analyzed 18 weeks posttransplantation. Flow cytometry analysis was performed on freshly collected BM cells. Cells were treated with 1× red blood cell lysis buffer (STEMCELL Technologies catalog number 20110), washed and stained with pacific blue–labeled anti-human CD45 (BioLegend catalog number 304029), APC-eFluor 780–labeled anti-mouse CD45 (eBioscience catalog number 47-0453-82), PE-labeled anti-human CD33 (BD Biosciences catalog number 555450), PE-Cy7 labeled anti-human CD19 (BD Biosciences catalog number 557835), and fluorescein isothiocyanate–labeled anti-human CD3 (BD Biosciences catalog number 555332) antibodies. Cells were then washed and analyzed using a FACSCanto II apparatus. BD FACSDiva software (BD Biosciences) was used to analyze the flow cytometry data.
RNA sequencing and data analysis
Cells (3-5 × 105) were sorted from CB-derived CD34+ HSPCs cultured for 7 days according to EPCR expression levels and preserved at −80°C in TRIzol Reagent (Thermo Fisher Scientific catalog number 15596026). Complimentary DNA libraries were constructed according to TruSeq Protocols (Illumina), and sequencing was performed using an Illumina HiSequation 2000 instrument. We used the Casava pipeline (Illumina) and RefSeq release 63 for subsequent mapping and quantification of gene expression. Reads per kilobase million (RPKM) values were loaded into R software, and differential expression was tested using Wilcoxon rank-sum statistics. We selected differentially expressed genes based on significance (P ≤ .01, Mann-Whitney U test), their mean expression values in at least 1 of the comparison groups (≥ 1 RPKM), and a minimum 2-fold expression difference. We converted all gene names using the HGNChelper package in R to facilitate comparisons with external datasets. Gene set enrichment analyses (GSEA) were done using the GSEA desktop application (Broad Institute). We performed analyses using the GSEA hallmark gene set collection and 2 curated HSC gene sets16,17 using standard settings. Overlap between genes associated with EPCR expression and published HSC signatures was determined using the intersect command in R. Heat maps were generated using GENE-E software (Broad Institute).
Gene Expression Omnibus accession number
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE77128; www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=cfmxesswhfwnhkb&acc=GSE77128).
Lentiviral transduction of CD34+ CB cells
CD34+ cells were prestimulated for 24 hours in HSC expansion media. Cells were then transduced for 16 hours on retronectin-coated (TaKaRa catalog number T100A) plates (20 µg/cm2) at a multiplicity of infection of 100 in HSC expansion media supplemented with polybrene (3 µg/mL). Transduced CD34+ cells were then washed and cultured in HSC expansion media in the presence of vehicle (DMSO) or UM171 (38 nM). Transduction efficiency (assessed by GFP expression) and expression of HSC cell surface markers were determined 3 (Figure 4) or 7 days (supplemental Figure 10) posttransduction.
Transplantation of transduced cells
The progeny of 5 × 103 CD34+ starting cells (d0 equivalent) cultured for 6 (Figure 4) or 7 days (supplemental Figure 10) posttransduction were transplanted by tail vein injection into sublethally irradiated (250 cGy, <24 hours before transplantation) 8- to 16-week-old female NSG mice. BM analysis was performed 24 weeks posttransplantation to determine human engraftment levels.
Statistical analysis
Statistical analysis of all in vitro experiments was done using GraphPad Prism version 6.01. The Mann-Whitney U test was used after confirming normal distribution to compare the fold change between the conditions tested. Bars and error bars represent means or medians and standard deviations (SD) or standard error of the mean, respectively, as specified. Extreme limiting dilution analysis software (http://bioinf.wehi.edu.au/software/elda/)15 was used to estimate LT-HSC frequencies with 95% confidence intervals. Statistical analysis of differences in LT-HSC frequencies was performed using the Mann-Whitney U test. P values < 0.05 were considered significant.
