Key Points
Combination of Ig/TCR and BCR-ABL1 genomic approach for MRD monitoring in childhood ALL reveals patients with CML-like disease.
Monitoring ALL using BCR-ABL1 genomic breakpoint is feasible and enables the most specific and sensitive MRD quantification.
Abstract
We used the genomic breakpoint between BCR and ABL1 genes for the DNA-based monitoring of minimal residual disease (MRD) in 48 patients with childhood acute lymphoblastic leukemia (ALL). Comparing the results with standard MRD monitoring based on immunoglobulin/T-cell receptor (Ig/TCR) gene rearrangements and with quantification of IKZF1 deletion, we observed very good correlation for the methods in a majority of patients; however, >20% of children (25% [8/32] with minor and 12.5% [1/8] with major-BCR-ABL1 variants in the consecutive cohorts) had significantly (>1 log) higher levels of BCR-ABL1 fusion than Ig/TCR rearrangements and/or IKZF1 deletion. We performed cell sorting of the diagnostic material and assessed the frequency of BCR-ABL1-positive cells in various hematopoietic subpopulations; 12% to 83% of non–ALL B lymphocytes, T cells, and/or myeloid cells harbored the BCR-ABL1 fusion in patients with discrepant MRD results. The multilineage involvement of the BCR-ABL1-positive clone demonstrates that in some patients diagnosed with BCR-ABL1-positive ALL, a multipotent hematopoietic progenitor is affected by the BCR-ABL1 fusion. These patients have BCR-ABL1-positive clonal hematopoiesis resembling a chronic myeloid leukemia (CML)–like disease manifesting in “lymphoid blast crisis.” The biological heterogeneity of BCR-ABL1-positive ALL may impact the patient outcomes and optimal treatment (early stem cell transplantation vs long-term administration of tyrosine-kinase inhibitors) as well as on MRD testing. Therefore, we recommend further investigations on CML-like BCR-ABL1-positive ALL.
Introduction
The BCR-ABL1 fusion gene, resulting from the reciprocal translocation t(9;22)(q34;q11), is a hallmark of chronic myeloid leukemia (CML) and is also present in a subset of acute lymphoblastic leukemia (ALL). According to the genomic breakpoint within the BCR gene, there are 2 common variants of the fusion, major (M) and minor (m) BCR-ABL1, that encode the p210BCR-ABL1 and p190BCR-ABL1 proteins, respectively.1 Almost all patients diagnosed with CML carry the major-BCR-ABL1, whereas minor-BCR-ABL1-positive CML is very rare (∼1% cases).2 In contrast, the minor-BCR-ABL1 is prevalent in ALL, particularly in children.3 Generally, BCR-ABL1-positive leukemia is rare among children; BCR-ABL1 fusion occurs in 2% to 4% of pediatric ALL cases,4,5 and CML represents 2% to 4% of all leukemia diagnosed in childhood.6
The overall survival of BCR-ABL1-positive pediatric ALL has improved significantly since the introduction of tyrosine kinase inhibitors (TKIs) into treatment protocols.3,7-9 Despite this improvement, BCR-ABL1-positive ALL has remained a high-risk subgroup with an unfavorable outcome. Therefore, the ongoing studies on childhood ALL aim to find an optimal chemotherapy backbone to the TKI treatment and to reduce the number of patients undergoing stem cell transplantation (SCT).3,9 In pediatric CML, treatment standardization and balancing the survival advantage vs side effects of long-lasting TKI administration are the most important tasks10-12 ; in the advanced phases of CML, SCT is still considered the treatment of choice.13,14
One of the tools that might enable finding optimal treatments for both BCR-ABL1-positive ALL and CML is minimal residual disease (MRD) monitoring. Two targets are routinely used for polymerase chain reaction (PCR)–based MRD monitoring in BCR-ABL1-positive ALL, clonal immunoglobulin/T-cell receptor (Ig/TCR) gene rearrangements, and BCR-ABL1 transcript levels. We have previously compared the quantification of these 2 targets in childhood ALL,15 demonstrating that ∼20% of Ig/TCR-negative samples are positive (sometimes at high levels) using BCR-ABL1 transcript, and we presented 1 patient in whom the discrepancy was caused by the presence of BCR-ABL1 fusion outside the B-lymphoid blast population.15 Recent data show that even among BCR-ABL1-positive childhood ALL patients with a very good treatment response assessed by Ig/TCR MRD monitoring, 25% to 35% of patients relapse.16 However, BCR-ABL1-based MRD data and their concordance with the Ig/TCR levels in relapsing vs nonrelapsing patients were not analyzed.