Results
UM171 induces EPCR expression on CD34+CD90+CD133+ CB cells
We previously showed that exposure of cord blood (CB)-derived CD34+ cells to UM171 leads to a rapid induction of EPCR expression both at the messenger RNA (mRNA) (12 hours) and protein (24 hours) level.6 This effect was further confirmed following expansion of CB CD34+ cells for 7 days with UM171 using different clones of the EPCR antibody (supplemental Figure 2) and different sources of human hematopoietic cells, such as mobilized peripheral blood and BM (supplemental Figure 3). Interestingly, although exposure of CB, mobilized peripheral blood, or BM cells to the aryl hydrocarbon receptor antagonist SR1, a small molecule promoting the ex vivo expansion of CD34+ cells,18 produces a small increase in the proportion of EPCR+ cells (most likely due to an overall expansion of the CD34+ population), the most substantial and consistent increase in EPCR expression for these cells was observed upon UM171 treatment (supplemental Figure 3). As expected, no significant additive effect was noted when cultures were treated with both UM171 and SR1 (supplemental Figure 3). Moreover, although both the EPCR−CD34+ and EPCR+CD34+ populations are comprised within the CD34+CD38− and CD49fMed subsets, which represent typical phenotypes of fresh HSCs, only the EPCR+CD34+ population expresses CD90 and CD133, indicating that EPCR positivity defines a subset of HSCs with a more primitive phenotype (ie, CD34+CD38CD49fmedCD90+CD133+) (supplemental Figure 4).
EPCR expression is strongly associated with HSC activity in culture
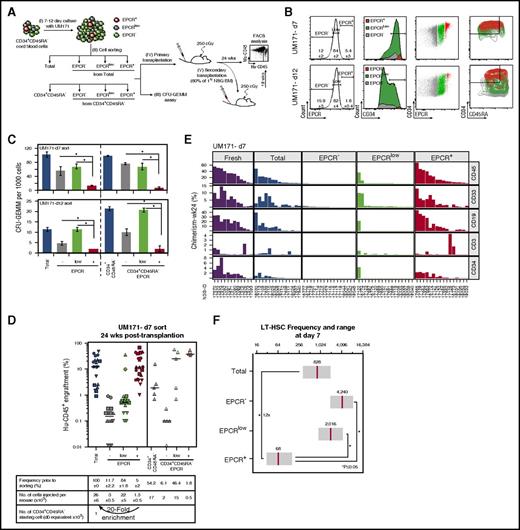
To determine if EPCR expression is a relevant marker for expanded CB HSCs, we sorted EPCR–, EPCRlow, and EPCR+ populations from CD34+CD45RA– CB cells expanded with UM171 for various time periods and assessed the ability of these cells to form colonies in semisolid cultures and to repopulate NSG mice (Figure 1A-B). The EPCR+ population was much less clonogenic than the EPCRlow and EPCR– populations in vitro (Figure 1C). In sharp contrast, the EPCR+ subpopulation included most of the long-term reconstitution activity (Figure 1D; supplemental Figure 5A), suggesting a potential value for this marker to identify expanded LT-HSCs in culture. In support of this, the relative enrichment of repopulating activity in the EPCR+ population increased from day 7 (20-fold relative enrichment, EPCR+ vs bulk) to day 12 (56-fold) of the culture (Figure 1D; supplemental Figure 5A; supplemental Table 1). In addition to their long-term reconstitution potential, EPCR-expressing cells exhibited multilineage engraftment (Figure 1E; supplemental Figure 5B). These results were reproduced in a second series of experiments, in which similar EPCR-based cell sorting was performed using CD34+CD45RA– cells (Figure 1D; supplemental Figure 5A, right panels).
The EPCR+population is enriched with LT-HSCs in culture. (A) Schematic representation of the experimental design to determine the impact of EPCR expression levels on HSPC activity in CB cultures expanded with UM171 (38 nM). (B) FACS profiles showing the percentage of EPCR subsets and their distribution in CD34+ and CD34+CD45RA– populations after culture (mean ± SD, 5 biological replicates). (C) CFU-GEMM counts for sorted EPCR subsets after culture (mean ± SD, 3 technical replicates, 2 biological replicates); *P < .05 (Mann-Whitney U test, 1-sided). Human (Hu) CD45 engraftment (D) and lineage potential (E) assessed for each NSG recipient (identified as NSG-ID) of the indicated sorted cells after 7 days of culture; n = 3 to 20 mice per condition (each geometric shape corresponds to a biological replicate; horizontal bars indicate median values). (F) Frequency of LT-HSCs within EPCR–, EPCRLow, and EPCR+ populations. Estimated LT-HSC frequencies (red lines) and 95% confidence intervals (gray boxes) presented as 1/number of sorted cells at day 7. Significance level *P < .05 (Mann-Whitney U test, 1-sided).