Discrepancies in MRD levels assessed using these 2 techniques could be ascribed to the different targets (Ig/TCR vs BCR-ABL1) and/or the different methods for complementary DNA (cDNA)–based vs DNA-based detection. The RNA/cDNA-based transcript quantification depends on the expression levels of BCR-ABL1, which may be heterogeneous within the blast population and between cell types and may be influenced by treatment. Therefore, the number of transcripts per cell may vary significantly within the BCR-ABL1-positive cell population, while DNA-based MRD tests measure 1 target molecule per cell. One way to resolve this issue is to also measure the levels of the genomic BCR-ABL1 fusion.
Several studies demonstrated the feasibility of characterizing the BCR-ABL1 fusion at the genomic level.17-33 The genomic fusion sequence was usually used for sensitive detection of rare (pre)leukemic cells or as an MRD target. In some studies, the DNA-BCR-ABL1-based MRD monitoring was compared with the transcript quantification, and the genomic approach was shown to be more sensitive.17,18,20,23-25,29,31 However, with a single exception,27 the published “genomic” MRD data are based on major-BCR-ABL1 monitoring, which is probably because the characterization of minor-BCR-ABL1 fusion is more demanding. The minor-BCR breakpoint region spans >70 kb, whereas the major-BCR breakpoint region is only ∼3 kb long.
Here, we present both minor- and major-BCR-ABL1 breakpoint identification in a large cohort of pediatric patients with BCR-ABL1-positive ALL. We used the patient-specific DNA breakpoint sequences for MRD quantification and compared the results with standard Ig/TCR MRD monitoring and BCR-ABL1 transcript levels. Moreover, to complement these approaches, we used IKZF1 deletions (present in two-thirds of childhood BCR-ABL1-positive ALL34 ) as an alternative MRD target. Finally, we investigated the cell lineages of BCR-ABL1-positive clones in several patients with large differences in the MRD results.
Methods
Patients and samples
This study included 67 patients diagnosed with either BCR-ABL1-positive childhood ALL (n = 64) or CML diagnosed in lymphoid blast crisis (LBC) (n = 3). Patients were treated according to various protocols (see Table 1 for details). Standard diagnostics were performed according to the practice of local diagnostic laboratories. Basic clinical/outcome data were collected from treating centers. Diagnostic and treatment procedures and protocols were approved by the local institutional review boards. Informed consent was obtained in accordance with the Declaration of Helsinki.
Quantification of the ALB gene35 by quantitative PCR (qPCR) was performed to measure the DNA concentration. In some diagnostic samples, the whole genome amplification using REPLI-g Midi Kit (Qiagen, Hilden, Germany) was performed to obtain sufficient DNA for BCR-ABL1 breakpoint characterization. The concentration of cDNA was measured using either B2M, GUSB, or ABL1 as a housekeeping gene.36,37
Genomic BCR-ABL1 breakpoint detection
Primers and their multiplexing in long-distance (LD) PCR for the BCR-ABL1 genomic breakpoint detection were based on previously published data27 with minor modifications and additional primers published elsewhere.22,25 The complete list of primers is shown in supplemental Table 1 (available on the Blood Web site); for schematic representation of primer positions and PCR conditions, see supplemental Figure 1.
The specific product sequences were obtained by Sanger sequencing. In 7 patients with minor-BCR-ABL1 fusion, the products were not successfully sequenced using the common Sanger approach. Here, we employed the GS Junior platform (454 next-generation sequencing technology; Roche Diagnostics, Rotkreuz, Switzerland)38 to find the BCR-ABL1 genomic breakpoint within the amplified PCR fragment. To annotate the sequencing results, BLAST,39 ENSEMBL,40 or University of California Santa Cruz BLAT41 tools were used.
IKZF1-deletions screening and monitoring
Patients were screened for 4 intragenic IKZF1 deletions (exons 2-7, 2-8, 4-7, and 4-8) using qPCR with KAPA Probe Fast mix (Kapa Biosystems, Wilmington, MA), as previously described,42,43 and using additional primers and probe (supplemental Table 2). Patients with high IKZF1 deletion levels at diagnosis were tested at later time points with the same assay and with their own standard curve to quantify MRD.
Ig/TCR and BCR-ABL1 MRD quantification
In 49/51 patients, clonal Ig/TCR rearrangement was available for MRD monitoring. Quantification of patient-specific Ig/TCR rearrangements was performed and interpreted according to the standards of the EuroMRD international network.44-49 MRD monitoring based on BCR-ABL1 transcript quantification, including normalization to control gene expression (B2M, GUSB, or ABL1) and interpretation, was performed as described previously.15,50
For MRD quantification based on the BCR-ABL1 genomic breakpoint, primers amplifying the fusion sequence were designed to produce the PCR product of 97 to 204 base pairs. First, the QuantiTect SYBR Green PCR (Qiagen) system was used. In 5 cases for which the optimization of SYBR Green system was not satisfactory, fluorescein/tetramethylrhodamine-labeled probe (preferentially covering the breakpoint sequence) was designed and TaqMan Universal Master Mix II (Thermo Fisher Scientific) was employed.
The MRD levels of all targets are measured relative to the reference diagnosis/relapse sample, which was set to 1 (100%). For statistical comparisons between MRD results, samples with low, nonquantifiable positivity (below quantitative range [QR]) were assigned an arbitrary level of 1 × 10−5. Negative samples were assigned a level of 5 × 10−7.
Definition of discordant MRD results
In this study, we sought to identify samples that gave clearly discordant MRD results using different tests. Normal variation is considered a <0.5 log difference. Consequently, when the variation between results was ≤1 log, the results were scored as concordant; discordant samples were defined as those for which the MRD levels differed by >1 log. Moreover, although this might artificially increase the number of concordant samples, with respect to QR and sensitivity, we considered 2 samples concordant if (1) 1 target was quantifiable at a level <1 log above QR/sensitivity of the other target and the other target was nonquantifiably positive/negative, respectively; or (2) 1 target was nonquantifiably positive, whereas the other target was negative, and sensitivity of the former was <1 log higher than the sensitivity of the latter.
Patients with more subsequent follow-up samples that were discordant for BCR-ABL1 DNA vs Ig/TCR were considered as patients with discordant MRD.
Cell sorting and fluorescence in situ hybridization (FISH)
Frozen viable cells from bone marrow or peripheral blood samples were sorted on slides using a BD FACS Aria III (BD, Franklin Lakes, NJ). Following cell sorting into T cells (CD3+), myeloid cells (CD33+/dim), malignant B-cell precursors (CD19+CD10+CD45dim), and nonmalignant B cells (CD19+CD10−CD45++) (all supplemented in some cases with additional markers to minimize possible contamination of non-ALL subpopulations by leukemic blasts), FISH analysis was performed. The cells were fixed on slides using modified blastomere HCl/Tween20 fixation.51 For BCR-ABL1 chromosomal fusion visualization BCR-ABL Translocation, the Dual Fusion LPH 007 probe (Aquarius Haematology Probes, Cytocell, Cambridge, United Kingdom) or Vysis LSI BCR/ABL Dual Color Dual Fusion Translocation Probe (Abbott Molecular, Des Plaines, IL) was used. In selected cases, qPCR targeted to patient-specific genomic BCR-ABL1 fusion and/or Ig/TCR rearrangements was performed using DNA isolated from the sorted subpopulations.
Twelve of the ALL patients with analyzed cell subpopulations represent unselected consecutive cases that were diagnosed/relapsed in the Czech Republic between June 2007 and December 2014 (only 1 patient from this period was not included because of a lack of material for cell sorting).
Statistical analysis
The correlation of individual methods was analyzed using the Spearman correlation rank. The double-negative samples were excluded from the analysis. For comparisons of groups, the Mann-Whitney U test and Kruskal-Wallis test were used. Initial WBC was compared by 2-way ANOVA to compensate for the effect of the BCR-ABL1 variant (minor vs major).
Results
Breakpoint detection
We examined leukemic DNA for 67 childhood BCR-ABL1-positive ALL/CML cases to find the genomic BCR-ABL1 breakpoint. We characterized the fusion sequence in 54 cases (81%), 16 with major and 38 with minor breakpoint variants. In 13 cases (4 major and 9 minor), we did not find the genomic breakpoint sequence; in 7 of these cases, the DNA quality was poor or suboptimal (control LD PCR amplified only products of <2 kb [3 cases] or <7 kb [4 cases]). In 1 patient, the DNA quality and integrity were not tested using control LD PCR because of a low amount of material for further analyses. In 3 cases, the LD PCR yielded a positive product; however, the sequencing (both Sanger and next-generation sequencing) was unsuccessful. In the remaining 2 patients, we did not obtain a specific LD PCR product despite the adequate DNA quality. The fusion gene sequences of the 54 successfully analyzed cases are listed in supplemental Table 3.
MRD samples and sensitivity
We analyzed MRD in 548 bone marrow samples from 48 ALL patients (minor-BCR-ABL1-positive ALL, 433 samples/36 patients; major-BCR-ABL1-positive ALL, 115 samples/12 patients) and 3 CML patients (34 samples) by quantifying the BCR-ABL1 genomic breakpoint and at least one of the other targets (Ig/TCR, 560 samples; IKZF1del, 194 samples; and BCR-ABL1 transcript, 410 samples). Moreover, in 143 samples, we assessed the MRD levels in the peripheral blood (supplemental Figure 2).
The sensitivity of the DNA approaches was generally comparable, with no statistically significant differences between Ig/TCR, BCR-ABL1, and IKZF1 tests with mean sensitivities of 4.4 vs 4.5 vs 4.3 logs and median values of 4 vs 5 vs 4 logs, respectively. On the other hand, the QR (ie, the range in which MRD can be accurately quantified) was significantly lower for Ig/TCR compared with BCR-ABL1 (P = .0021, mean 3.7 logs/median 4 logs vs mean 4.1 logs/median 4 logs). The QR for IKZF1 assays was intermediate and not significantly different from either BCR-ABL1 or Ig/TCR tests. The quantitative reverse transcription PCR for BCR-ABL1 transcript detection reliably detected ≤10 copies of cDNA. As 1 standard quantitative reverse transcription PCR consisted of cDNA from ∼100 000 cells, the sensitivity of this method is generally comparable to the DNA-based assays (4-5 logs).