The EPCR+population is enriched with LT-HSCs in culture. (A) Schematic representation of the experimental design to determine the impact of EPCR expression levels on HSPC activity in CB cultures expanded with UM171 (38 nM). (B) FACS profiles showing the percentage of EPCR subsets and their distribution in CD34+ and CD34+CD45RA– populations after culture (mean ± SD, 5 biological replicates). (C) CFU-GEMM counts for sorted EPCR subsets after culture (mean ± SD, 3 technical replicates, 2 biological replicates); *P < .05 (Mann-Whitney U test, 1-sided). Human (Hu) CD45 engraftment (D) and lineage potential (E) assessed for each NSG recipient (identified as NSG-ID) of the indicated sorted cells after 7 days of culture; n = 3 to 20 mice per condition (each geometric shape corresponds to a biological replicate; horizontal bars indicate median values). (F) Frequency of LT-HSCs within EPCR–, EPCRLow, and EPCR+ populations. Estimated LT-HSC frequencies (red lines) and 95% confidence intervals (gray boxes) presented as 1/number of sorted cells at day 7. Significance level *P < .05 (Mann-Whitney U test, 1-sided).
We next examined if EPCR is also a reliable marker for HSCs cultured in the absence of UM171. For these experiments, CD34+CD45RA– CB cells were sorted based on EPCR expression after 3 days in culture when EPCR levels are at their highest (supplemental Figure 6A). Consistent with our findings with UM171-treated cultures, EPCR+ cells from DMSO-supplemented cultures were characterized by long-term repopulating activity similar to that found in unpurified cells (supplemental Figure 6C-D; supplemental Table 2), suggesting that EPCR identifies expanded CB cells with LT reconstitution potential in culture, irrespective of the presence of UM171.
EPCR-positive CB populations are enriched with LT-HSCs
Using limiting dilution analysis, we estimated that 1 in 68 EPCR+ cells is a multipotent LT-HSC in UM171-supplemented cultures harvested at day 7. LT-HSC frequencies were lower in all other fractions, ranging from 1 in 2016 cells in EPCRlow to 1 in 4240 cells in EPCR– subpopulations (Figure 1F). Considering the LT-HSC frequency measured in the unsorted expanded CD34+CD45RA– culture, we calculated that sorting on EPCR expression alone provides a 12-fold net enrichment of LT-HSCs (Figure 1F; supplemental Figure 7; supplemental Table 3), supporting the idea that EPCR identifies LT-HSCs in expanded CB cultures.
EPCR+ cells show extensive self-renewal potential
To further evaluate the repopulation potential of each of the aforementioned EPCR subsets, we performed secondary transplantation experiments in which cells collected 24 weeks posttransplantation from primary mice were transplanted into secondary recipients and monitored for an additional 18 weeks. Reconstitution of secondary recipients was limited to mice transplanted with grafts originating from either unsorted cells or EPCR+ cells (supplemental Figure 8; supplemental Table 4). Secondary recipients of EPCR– or EPCRlow BM showed very low levels of engraftment, which was frequently undetectable. In similar experiments, CD34+ cells from primary recipients (previously exposed or not to UM171 in culture) were also sorted based on EPCR expression before being injected into secondary animals (supplemental Figure 9). Again, repopulation activity was restricted to the EPCR-positive fraction. These findings suggest that only EPCR+ cells possess the required self-renewal potential to provide long-term multipotent reconstitution in serial transplantation settings.
EPCR expression defines hierarchical organization of cultured HSCs
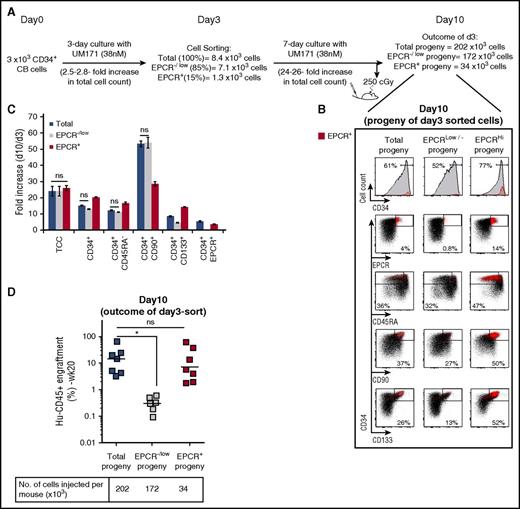
To further characterize the EPCR+ population, CD34+ CB cells expanded for 3 days were sorted based on EPCR expression, and subsets were expanded for 7 days, at which time phenotypical and in vivo functional analyses were conducted (Figure 2). EPCR expression dictated phenotypical hierarchy in vitro because only EPCR+ cells gave rise to EPCR+ progeny (Figure 2B, right). Accordingly, EPCR– cells (supplemental Figure 10) could not generate EPCR+ cells (Figure 2B, middle). In line with these results, long-term repopulation activity was mostly observed in recipients transplanted with the output of cultures initiated with EPCR+ cells (Figure 2D). Most interestingly, we noticed a substantial expansion of the CD34+CD90+ and CD34+CD133+ populations (which represent phenotypes reminiscent of HSCs) in cultures originating from CD34+EPCR–/low cells (Figure 2C). These populations contributed poorly to in vivo reconstitution (Figure 2D), suggesting that the EPCR mark is required to identify repopulating units among the CD34+CD90+CD133+CD45RA– population and hence defines HSC hierarchy in UM171-supplemented cultures.
Expanded EPCR+progeny retains most of the HSPC activity. (A) Schematic representation of EPCR sorting and transplantation strategy to characterize the progeny of the EPCR+ population. (B) FACS profiles showing expression of the indicated markers on cultured EPCR subsets. (C) Fold expansion of the indicated HSPC populations from day 3 to day 10 of the culture (mean ± SD, 3 technical replicates). Differences between conditions are statistically significant unless labeled as not significant (ns; Mann-Whitney U test, 1-sided). (D) Engraftment levels of EPCR subset progeny in NSG mice at day 10 of the culture. Horizontal bars indicate median values (n = 6 to 7 mice per condition, technical replicates). *P < .05 (Mann-Whitney U test, 1-sided).
Expanded EPCR+progeny retains most of the HSPC activity. (A) Schematic representation of EPCR sorting and transplantation strategy to characterize the progeny of the EPCR+ population. (B) FACS profiles showing expression of the indicated markers on cultured EPCR subsets. (C) Fold expansion of the indicated HSPC populations from day 3 to day 10 of the culture (mean ± SD, 3 technical replicates). Differences between conditions are statistically significant unless labeled as not significant (ns; Mann-Whitney U test, 1-sided). (D) Engraftment levels of EPCR subset progeny in NSG mice at day 10 of the culture. Horizontal bars indicate median values (n = 6 to 7 mice per condition, technical replicates). *P < .05 (Mann-Whitney U test, 1-sided).
In vivo HSPC activity is impaired by EPCR knockdown
To determine if EPCR expression is essential for HSPC function, we generated 2 EPCR-targeting short hairpin RNA vectors, which produced different knockdown levels (supplemental Figure 11A-B). EPCR silencing had no impact on the expression of other HSC markers, such as CD90 or CD34, or on cell proliferation in UM171-supplemented media (supplemental Figure 11C). In contrast, HSPC activity was found to be inversely proportional to the magnitude of EPCR knockdown (supplemental Figure 11D). As a complementary approach to evaluate the effects of manipulating EPCR expression on HSPC activity, we overexpressed EPCR in CB cells. Ectopic EPCR expression did not influence CD34 and CD45RA expression at the surface of CB cells or their ability to repopulate NSG mice at early and late time points (supplemental Figure 12).
To assess if impaired homing contributes to the effects of EPCR downregulation on HSPC engraftment, we transplanted CellTrace Violet–labeled CB cells sorted for EPCR expression in NSG mice and observed that EPCR+ and EPCR– cells are equally found in the BM shortly after transplantation. Prior exposure of EPCR+ cells to EPCR neutralizing/blocking antibody did not affect this process, suggesting that EPCR expression does not influence homing of HSPCs (supplemental Figure 13). Altogether, these results indicate that EPCR does not operate by modulating the expression of HSC markers or by affecting cell proliferation, but rather is required for intrinsic HSPC activity in vivo.