Comparison of MRD by different targets
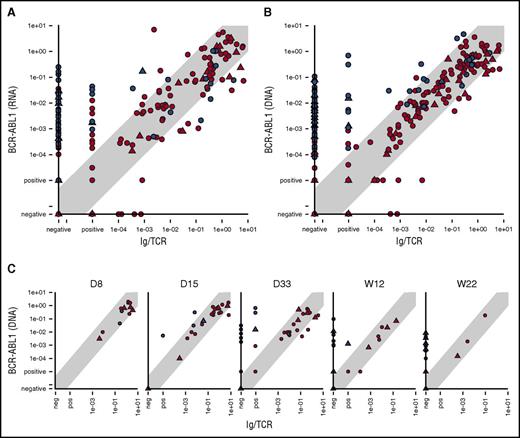
Comparison of BCR-ABL1 RNA with Ig/TCR MRD levels confirmed our previous data15 that showed a poor correlation (Spearman correlation coefficient, 0.63) and significant number of samples (23%) with quantifiable BCR-ABL1 levels, while Ig/TCR MRD negative (Figure 1A).
Comparison of the MRD levels in ALL patients. Comparison of the MRD levels in ALL patients measured by Ig/TCR vs BCR-ABL1 transcript quantification (A) as well as vs BCR-ABL1 genomic breakpoint quantification in all samples (B) and separately in selected time points during frontline treatment (C). Samples from patients with major-BCR-ABL1 fusion variant are shown as triangles and minor-BCR-ABL1 as circles. Samples from patients with concordant MRD course are in red, and samples from patients with discordant MRD are in blue. The light gray diagonal shape represents the area of concordance ±1 log. D, day; W, week (from the start of treatment).
Comparison of the MRD levels in ALL patients. Comparison of the MRD levels in ALL patients measured by Ig/TCR vs BCR-ABL1 transcript quantification (A) as well as vs BCR-ABL1 genomic breakpoint quantification in all samples (B) and separately in selected time points during frontline treatment (C). Samples from patients with major-BCR-ABL1 fusion variant are shown as triangles and minor-BCR-ABL1 as circles. Samples from patients with concordant MRD course are in red, and samples from patients with discordant MRD are in blue. The light gray diagonal shape represents the area of concordance ±1 log. D, day; W, week (from the start of treatment).
The results of the genomic BCR-ABL1 quantification also showed differences with the Ig/TCR levels (Figure 1B). Despite using the same DNA-based methodology, the Spearman correlation was low (0.62) for Ig/TCR vs BCR-ABL1 DNA overall (with 0.61 for minor- and 0.65 for major-BCR-ABL1) and consistent with Ig/TCR vs BCR-ABL1 RNA correlation. In contrast, the correlation coefficient was significantly better (0.85) between the BCR-ABL1 DNA and RNA MRD levels. The comparison of 2 DNA targets also confirmed our observation that although the MRD results assessed by the different techniques correlated well in the majority of patients (“patients with concordant MRD”), there were some “patients with discordant MRD” and several consecutive samples with significantly higher (>1 log) BCR-ABL1 levels compared with Ig/TCR. There was no significant difference in the frequency of patients with discordant MRD among minor- vs major-BCR-ABL1 patients (10/34 vs 5/12 patients; P = .49).
In patients with concordant MRD, only rare individual samples (12/386 all samples, 3% or 12/255 non–double-negative samples, 4.7%) did not fit into the 1 log difference interval, and all divergences were only slightly higher, well within 2 logs. By contrast, in patients with discordant MRD, 50% of all and 60% of the non–double-negative samples (71/141 or 71/120, respectively) had BCR-ABL1 levels higher than Ig/TCR by >1 log and 50% of those (n = 35) by >2 logs.
In patients with discordant MRD, the proportion of samples with significantly higher BCR-ABL1 compared with Ig/TCR levels increased with the treatment time point from 0% (0/3 patients analyzed after 1 week of treatment) to 50% after 2 weeks (2/4 patients analyzed) and 80% to 90% after 1, 3, and 5 months of treatment (8/9, 8/10, and 8/10 patients, respectively) (Figure 1C).
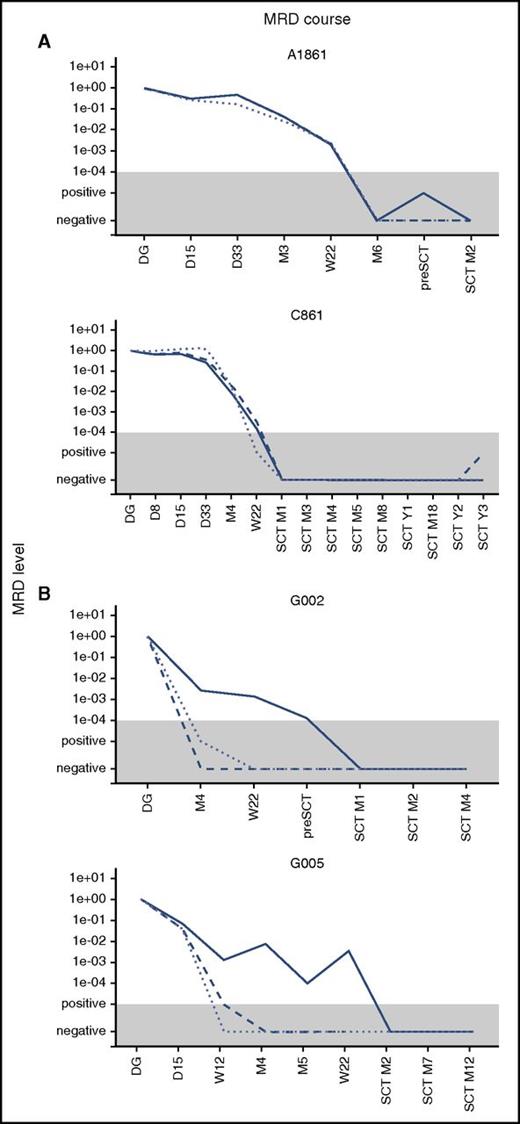
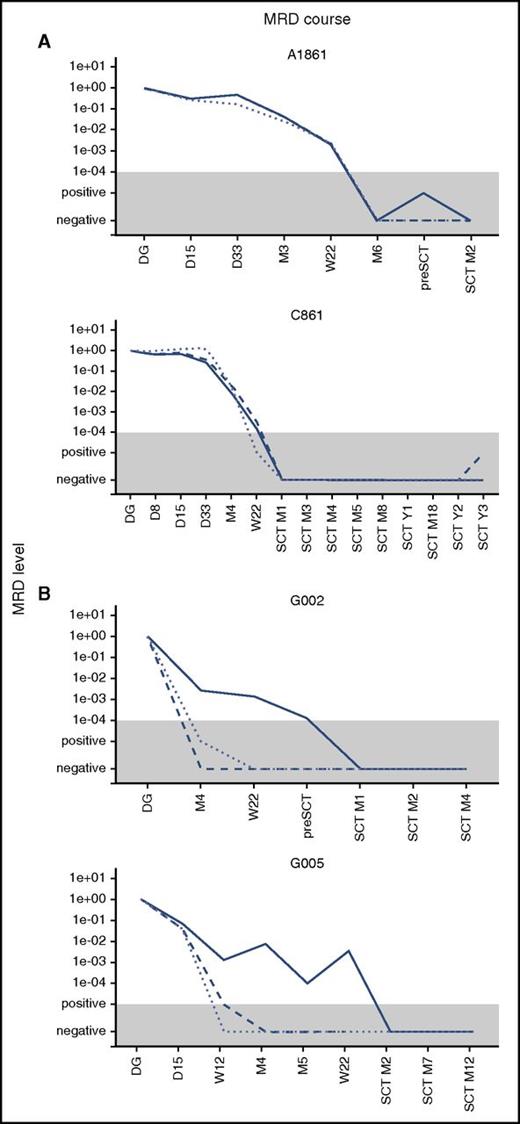
We used IKZF1 deletions as an alternative target for MRD monitoring. In patients with concordant MRD, the data from IKZF1 quantification correlated well with the BCR-ABL1 and Ig/TCR targets. In patients with discordant MRD, the levels of IKZF1 deletion mimicked the Ig/TCR levels, which were significantly lower than BCR-ABL1 MRD. The overall correlation is shown in supplemental Figure 2, and examples of the MRD course are shown in Figure 2.
Examples of the MRD course. Examples of the MRD course measured by Ig/TCR (dashed line), BCR-ABL1 genomic breakpoint (full line), and IKZF1 deletion (dotted line) quantification in 2 illustrative patients with concordant MRD (A) and 2 with discordant MRD (B). The gray area represents the level of sensitivity of the Ig/TCR quantification. M, month (from the start of treatment or from SCT).
Examples of the MRD course. Examples of the MRD course measured by Ig/TCR (dashed line), BCR-ABL1 genomic breakpoint (full line), and IKZF1 deletion (dotted line) quantification in 2 illustrative patients with concordant MRD (A) and 2 with discordant MRD (B). The gray area represents the level of sensitivity of the Ig/TCR quantification. M, month (from the start of treatment or from SCT).
BCR-ABL1 presence in hematopoietic lineages
To investigate the biological basis of discordant MRD, we analyzed hematopoietic cell subpopulations in selected patients and searched for the presence of BCR-ABL1 fusion in distinct cell types. We analyzed sorted ALL B-cell precursors, non-ALL B cells, T cells, and myeloid cells from the diagnostic samples as well as performed FISH with BCR-ABL1 probe in 12 patients (10 ALL with BCR-ABL1 fusion and 2 CML) and qPCR BCR-ABL1 and Ig/TCR detection in 1 additional CML and 3 BCR-ABL1-positive ALL cases (1 was previously published15 ). In 2 ALL patients, one of the target sequences for MRD monitoring was not successfully identified; therefore, in this analysis, we used BCR-ABL1 transcript quantification instead of the genomic BCR-ABL1 fusion and flow-cytometric MRD assessment instead of Ig/TCR quantification, respectively, to assess the MRD concordance in these 2 patients.
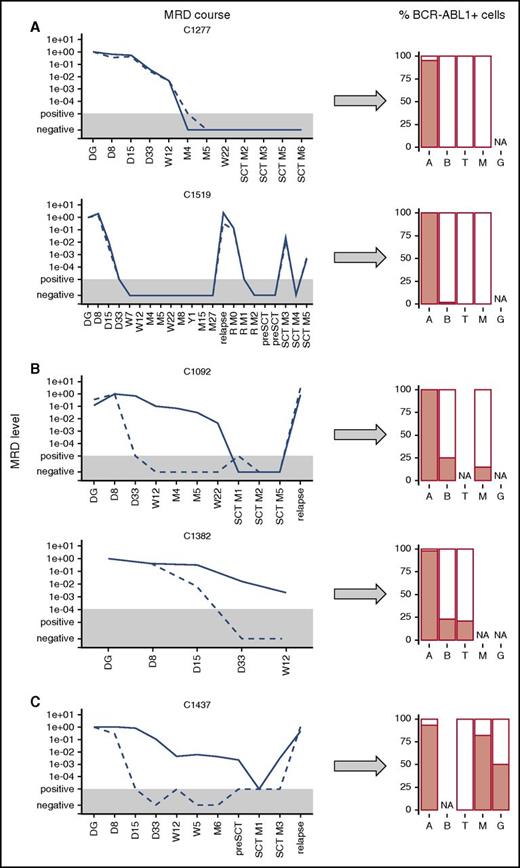
In 7 patients with concordant MRD, the BCR-ABL1 fusion was only found in the ALL B-cell precursors (93% to 100%), whereas non-ALL B cells, T cells, and myeloid cells were negative (<2%); 2 cases are shown in Figure 3A. Two patients with concordant MRD were diagnosed as ALL with significant aberrant expression of CD33 (40% of ALL blasts at diagnosis and 100% at subsequent relapse in 1 patient and bilineal B/Myelo-leukemia in the other). In addition to the ALL blasts, these 2 patients also had BCR-ABL1-positive cells in the CD33pos-sorted myeloid subpopulation. Consequently, qPCR revealed that the sorted CD33-positive cells harbored both BCR-ABL1 and Ig/TCR rearrangements, whereas non-ALL B cells and T lymphocytes were BCR-ABL1-negative.
Presence of BCR-ABL1-positive cells in hematopoietic lineages. Presence of BCR-ABL1-positive cells in hematopoietic lineages (A, ALL blasts; B, non-ALL B cells; G, granulocytes; M, myeloid cells/monocytes; T, T cells) at diagnosis in ALL patients with concordant (A) and discordant (B) MRD courses and in CML patient (C). The MRD levels are shown for Ig/TCR (dashed line) and BCR-ABL1 genomic breakpoint (full line); the gray area represents the level of sensitivity of the Ig/TCR quantification. NA, not available.
Presence of BCR-ABL1-positive cells in hematopoietic lineages. Presence of BCR-ABL1-positive cells in hematopoietic lineages (A, ALL blasts; B, non-ALL B cells; G, granulocytes; M, myeloid cells/monocytes; T, T cells) at diagnosis in ALL patients with concordant (A) and discordant (B) MRD courses and in CML patient (C). The MRD levels are shown for Ig/TCR (dashed line) and BCR-ABL1 genomic breakpoint (full line); the gray area represents the level of sensitivity of the Ig/TCR quantification. NA, not available.
In 4 patients with discordant MRD results, we detected BCR-ABL1 fusion not only in the malignant B-cell precursors but also in non-ALL B cells (15% to 83%), T cells (12% to 21%), and myeloid cells (15% to 80%). Importantly, the populations other than ALL B cells were also tested by qPCR and all were positive for the BCR-ABL1 genomic fusion, whereas they were low/negative (<5 × 10−2, representing maximal expected level of possible sort contamination by ALL blasts) for the patient-specific Ig/TCR rearrangements (5 populations tested).
In the control analysis of sorted populations from 3 patients with typical CML (1 in LBC, 2 in the chronic phase) tested for BCR-ABL1 fusion, we found that the proportions of BCR-ABL1-positive cells were 93% and 100% of positive lymphoid blasts; 82%, 100%, and 34% of monocytes; 50%, 100%, and 34% of granulocytes; and 0%, 0%, and 0% of T cells, respectively, in the 3 CML cases (1 patient shown in Figure 3C).
Initial WBC count and diagnostic BCR-ABL1 expression
A very high WBC count (usually >100 × 109/L) is characteristic of CML LBC. Indeed, the 3 patients with CML LBC had higher WBC levels compared with ALL patients (P = .015), and ALL patients with the major-BCR-ABL1 variant had a higher initial WBC count compared with patients with minor-BCR-ABL1 (P = .002). With respect to the Ig/TCR vs BCR-ABL1 correlation, the ALL patients with discordant MRD had a lower initial WBC count compared with patients with concordant MRD (P = .058).
The expression of the BCR-ABL1 fusion transcript at diagnosis varied significantly between the patients. When normalized to the expression of a housekeeping gene (B2M or GUSB), the diagnostic levels differed by >2 logs for the major-BCR-ABL1 variant and by >3 logs for the minor-BCR-ABL1 variant. In all patients with major-BCR-ABL1, the minor-BCR-ABL1 transcript was also detected at levels that were ∼3 logs lower compared with the dominant major transcript. Patients with concordant MRD tended to have higher BCR-ABL1 expression compared with patients with discordant results (P = .098).
Effect of discordant vs concordant MRD on outcomes
There was no significant difference in the outcome between the patients with concordant and discordant MRD in our cohort in which 80% of patients underwent SCT and all but 3 children received TKI therapy (albeit another 4 only after relapse). For patients with discordant MRD, 10 out of the 12 transplanted patients are alive with a median follow-up 10 years, whereas only 1 out of the 3 nontransplanted patients is alive (40 months from diagnosis; log-rank test for overall survival, P = .019). The outcome of the patients with concordant MRD who were transplanted did not differ significantly from those who received chemotherapy alone (10/25 and 1/6 died, respectively; log-rank test for overall survival, P = .48). For details on the treatment and outcome, see Table 1.
Discussion
Comparison of the MRD levels measured by quantification of DNA vs RNA can always be challenged; although the number of target DNA copies per cell is usually constant at 1 or 2 per genome, the expression levels of both the target and housekeeping gene used for normalization can vary significantly (eg, in different patients, among different cell types, and during treatment). Therefore, we aimed to investigate our previous data for BCR-ABL1-positive childhood ALL, which showed a poor correlation of 2 routine MRD approaches in some patients (DNA-based Ig/TCR quantification and RNA-based BCR-ABL1 transcript quantification) by complementing it with MRD analysis using BCR-ABL1 fusion at the DNA level.
We found the genomic breakpoint in most patients (>80%) with no prior selection in terms of the DNA quality. In the remaining cases, poor DNA quality for the LD PCR was usually the limiting factor; moreover, in rare cases, the fusion might be more complex at the genomic level, precluding successful analysis with this approach. In particular, we detected cases in which an inverted part of the ABL1 gene was inserted into the fusion and deduced that if the size of such insertion/inversion was longer (approximately >10 kb), we would probably be unsuccessful using our LD-PCR approach. Moreover, the affected introns harbor many repetitive sequences that hamper routine sequencing. We wanted to establish if our approach could be used for routine MRD monitoring; therefore, we did not perform any additional experiments beyond standard testing to find the breakpoint in all samples subjected to analysis.
Our results confirmed and further extended our previous data from the analysis of BCR-ABL1 transcript quantification; although in the majority of ALL patients the MRD levels correlated very well (within 1 log), other patients had several consecutive samples showing significantly higher BCR-ABL1 levels compared with Ig/TCR. Our present study was artificially enriched for such samples, as we selected some cases based on already known discordance between the Ig/TCR and BCR-ABL1 transcript levels. In the unselected consecutive cohorts that were analyzed within this study, the incidence of patients with discordant MRD was 22.5% (9/40 patients; 8/32 [25%] with the minor- and 1/8 [12.5%] with major-BCR-ABL1 variant). These data also show that the poor correlation is not limited to the cases with major-BCR-ABL1 fusion; in contrast, we detected more discordant cases among minor-BCR-ABL1-positive patients. Only ALL blasts were BCR-ABL1-positive in patients with concordant MRD, although 2 patients diagnosed with ALL with myeloid markers had BCR-ABL1 positivity in the myeloid fraction. In all patients with discordant MRD in whom the sorted cell populations were analyzed (as well as in our control CML cases), we established that the source of the poor correlation was the presence of BCR-ABL1 in cells that were not derived from ALL lymphoid blast clones.
These experiments provide evidence that a multipotent hematopoietic progenitor is affected by BCR-ABL1 fusion in some cases and that the ALL patients with discordant MRD have a “CML-like” disease background. The absence of clonal Ig/TCR rearrangements in the BCR-ABL1-positive cells detected in sorted cell subpopulations other than ALL blasts and during the follow-up of patients with discordant MRD also rules out the possibility that these cells originate by dedifferentiation or transdifferentiation of the original ALL cell. IKZF1 gene deletions, considered to be a subsequent hit cooperating with BCR-ABL1 in the ALL pathogenesis52 and known to emerge in LBC after being originally negative during the chronic CML phase,53,54 correlated in ALL patients with the lymphoid Ig/TCR clone and were absent in Ig/TCR-negative BCR-ABL1-positive cells detected during treatment, which further supports the CML-like pathogenesis with IKZF1 deletion acquired in progression to ALL. Moreover, the patients with discordant MRD, both ALL and CML patients, tend to have lower fusion transcript expression at diagnosis compared with patients with “typical” BCR-ABL1-positive ALL.
In contrast, the initial WBC level tended to be lower in the ALL cases with discordant MRD than both the concordant ALL patients and typical CML cases. Although we found BCR-ABL1 in other cell types, including the myeloid lineage, in patients with discordant MRD, the proportion of positive cells was lower than in the classical CML-LBC patients. Moreover, unlike the CML patients, ALL cases with discordant MRD also harbored BCR-ABL1 fusion in T cells, whereas putative stem cells (CD34+CD38−CD133+) were not conclusively BCR-ABL1 positive (supplemental Figure 3). These data suggest that ALL cases with discordant MRD differ from both “typical ALL” and classical CML. In our experience, the multilineage involvement of the primary leukemogenic aberration is limited to BCR-ABL1-positive ALL and rare MLL-rearranged leukemia.55 Bilineal leukemia cases have identical genetic aberrations, including the same Ig/TCR rearrangements present in both lymphoid and myeloid compartments56 ; therefore, they are similar to our 2 BCR-ABL1-positive ALL cases with myeloid markers and Ig/TCR detected in CD33pos-sorted subpopulation, which we interpret as being the result of cell plasticity57 instead of a “CML-like” pathogenesis.
The impact of the specific disease biology in patients with discordant MRD on the prognosis and optimal treatment needs to be systematically analyzed in contemporary protocols to draw definitive conclusions. Patients in our study were treated with several trials over an extended period. However, based on small numbers, our data suggest that patients with discordant MRD benefited from SCT in contrast to patients with concordant MRD who did well on chemotherapy alone. Considering that early and prolonged use of TKI is now often preferred to SCT in BCR-ABL1-positive ALL, whereas SCT is still the treatment of choice for advanced phases of childhood CML, this perspective should be considered carefully in future protocols for children diagnosed as BCR-ABL1-positive ALL, and the patients with discordant MRD should be identified.
In conclusion, BCR-ABL1 monitoring at the genomic DNA level is a feasible approach that provides the most accurate and sensitive quantification of BCR-ABL1-positive cells during leukemia treatment. Moreover, used in combination with standard Ig/TCR monitoring, it can unmask cases that have discordant results associated with different disease biology, originating from a multipotent hematopoietic progenitor. A systematic, protocol-based study is needed to precisely define the prognosis and optimal treatment of MRD-discordant cases and the role of early SCT vs prolonged TKI therapy in these patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank to all centers of the Czech Pediatric Hematology Working Group (CPH), Australian and New Zealand Children’s Oncology Group, and German ALL-BFM Group for taking care of the children included in this study and for providing outcome data. The authors would also like to thank Pavel Semerak for his expertise and assistance in sorting experiments.
This work was supported by the “Kapka nadeje” Foundation, grants from the Czech Health Research Council (16-30186A and 15-31540A), the Grant Agency of Charles University (GAUK 554214), the project for conceptual development of research organization 00064203 (University Hospital Motol, Prague, Czech Republic), NPU I nr.LO1604, and ERDF OPPK CZ.2.16/3.1.00/28007 and CZ.2.16/3.1.00/24022. In Australia, research was funded by grants from National Health and Medical Research Council Australia and the Cancer Council NSW. These bodies had no role in the study design, collection, and analysis of data or the decision to publish.
Authorship
Contribution: L.H., M.Z., N.C.V., K.B., M.T., E.P., M.V., J.L., K.M.P., E.F., W.M., and J.E.G. performed research and analyzed and interpreted data; P.JS., G.C., R.S., J.S., and J.T. collected and interpreted data; J.Z. designed the research; L.H. and J.Z. wrote the manuscript; and all authors revised the manuscript and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Zuna, Childhood Leukemia Investigation Prague (CLIP), Department of Pediatric Oncology and Oncology, Second Faculty of Medicine, Charles University and University Hospital Motol, V Uvalu 84, 150 06 Prague, Czech Republic; e-mail: jan.zuna@lfmotol.cuni.cz.