EPCR+ cells show a HSPC signature
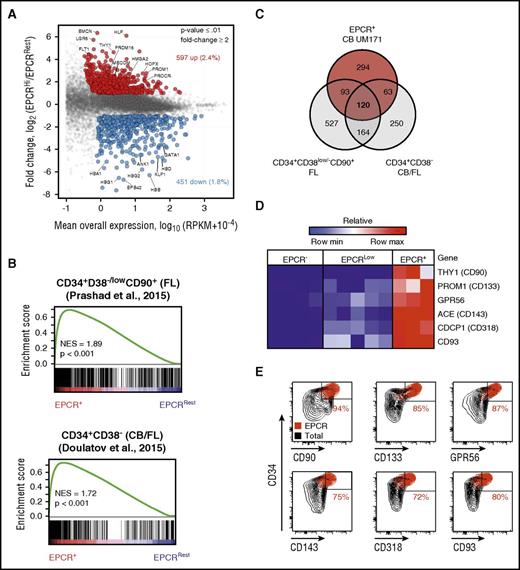
To determine the transcriptional identity of EPCR+ cells, we performed mRNA sequencing experiments using EPCR–, EPCRlow, and EPCR+ populations sorted from CD34+CD45RA– CB cells expanded for 7 days with UM171 (Figure 1B). Using a minimum number of stringent selection criteria (≥1 RPKM expression; ≥ twofold change in expression and P ≤ .01), a total of 1048 differentially expressed genes (597 upregulated and 451 downregulated) were identified between the EPCR+ and EPCR–/low populations (Figure 3A; supplemental Table 5). Gene set enrichment analysis (GSEA) revealed that published HSC-associated genes identified in uncultured human CB and fetal liver cells16,17,19 were significantly enriched in the EPCR+ population (Figure 3B; supplemental Figure 14). In contrast, GSEA identified a marked underrepresentation of differentiation-related genes in the EPCR+ population, namely genes associated with erythroid and myeloid differentiation (Figure 3B; supplemental Figure 14B-D). Intersecting our data with that of others,16,17 we were able to define a refined 120-gene HSPC signature (Figure 3C; supplemental Figure 14A). This signature is mostly comprised of either well-accepted transcriptional regulators of HSC self-renewal/function, such as HLF, PRDM16, and MECOM, or genes encoding cell surface markers typically used to purify HSCs (CD90, CD133, CD143, CD318, CD93, and GPR56)20,,,,-25 (Figure 3A,D), markers for which overlapping expression with EPCR on CB cells was confirmed by flow cytometry (Figure 3E). These results suggest that the gene expression profile of EPCR+ cells is reminiscent of a HSPC signature.
Gene expression profile of cultured EPCR+population is enriched with HSC-related genes. (A) Scatter plot of global mRNA profiling showing upregulated (red) and downregulated (blue) genes in EPCR+ versus EPCRRest (EPCR– and EPCRlow) populations (selection criteria: P ≤ .01, mean expression ≥1 RPKM; ≥ 2-fold difference). (B) GSEA plot showing enrichment for HSC-related genes in the EPCR+ population. Gene expression profiles of uncultured purified human HSCs derived from CB and fetal liver (FL) from 2 independent data sets were used for the GSEA analysis. All comparisons were significant. (C) Venn diagram showing the number of common and distinct upregulated genes in the EPCR+ population in CD34+CD38low/–CD90+ FL cells and in CD34+CD38– (FL and CB) cells. mRNA (D) and protein (E) levels of known HSC surface markers in various CB cell populations. GPR56, G protein-coupled receptor 56; NES, normalized enrichment score.
Gene expression profile of cultured EPCR+population is enriched with HSC-related genes. (A) Scatter plot of global mRNA profiling showing upregulated (red) and downregulated (blue) genes in EPCR+ versus EPCRRest (EPCR– and EPCRlow) populations (selection criteria: P ≤ .01, mean expression ≥1 RPKM; ≥ 2-fold difference). (B) GSEA plot showing enrichment for HSC-related genes in the EPCR+ population. Gene expression profiles of uncultured purified human HSCs derived from CB and fetal liver (FL) from 2 independent data sets were used for the GSEA analysis. All comparisons were significant. (C) Venn diagram showing the number of common and distinct upregulated genes in the EPCR+ population in CD34+CD38low/–CD90+ FL cells and in CD34+CD38– (FL and CB) cells. mRNA (D) and protein (E) levels of known HSC surface markers in various CB cell populations. GPR56, G protein-coupled receptor 56; NES, normalized enrichment score.
Discussion
Our data shows that EPCR represents a novel robust marker for expanded human LT-HSCs. In UM171-treated cultures, positivity for EPCR defines a highly HSC-enriched population, which includes most repopulating units. EPCR+ cells are uniformly positive for CD34, CD90, and CD133 and represent a subset of this triple positive population. EPCR is thus the most reliable indication of expanded human CB HSCs, especially in the presence of UM171. The utility of this marker to detect expanded HSCs is less obvious when using fresh CB cells because EPCR levels are lower in these cells (supplemental Figure 15; supplemental Table 6). Nonetheless, nonexpanded CB CD34+EPCR+ cells express HSC surface markers, such as CD90, CD49f, CD133, and ckit (supplemental Figure 16), and can repopulate secondary recipient mice (supplemental Figure 15), suggesting that EPCR also identifies LT-HSCs under these conditions. Using the EPCR marker as a tool to monitor ex vivo HSPC activity, we determined that EPCR-positive cells were ∼3 to 4 times more frequent at day 7 than at day 12 of the culture (Figure 1B, compare percentage of EPCR+ cells at day 7 and day 12). Accordingly, the HSC activity measured in expanded cultures was >4 times greater when CB cells were expanded for 7 days compared with 12 days (compare human engraftment of total culture in Figure 1D [7 days of expansion] and supplemental Figure 5A [12 days]). These results suggest that EPCR monitoring also represents a new method to predict expansion levels of human CB-derived HSPCs.
EPCR-positive HSCs are competent for both short-term (3 weeks: supplemental Figure 17) and long-term (24 weeks) reconstitution, and therefore, distinguishing cells with LT from cells with short-term repopulation potential within the EPCR+ subset is the next step for future investigation. Surprisingly, our results show that although the CD34+EPCR–/Low population is enriched for CFU-GEMM content (Figure 1C), it is devoid of short-term reconstitution potential (supplemental Figure 17). This discrepancy might be explained by the fact that the NSG xenograft mouse model used in our experiments is not ideal to measure multipotent progenitor output. The NSG-W4126 or W41-NRG mouse27 models, which are known to better support multilineage human cell chimerism, could represent a more reliable system to monitor progenitors as well as short-term– and LT-HSCs, and should thus be tested in future studies.
Functional studies performed with short hairpin RNA vectors suggest that EPCR expression is crucial for human HSC activity in vivo. This observation is consistent with reports demonstrating that EPCR/PAR1 signaling regulates apoptosis in HSCs.11 Although recent findings showed that mice genetically modified to express low levels of EPCR display defects in HSC BM homing, we did not observe significant differences in homing capacity between sorted EPCR– and EPCR+ CB cells. This finding suggests that EPCR silencing disrupts HSC function by affecting HSC self-renewal rather than homing ability.
Transcriptional profiling of EPCR–/low and EPCR+ populations sorted from expanded CB shows striking molecular differences between these populations, with the genetic makeup governing HSC specification and function being reflected in the EPCR+ fraction. Our analysis uncovered genes preferentially expressed in the EPCR+ fraction, including EMCN28 and LGR5,29 which represent potential stem cell surface markers (supplemental Figure 14). The use of these markers in stem cell purification procedures could further enrich the EPCR+ population with LT-HSCs and possibly contribute to achieve LT-HSC detection at single-cell resolution. Studying transcription factors upregulated in the extensively self-renewing EPCR+ population could possibly accelerate the identification of the genetic pathways governing HSC “stemness.” Inducing these transcriptional programs in a variety of human cells, including hematopoietic progenitor cells, embryonic stem cell, and induced pluripotent cells, has the potential of converting their fate to fully functional HSCs that are compatible with therapeutic applications.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE77128).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank M.-E. Bordeleau and K. Humphries for critical reading of this manuscript. The authors also thank D. Gagné at the Institute of Research in Immunology and Cancer for technical support with flow cytometry sorts and M. Frechette and V. Blouin-Chagnon for assistance with in vivo experiments. The authors also thank Héma-Québec and Charles-Le Moyne Hospital for providing human umbilical CB units and our research assistants, S. Corneau and I. Boivin, for purifying the CD34+ CB cells.
This work was supported by grants from the Canadian Institutes of Health Research and the Stem Cell Network of Canada (G.S.).
Authorship
Contribution: I.F. designed and performed the experiments, generated the figures, and cowrote the manuscript; J.C. helped in experiment design and with EPCR knockdown experiments; T.M. generated the EPCR-targeting short hairpin RNA vectors plasmids; B.L. performed the RNA sequencing analysis; I.F. and E.T. generated in vivo data for short-term engraftment; L.A. and P.P.R. confirmed EPCR induction upon UM171 treatment in AML5 cells by surface biotinylation and western blot analysis; N.M. assisted I.F. with the in vivo studies; and G.S. provided project coordination and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guy Sauvageau, Institute of Research in Immunology and Cancer, University of Montreal, P.O. Box 6128, Station Centre-ville, Montreal, QC H3C 3J7, Canada; e-mail: guy.sauvageau@umontreal.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal