Key Points
Binding to fibrinogen is mediated by the central region of Aβ42 and is enhanced by its C-terminal residues.
Aβ42 binds the αC region of fibrinogen, delaying plasmin-mediated fibrin cleavage and generating a persistent αC degradation product.
Abstract
The majority of patients with Alzheimer disease (AD) suffer from impaired cerebral circulation. Accumulating evidence suggests that fibrinogen, the main protein component of blood clots, plays an important role in this circulatory dysfunction in AD. Fibrinogen interacts with β-amyloid (Aβ), forming plasmin-resistant abnormal blood clots, and increased fibrin deposition is found in the brains of AD patients and mouse models. In this study, we investigated the biochemical and structural details of the Aβ-fibrinogen interaction. We identified the central region of Aβ42 as the most critical region for the interaction, which can be inhibited by specific antibodies against the central region of Aβ and by naturally occurring p3 peptides, Aβ17-40 and Aβ17-42. X-ray crystallographic analysis revealed that Aβ42 binding to fragment D of fibrinogen induced a structural change in the C-terminal region of the fibrinogen β-chain (β384-393). Furthermore, we identified an additional Aβ-binding site within the αC region of fibrinogen. Aβ binding to this αC region blocked plasmin-mediated fibrin cleavage at this site, resulting in the generation of increased levels of a plasmin-resistant fibrin degradation fragment. Overall, our study elucidates the Aβ-fibrinogen interaction and clarifies the mechanism by which Aβ-fibrinogen binding delays fibrinolysis by plasmin. These results may facilitate the development of effective therapeutics against the Aβ-fibrinogen interaction to treat cerebrovascular abnormalities in AD.
Introduction
Accumulating evidence implicates fibrin(ogen), the main protein component of blood clots, in Alzheimer disease (AD) pathogenesis.1-3 Activation of the coagulation cascade results in the cleavage of soluble fibrinogen to fibrin, which polymerizes to form an insoluble network. Because fibrin is occlusive4 and proinflammatory,5 its clearance (fibrinolysis) by plasmin is a tightly regulated process. Disturbances to fibrinolysis may therefore have significant consequences for occlusive and inflammatory pathology in various diseases, including AD. Indeed, increased fibrin accumulation in the brains of AD patients and mouse models is correlated with areas of neuronal dysfunction.6
We have previously identified the AD-related peptide, β-amyloid (Aβ), as a factor capable of modulating fibrin clot structure and stability.7,8 Aβ42 binds fibrinogen with a Kd of 26.3 ± 6.7 nM,7 and fibrin clots formed in the presence of Aβ42 are structurally altered and more resistant to fibrinolysis. Aβ42 can also bind to preformed fibrin and block the access of plasmin to fibrin.8 Fibrinogen, which is composed of 2 fragment D domains and 1 fragment E domain, is a heterodimer composed of pairs of α, β, and γ chains.9 Aβ42 binds β-chain residues β366-414 within fragment D.7 This region is in close proximity to the b-hole of fibrinogen,10 which is involved in the lateral aggregation of fibrin protofibrils.11,12
Two different types of therapeutics targeting Aβ-fibrinogen association have been investigated.2,13 The root extract of Aristolochia indica efficiently degrades fibrin-Aβ coaggregates in vitro and in a rat model.13 Furthermore, long-term treatment with RU-505, a specific inhibitor of the Aβ-fibrinogen interaction, results in reduced thrombosis, decreased AD pathology, and improved cognitive performance in a mouse model of AD.2 Although both novel therapeutics targeting the Aβ-fibrinogen interaction in AD are effective in vitro and in vivo, low selectivity of the enzyme from A indica and micromolar half-maximal inhibition (IC50) levels of RU-505 limit their capabilities for clinical development. To improve selectivity and potency of therapeutics against the Aβ-fibrinogen interaction, a better understanding of the Aβ-fibrinogen interaction is needed.
Here, we analyzed the region within Aβ responsible for Aβ-fibrinogen binding using biochemical approaches and examined the structural aspects of binding between Aβ and fragment D of fibrinogen using X-ray crystallography. In addition, we further investigated the mechanism by which Aβ-fibrinogen binding delays fibrinolysis by plasmin.
Methods
Preparation of Aβ42 and fibrinogen fragment D
Aβ42 (Anaspec) was reconstituted in a minimal volume of 0.1% NH4OH and then diluted to the desired concentration with 50 mM Tris or phosphate-buffered saline (pH 7.4). Solubilized Aβ42 was spun at 12 000 × g for 15 minutes to remove aggregated material14 and the concentration was established by bicinchoninic acid assay (Thermo Scientific). Fibrinogen fragment D was prepared and purified as previously described.15
Identification of fibrinogen-binding domains on Aβ
Fibrinogen- or fragment D–binding assay using biotinylated Aβ fragments.
Synthetic N-terminally biotinylated Aβ fragments 1 to 16, 15 to 25, 22 to 41, and 1 to 42 (50 nM; Anaspec) were incubated with fibrinogen (5 nM; Calbiochem) or fragment D (100 nM) for 1 hour at room temperature (RT) in 50 mM Tris pH 7.4 containing 500 mM NaCl, 0.01% bovine serum albumin (BSA), protease inhibitor cocktail (Roche), and 0.01% NP-40 or 0.05% Tween-20. Streptavidin-coated magnetic beads (Dynabeads M-280; Thermo-Fisher) were added for 30 minutes, washed, and eluted with nonreducing 1× lithium dodecyl sulfate sample buffer (Thermo Fisher Scientific). Eluates were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on a 4% to 20% Tris-glycine gradient gel (Life Sciences) followed by western blot using a polyclonal antibody against fibrinogen (Dako). For the AlphaLISA assay, various concentrations (0.02-20 µM) of N-terminally biotinylated Aβ fragments 1 to 16, 15 to 25, 22 to 41, and 1 to 42 (50 nM, Anaspec) were incubated with 1 nM fibrinogen for 30 minutes at RT in a final volume of 10 µL of assay buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.05% Tween-20, 0.1% BSA) in white 384-well plates (Greiner). The mixture was incubated with the anti-fibrinogen antibody, 20 µg/mL streptavidin-conjugated donor, and protein A–conjugated acceptor beads (PerkinElmer) for 90 minutes at RT. Samples were read by a PerkinElmer EnVision plate reader.
Aβ-fibrinogen interaction inhibition assay using nonbiotinylated Aβ fragments.
Various concentrations (0.05-20 µM) of 16 nonbiotinylated Aβ fragments listed in supplemental Figure 1 (available on the Blood Web site) (Anaspec or rPeptide) were plated in white 384-well plates and incubated with 10 nM biotinylated Aβ42 and 1 nM fibrinogen for 30 minutes at RT. The rest of the AlphaLISA assay was performed as described in “Fibrinogen- or fragment D–binding assay using biotinylated Aβ fragments,” and the data fitted to a sigmoidal dose-response equation (Y= bottom + (top − bottom)/1 + 10(logIC50 − X) × Hill coefficient) using GraphPad Prism 4 to calculate IC50. For pulldown experiments, various concentrations of nonbiotinylated Aβ17-42 were incubated with fragment D (200 nM) and biotinylated Aβ42 (50 nM) for 30 minutes. The rest of the pulldown assay was performed as described in “Fibrinogen- or fragment D–binding assay using biotinylated Aβ fragments.”
Five alanine-scanning peptides, L17A, V18A, F19A, F20A, and D23A, were synthesized by replacing L17, V18, F19, F20, or D23 in Aβ17-42 with alanine (Chinese Peptide Company). Various concentrations (0.01-20 µM) of alanine-scanning Aβ17-42 peptides were incubated with 10 nM biotinylated Aβ42 and 1 nM fibrinogen for 30 minutes at RT. The rest of the AlphaLISA assay was performed as described in “Fibrinogen- or fragment D–binding assay using biotinylated Aβ fragments.”
Aβ-fibrinogen interaction inhibition assay using antibodies raised against specific regions of Aβ.
Biotinylated Aβ42 (50 nM) and fibrinogen (5 nM) were incubated with anti-Aβ antibodies 3D6 (50 nM; Elan), 6F/3D (50 nM; Dako), 4G8 (50 nM; Covance), ab62658 (50 nM; Abcam), or G2-11 (50 nM; Abcam) in 50 mM Tris pH 7.4 containing 150 mM NaCl, 0.01% NP-40, 0.01% BSA, and protease inhibitor cocktail; pulldown assays were performed as described in “Fibrinogen- or fragment D–binding assay using biotinylated Aβ fragments.”
SDS-stable complex formation
Fragment D (126 μM) was incubated with Aβ42 or Aβ42 G37D (111 μM) in 50 mM Tris pH 7.6 with or without 1 mM EDTA for 5 days at 37°C. Fibrinogen (14.7 μM) was incubated with Aβ42 mutant G37D (30 μM; Anaspec) in 50 mM Tris pH 7.6 for 24 hours at 37°C. SDS denaturation was carried out at 100°C for 5 minutes under nonreducing conditions; samples were analyzed by western blot using antibodies against Aβ (6E10; Covance) and fibrinogen (Dako).
Crystallization of fibrinogen fragment D and of the fragment D-Aβ42 complex
Fragment D crystals were obtained with the assistance of The Rockefeller University Structural Biology Resource Center as described in Everse et al.15 Briefly, crystals were obtained by sitting drop vapor diffusion at 4°C from 50 mM Tris, pH 8.5, 70 mM CaCl2, 2 mM sodium azide, 12% to 17% polyethylene glycol 3350 in drops of 10 to 20 μL, with fragment D at 15 mg/mL. Aβ42 or 5-carboxy-tetramethylrhodamine (TAMRA)–Aβ42 (Anaspec) were reconstituted to 0.7 to 1.5 mg/mL in 50 mM Tris, pH 8.5 with 0.1% NH4OH, then diluted twofold in 2× reservoir buffer (50 mM Tris pH 8.5, 4 mM sodium azide, 140 mM CaCl2, 34% polyethylene glycol 3350) to yield Aβ peptides in 1× reservoir buffer. Fragment D crystals were then soaked in the Aβ solution (containing an excess of Aβ over fragment D) at 4°C. Crystals soaked for 6 hours with TAMRA-Aβ42 were washed 3 times with reservoir buffer and imaged using a fluorescence microscope (Zeiss). Crystals soaked for 2 weeks with unlabeled Aβ42 were frozen in the N2 cryostream in reservoir buffer without cryoprotection and diffracted to 3.3 Å at the National Synchrotron Light Source, Brookhaven National Laboratory (beam line, ×25; wavelength = 1.1Å). Data were also obtained to 2.9 Å for crystals not subjected to soaking (native crystals). Data reduction was performed using HKL2000 software. Molecular replacement and subsequent refinement were performed with PHENIX software (PHENIX-dev-1555)16 using the Protein Data Bank (PDB) entry 1FZA for fragment D.17
Analysis of Aβ-induced delayed fibrinolysis
The effect of Aβ on fibrin degradation by plasmin was analyzed by an in vitro fibrin clot formation and degradation assay.8 Briefly, fibrinogen (1.5μM) with or without Aβ42 (3μM) was mixed with plasminogen (250 nM) in 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) buffer (pH 7.4) with 137 mM NaCl. Fibrin clot formation and degradation were initiated by adding thrombin (0.5 U/mL; Sigma), tissue plasminogen activator (tPA) (0.15 nM; kindly provided by Genentech), and CaCl2 (5 mM) in a final volume of 150 μL. Assays were performed at RT in 96-well plates (Fisher Scientific) in triplicate, and fibrin clot formation and degradation were monitored at 450 nm using a Molecular Devices Spectramax Plus384 reader. Reactions were stopped after 8 hours by adding reducing 4× lithium dodecyl sulfate sample buffer containing 100 mM dithiothreitol. Fibrin degradation products were analyzed by SDS-PAGE on a 4% to 20% Tris-glycine gradient gel and visualized by colloidal blue stain (Invitrogen).
Mass spectrometric analysis of fibrin degradation products.
A protein gel band resistant to degradation in the presence of Aβ42 was excised and submitted to The Rockefeller University Proteomics Resource Center for mass spectrometric analysis (see supplemental Methods).
Edman sequencing of fibrin degradation products.
See supplemental Methods.
Identification of Aβ42-binding fibrinogen degradation products.
Fibrinogen (15 μM in 50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM iodoacetamide) was incubated with 140 nM plasminogen and 20 nM tPA for 6 hours at 37°C to generate fibrinogen degradation products (FDPs). Digestions were stopped with aprotinin (Sigma). FDPs (500 μL) were incubated for 3 hours at RT with synthetic N-terminally biotinylated Aβ42 (2μM) in phosphate-buffered saline adjusted to contain 500 mM NaCl, 0.01% NP-40, and protease inhibitor cocktail, the Aβ-interacting peptides pulled down with streptavidin-sepharose beads (Invitrogen) for 1 hour at RT, and the peptides eluted with sample loading buffer and analyzed by SDS-PAGE on a 4% to 20% Tris-glycine gradient gel. FDPs bound to biotin-labeled Aβ42 were visualized by colloidal blue stain. Incubations that did not contain biotinylated Aβ42 served as a control for nonspecific binding to the streptavidin-sepharose beads. Mass spectrometric analysis on bands pulled down by Aβ42 was performed by The Rockefeller University Proteomics Resource Center as described in supplemental Methods.
Results
Identification of the fibrinogen-binding region within Aβ42
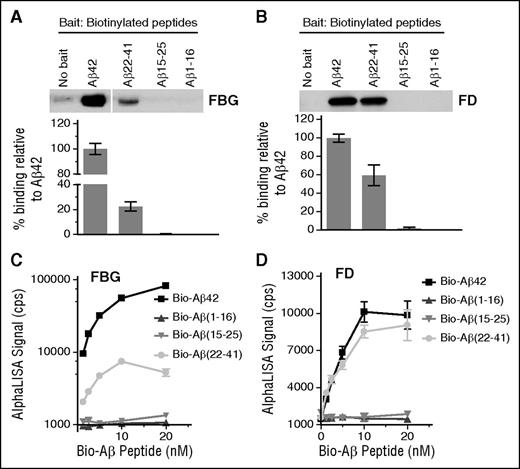
To determine which region of Aβ42 is responsible for fibrinogen binding, 3 biotinylated Aβ fragments (Aβ1-16, Aβ15-25, and Aβ22-41) were analyzed for their ability to bind fibrinogen and fragment D by pulldown assay. Among the 3 fragments, only Aβ22-41 showed binding to both fibrinogen (Figure 1A) and fragment D (Figure 1B). We next analyzed the binding affinity of these fragments to fibrinogen (Figure 1C) and fragment D (Figure 1D) using AlphaLISA. Aβ42 and Aβ22-41 dose-dependently bound to fibrinogen and fragment D (Figure 1C-D), whereas Aβ1-16 or Aβ15-25 showed no binding.
Aβ22-41 binds to fibrinogen and fragment D. (A-B) Biotin-labeled Aβ42, Aβ1-16, Aβ15-25, and Aβ22-41 were incubated with fibrinogen (FBG) or fragment D (FD), and pulldown assays were carried out using streptavidin-coated magnetic beads. All samples were analyzed by western blot in unreduced condition using an anti-fibrinogen antibody. Only Aβ22-41 showed binding to both fibrinogen (A) and fragment D (B). When no Aβ peptides were added, the level of bound fibrinogen or fragment D was negligible. Images and graphs are representative of 4 experiments. (C-D) The binding between biotin-labeled Aβ42 or Aβ fragments with fibrinogen or fragment D was determined by AlphaLISA (n = 3). Controls and other lanes in panel A are from the same gel with some lanes omitted for clarity. Results presented in graphs are mean ± standard error of the mean (SEM).
Aβ22-41 binds to fibrinogen and fragment D. (A-B) Biotin-labeled Aβ42, Aβ1-16, Aβ15-25, and Aβ22-41 were incubated with fibrinogen (FBG) or fragment D (FD), and pulldown assays were carried out using streptavidin-coated magnetic beads. All samples were analyzed by western blot in unreduced condition using an anti-fibrinogen antibody. Only Aβ22-41 showed binding to both fibrinogen (A) and fragment D (B). When no Aβ peptides were added, the level of bound fibrinogen or fragment D was negligible. Images and graphs are representative of 4 experiments. (C-D) The binding between biotin-labeled Aβ42 or Aβ fragments with fibrinogen or fragment D was determined by AlphaLISA (n = 3). Controls and other lanes in panel A are from the same gel with some lanes omitted for clarity. Results presented in graphs are mean ± standard error of the mean (SEM).
Although the binding affinity of Aβ22-41 to fragment D was >50% of Aβ42 (Figure 1B,D), the binding affinity of Aβ22-41 to fibrinogen was only 10% to 20% of Aβ42 (Figure 1A,C), indicating that an additional Aβ42 region may be involved in Aβ42-fibrinogen binding. These results also suggest that there are additional Aβ42-binding sites on fibrinogen outside of fragment D and that the decrease in binding affinity of Aβ22-41 to fibrinogen may be due to a loss of affinity of Aβ22-41 for those binding sites. Overall, both the pulldown and AlphaLISA results indicate that the C-terminal two-thirds of Aβ are involved in binding fibrinogen and fragment D.
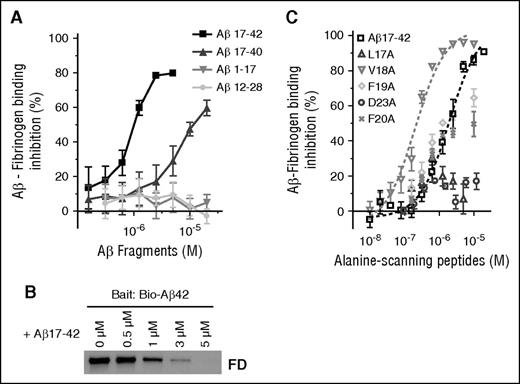
We further narrowed down the fibrinogen-binding region within Aβ42 using 16 nonbiotinylated Aβ peptide fragments spanning the entire length of Aβ42 (supplemental Figure 1). Of the 16 Aβ fragments, only Aβ17-40 and Aβ17-42 inhibited Aβ-fibrinogen binding by AlphaLISA (Figure 2A), with Aβ17-42 having 10-fold higher inhibitory efficacy (IC50 = 1.03 µM) compared with Aβ17-40 (IC50 = 13.4 µM). All other Aβ fragments, including Aβ1-17 and Aβ12-28, had no inhibitory activity (Figure 2A), and a combination of Aβ1-16 and Aβ17-42 did not have a higher inhibitory effect than Aβ17-42 alone (supplemental Figure 2). The inhibitory efficacy of Aβ17-42 was confirmed via pulldown assay, where pulldown of fragment D by biotinylated Aβ42 was dose-dependently decreased in the presence of nonbiotinylated Aβ17-42 (Figure 2B). Our AlphaLISA and pulldown results indicate that Aβ17-40 and Aβ17-42, naturally occurring Aβ fragments known as p3 peptides,18 inhibit the Aβ42-fibrinongen interaction, suggesting that these peptides may play a physiological role in modulating Aβ42-mediated effects on fibrin clots.
Naturally occurring p3 peptides, Aβ17-40 and Aβ17-42, inhibit the Aβ-fibrinogen interaction. (A) Biotinylated Aβ42 was incubated with fibrinogen in the presence of various concentrations (0.05-20 µM) of 16 nonbiotinylated Aβ fragments listed in supplemental Figure 1. The inhibitory efficacy of the Aβ fragments on the Aβ42-fibrinogen interaction was analyzed using AlphaLISA. Of the 16 Aβ fragments tested, only Aβ17-40 (IC50 = 13.4 µM) and Aβ17-42 (IC50 = 1.03 µM) showed inhibitory efficacy (n = 3). (B) Western blot analysis with anti-fibrinogen antibody shows that Aβ17-42 blocks the ability of biotinylated Aβ42 to pull down fragment D (FD) in a dose-dependent manner. (C) Various concentrations (0.01-20 µM) of 5 alanine-scanning Aβ peptides (L17A, V18A, F19A, F20A, and D23A) were incubated with biotinylated Aβ42 and fibrinogen, and their ability to inhibit the Aβ42-fibrinogen interaction was analyzed using AlphaLISA (n = 3-6). Aβ L17A and D23A had almost no inhibitory activity (IC50 > 20 µM), whereas F19A (IC50 = 3.7 µM) and F20A (IC50 = 6.8 µM) showed a compatible inhibitory efficacy to original Aβ17-42. Interestingly, V18A (IC50 = 0.26 µM) had fivefold greater inhibitory efficacy than Aβ17-42. Results presented in graphs are mean ± SEM.
Naturally occurring p3 peptides, Aβ17-40 and Aβ17-42, inhibit the Aβ-fibrinogen interaction. (A) Biotinylated Aβ42 was incubated with fibrinogen in the presence of various concentrations (0.05-20 µM) of 16 nonbiotinylated Aβ fragments listed in supplemental Figure 1. The inhibitory efficacy of the Aβ fragments on the Aβ42-fibrinogen interaction was analyzed using AlphaLISA. Of the 16 Aβ fragments tested, only Aβ17-40 (IC50 = 13.4 µM) and Aβ17-42 (IC50 = 1.03 µM) showed inhibitory efficacy (n = 3). (B) Western blot analysis with anti-fibrinogen antibody shows that Aβ17-42 blocks the ability of biotinylated Aβ42 to pull down fragment D (FD) in a dose-dependent manner. (C) Various concentrations (0.01-20 µM) of 5 alanine-scanning Aβ peptides (L17A, V18A, F19A, F20A, and D23A) were incubated with biotinylated Aβ42 and fibrinogen, and their ability to inhibit the Aβ42-fibrinogen interaction was analyzed using AlphaLISA (n = 3-6). Aβ L17A and D23A had almost no inhibitory activity (IC50 > 20 µM), whereas F19A (IC50 = 3.7 µM) and F20A (IC50 = 6.8 µM) showed a compatible inhibitory efficacy to original Aβ17-42. Interestingly, V18A (IC50 = 0.26 µM) had fivefold greater inhibitory efficacy than Aβ17-42. Results presented in graphs are mean ± SEM.
To analyze which amino acids within Aβ17-42 are important for the Aβ-fibrinogen interaction, we tested the ability of 5 alanine-scanning peptide analogs of Aβ17-42, where L17, V18, F19, F20, or D23 were replaced with alanine (L17A, V18A, F19A, F20A, and D23A), to inhibit Aβ42-fibrinogen binding by AlphaLISA. Analog peptides L17A and D23A exhibited almost no inhibitory activity, whereas F19A and F20A showed comparable inhibitory efficacy to original Aβ17-42. Interestingly, Aβ V18A (IC50 = 0.26 µM) showed fivefold higher inhibitory efficacy than Aβ17-42 (Figure 2C). This alanine-scanning experiment indicates that L17 and D23 are crucial for Aβ-fibrinogen binding.
Fragments of Aβ42 may adopt a different tertiary structure compared with Aβ42, which may affect their ability to bind to fibrinogen. We therefore examined the ability of biotinylated Aβ42 to bind fibrinogen in the presence of antibodies raised against specific regions of Aβ (Figure 3A). Antibody 6F/3D (against Aβ8-17) and 4G8 (against Aβ17-24) blocked pulldown of fibrinogen by biotinylated Aβ42 (Figure 3B), whereas antibodies raised against Aβ1-5, Aβ22-35, and Aβ33-42 had no effect on Aβ42-fibrinogen binding. The epitopes of antibodies 6F/3D and 4G8 were confirmed by enzyme-linked immunosorbent assay, which showed that 6F3D bound Aβ1-16 and 1-17 but not Aβ17-42, whereas 4G8 only bound Aβ17-42 (supplemental Figure 3). Thus, the central region of tertiary structured Aβ42 is critical for Aβ-fibrinogen interaction.
Specific antibodies against the central region of Aβ block the Aβ-fibrinogen interaction. (A) The epitopes for several antibodies against Aβ are illustrated in the schematic and include epitopes 1 to 5 (3D6; Elan), 8 to 17 (6F/3D; Dako), 17 to 24 (4G8; Covance), 22 to 35 (ab62658; Abcam), and 33 to 42 (G2-11; Abcam). (B) Antibodies at concentrations listed in “Methods” were incubated with fibrinogen and biotinylated Aβ42. Pulldown of biotinylated Aβ42 revealed that antibodies 6F/3D and 4G8 are able to interfere with the Aβ-fibrinogen interaction. Results presented in graphs are mean ± SEM, and statistical significance was determined using 1-way analysis of variance (ANOVA) and Bonferroni post hoc test (**P < .01; n = 3).
Specific antibodies against the central region of Aβ block the Aβ-fibrinogen interaction. (A) The epitopes for several antibodies against Aβ are illustrated in the schematic and include epitopes 1 to 5 (3D6; Elan), 8 to 17 (6F/3D; Dako), 17 to 24 (4G8; Covance), 22 to 35 (ab62658; Abcam), and 33 to 42 (G2-11; Abcam). (B) Antibodies at concentrations listed in “Methods” were incubated with fibrinogen and biotinylated Aβ42. Pulldown of biotinylated Aβ42 revealed that antibodies 6F/3D and 4G8 are able to interfere with the Aβ-fibrinogen interaction. Results presented in graphs are mean ± SEM, and statistical significance was determined using 1-way analysis of variance (ANOVA) and Bonferroni post hoc test (**P < .01; n = 3).
SDS-stable Aβ-fibrinogen complex formation
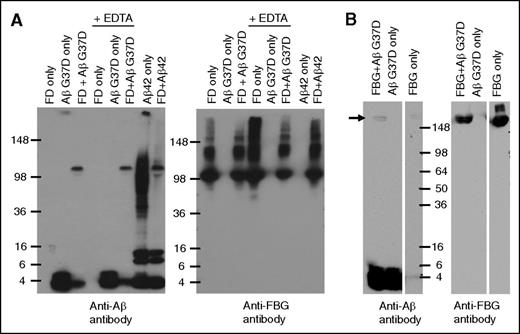
Our analysis of the Aβ-fibrinogen interaction suggested that prolonged incubation of Aβ and fragment D resulted in the formation of an SDS-stable complex. Because Aβ42 aggregates rapidly, we first examined this phenomenon using Aβ42 mutant G37D, which does not readily aggregate.19 Prolonged incubation of fragment D with Aβ G37D followed by western blot analysis with an anti-Aβ antibody showed Aβ42 G37D migrating at ∼4.5 kDa (corresponding to monomeric Aβ) as well as at ∼100 kDa (Figure 4A). The ∼100-kDa band was not detected in the Aβ42 G37D alone lane, indicating that it is a specific product of Aβ42 G37D-fragment D interaction. Anti-fibrinogen antibodies detected fragment D at ∼100 kDa, suggesting that the ∼100-kDa band detected by anti-Aβ antibodies represents Aβ42 G37D that remained in complex with fragment D during SDS-PAGE. SDS-stable complex formation could be mediated by the transglutaminase factor XIII (FXIII), which may contaminate fragment D preparations and lead to the formation of covalent crosslinks between Aβ and fragment D. FXIII activity is dependent on calcium,20 and incubation of fragment D with Aβ G37D in the presence of EDTA, a calcium chelator, still resulted in SDS-stable complexes, indicating that FXIII-mediated crosslinking is not involved in SDS-stable complex formation.
Long-term incubation of fibrinogen/fragment D with Aβ42 forms a SDS-stable complex. (A) Aβ42 G37D or Aβ42 was incubated with fragment D (FD) for 5 days at 37°C in the presence or absence of EDTA. Western blots were analyzed with antibody 6E10 against Aβ (left panel) and an antibody against fibrinogen (Dako; right panel). Aβ-fragment D SDS-stable complex was detected by 6E10. (B) Aβ42 G37D was incubated with fibrinogen (FBG) for 24 hours at 37°C. Western blots were analyzed with antibody 6E10 against Aβ (left panel) and an antibody against fibrinogen (right panel). Aβ-fibrinogen SDS-stable complex was also detected by 6E10 (arrow). Controls and other lanes in panel B are from the same gel with some lanes omitted for clarity.
Long-term incubation of fibrinogen/fragment D with Aβ42 forms a SDS-stable complex. (A) Aβ42 G37D or Aβ42 was incubated with fragment D (FD) for 5 days at 37°C in the presence or absence of EDTA. Western blots were analyzed with antibody 6E10 against Aβ (left panel) and an antibody against fibrinogen (Dako; right panel). Aβ-fragment D SDS-stable complex was detected by 6E10. (B) Aβ42 G37D was incubated with fibrinogen (FBG) for 24 hours at 37°C. Western blots were analyzed with antibody 6E10 against Aβ (left panel) and an antibody against fibrinogen (right panel). Aβ-fibrinogen SDS-stable complex was also detected by 6E10 (arrow). Controls and other lanes in panel B are from the same gel with some lanes omitted for clarity.
SDS-stable complex formation was also observed when Aβ42 G37D was incubated with fibrinogen instead of fragment D (Figure 4B) and when Aβ42 was used instead of Aβ42 G37D (Figure 4A). Interestingly, incubation of nonmutant Aβ42 with fragment D resulted in the formation of fewer Aβ42 oligomers (∼36-100 kDa) compared to Aβ42 incubation alone. Previous results show that the interaction between Aβ42 and fibrinogen or fragment D promotes Aβ42 fibrillization,7 which could account for the conversion of the oligomeric species of Aβ42 seen when incubated alone into fibrils in the presence of fragment D.
Aβ binding to fragment D induces structural change in C-terminal region of β-chain
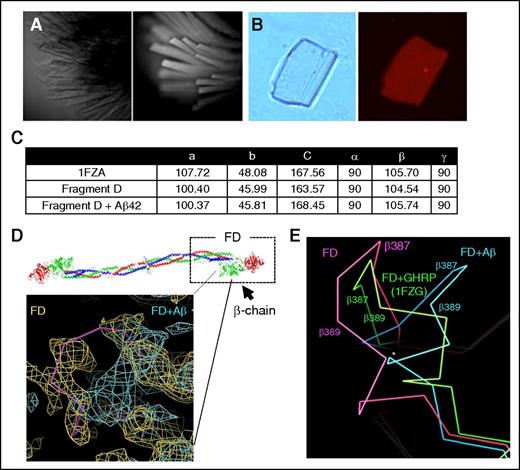
Structural details of the Aβ42-fibrinogen interaction were investigated via X-ray crystallography. We sought to obtain a crystal structure of the Aβ42-fragment D complex because fragment D is the major binding region for Aβ on fibrinogen, is smaller than fibrinogen, easier to manipulate, and has a published, reproducible crystal structure. Because large solvent channels are present in the fragment D structure, our strategy was to soak Aβ42 into preformed fragment D crystals. Fragment D crystals were obtained as described in “Methods” (Figure 5A). To test the ability of Aβ42 to penetrate into the crystal lattice, we soaked fragment D crystals with fluorescently labeled TAMRA-Aβ42, and nonspecifically bound TAMRA-Aβ42 was removed by repeatedly washing the crystals until no decrease in fluorescence was observed. Persistent fluorescence visualized by fluorescence microscopy confirmed binding of Aβ42 within the crystals (Figure 5B). Fragment D crystals were then soaked with unlabeled Aβ42, and data sets from soaked and unsoaked crystals were collected at the National Synchrotron Light Source. The structure was solved by molecular replacement as described in “Methods.” Soaking of Aβ42 did not damage fragment D crystals because the space groups and unit cell dimensions obtained were mostly isomorphous with dimensions found in unsoaked (native) crystals, and agreed relatively well with published dimensions for fragment D (Figure 5C).15
Fragment D structure is altered in the presence of Aβ42. The fragment D crystals soaked with Aβ42 were analyzed by X-ray crystallography. (A) Brightfield (left) and UV fluorescence (right) images of fragment D crystals, indicating crystals are proteinaceous. (B) Left, Brightfield image of a fragment D crystal that had been subjected to soaking in TAMRA-Aβ42 followed by extensive washing. Right, Persistent red fluorescence after washing indicated that TAMRA-Aβ42 was binding within the crystal. (C) Unit cell dimensions of published (1FZA), nonsoaked, and Aβ42-soaked fragment D crystals. (D) Diagram of human fibrinogen with fragment D (FD) marked with a box. The location of altered structure in Aβ42-soaked fragment D crystals is indicated by the solid pink line. Superimposed 2Fo-Fc maps from nonsoaked (yellow; Rwork/Rfree = 0.24/0.33) and Aβ42-soaked (teal; Rwork/Rfree = 0.28/0.39) fragment D crystals with coordinates of nonsoaked crystals. Human fibrinogen schematic was generated from PDB file 3GHG.21 (E) Protein backbone diagram showing the shift of the β384-393 loop from nonsoaked fragment D (pink) to b-hole peptide (GHRP)-bound (1FZG; green) fragment D and Aβ42-soaked fragment D (blue). GHRP, Gly-His-Arg-Pro-amide peptide.
Fragment D structure is altered in the presence of Aβ42. The fragment D crystals soaked with Aβ42 were analyzed by X-ray crystallography. (A) Brightfield (left) and UV fluorescence (right) images of fragment D crystals, indicating crystals are proteinaceous. (B) Left, Brightfield image of a fragment D crystal that had been subjected to soaking in TAMRA-Aβ42 followed by extensive washing. Right, Persistent red fluorescence after washing indicated that TAMRA-Aβ42 was binding within the crystal. (C) Unit cell dimensions of published (1FZA), nonsoaked, and Aβ42-soaked fragment D crystals. (D) Diagram of human fibrinogen with fragment D (FD) marked with a box. The location of altered structure in Aβ42-soaked fragment D crystals is indicated by the solid pink line. Superimposed 2Fo-Fc maps from nonsoaked (yellow; Rwork/Rfree = 0.24/0.33) and Aβ42-soaked (teal; Rwork/Rfree = 0.28/0.39) fragment D crystals with coordinates of nonsoaked crystals. Human fibrinogen schematic was generated from PDB file 3GHG.21 (E) Protein backbone diagram showing the shift of the β384-393 loop from nonsoaked fragment D (pink) to b-hole peptide (GHRP)-bound (1FZG; green) fragment D and Aβ42-soaked fragment D (blue). GHRP, Gly-His-Arg-Pro-amide peptide.
Analysis of electron density revealed that there were structural changes in Aβ42-soaked fragment D compared with data obtained from an unsoaked crystal. Specifically, a loop in the β-chain (β384-393) of fragment D, which is a part of the Aβ42-binding region identified previously,7 was shifted away from the coiled coil region in Aβ42-soaked fragment D crystals (Figure 5D). Fragment D structures crystallized in the presence of b-hole binding peptides, GHRP, (as in PDB 1FZG,22 among others; Figure 5E) also showed that this loop is shifted in a similar but less dramatic manner compared with the Aβ42-fragment D complex. Peptides binding the b-hole also induced a flip in a nearby loop (β395-400), but this flip was not observed in our structure, suggesting that Aβ may not fit all the way into the b-hole as the peptides do. Both Aβ42-soaked and unsoaked fragment D crystals were grown in identical conditions, and binding of Aβ42 within Aβ42-soaked crystals was confirmed using TAMRA-labeled Aβ42, suggesting that Aβ42 binding to fragment D induces the structural shift in the loop encompassing β384-393.
We also observed patches of density unaccounted for by the fragment D coordinates possibly corresponding to Aβ42 in Aβ42-soaked but not unsoaked crystals. These patches of density were found in the region of fragment D in spatial proximity to the Aβ-binding site on the β-chain,7 but the density was not sufficient to conclusively define Aβ42 placement (not shown). Based on the size of the water channels present in fragment D crystals and the size of the pocket near the β-chain region shown to bind Aβ42 (β384-393),7 both Aβ42 monomers and dimers may enter fragment D crystals during soaking. This nonhomogeneous Aβ population may have resulted in the indiscernible Aβ42 electron density map that we observed.
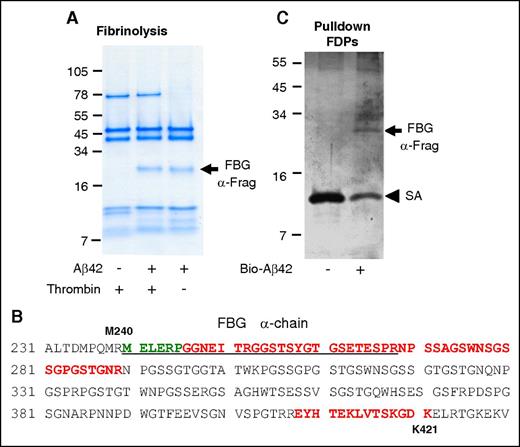
Binding of Aβ to the fibrinogen α-chain blocks its cleavage by plasmin
We previously found that Aβ42 binding to fibrin(ogen) interferes with plasmin(ogen) binding to fibrin and delays fibrin clot lysis.1,8 If Aβ42 binding to fibrin(ogen) interferes with plasmin(ogen)’s access to fibrin, then regions of fibrin(ogen) bound to Aβ42 would be expected to be protected from degradation. To investigate this idea, fibrin was formed in the presence and absence of Aβ42 and subjected to plasmin-mediated degradation. Analysis of the fibrin degradation products revealed a plasmin-resistant fibrin fragment (PRFF) migrating at ∼20 kDa (arrow in Figure 6A) that was observed only when fibrin was formed in the presence of Aβ42. Mass spectrometry analysis and N-terminal sequencing of the PRFF identified it as a fibrin α-chain fragment that includes residues Aα239-421 (Figure 6B). Fibrin formed in the presence of Aβ42 is thinner, arranged in a denser network,8 and interrupted by abnormal aggregates.1 To control for the potentially confounding effects of this abnormal fibrin network on plasmin-mediated degradation, we performed the same experiment without thrombin. Fibrinogen incubated with Aβ42 and subjected to plasmin-mediated digestion also yielded the same degradation-resistant fragment (Figure 6A), indicating that PRFF formation is independent of changes to fibrin network structure induced by Aβ42.
Aβ interacts with the α-chain of fibrinogen, producing a plasmin-resistant fibrin fragment during fibrinolysis. (A) Fibrin was digested with plasmin in the presence or absence of Aβ42. A plasmin-resistant fragment was observed only in the presence of Aβ42 (arrow). The same experiment was done without thrombin. In the absence of thrombin, plasmin degradation-resistant PRFF was also observed in the presence of Aβ42. Images are representative of ≥3 experiments. (B) Mass spectrometry analysis of the fragments in panels A and C showed they were derived from the α-chain of fibrinogen. Green residues were identified by N-terminal sequencing of band in panel A, red residues were identified by mass spectrometry analysis of band in panel A, and underlined residues were identified by mass spectrometry analysis of band in panel C. (C) Fibrinogen was partially digested with plasmin, incubated with biotinylated Aβ42, and Aβ42 was pulled down with streptavidin (SA)-coated beads. A fibrinogen fragment that bound to Aβ was observed (top arrow).
Aβ interacts with the α-chain of fibrinogen, producing a plasmin-resistant fibrin fragment during fibrinolysis. (A) Fibrin was digested with plasmin in the presence or absence of Aβ42. A plasmin-resistant fragment was observed only in the presence of Aβ42 (arrow). The same experiment was done without thrombin. In the absence of thrombin, plasmin degradation-resistant PRFF was also observed in the presence of Aβ42. Images are representative of ≥3 experiments. (B) Mass spectrometry analysis of the fragments in panels A and C showed they were derived from the α-chain of fibrinogen. Green residues were identified by N-terminal sequencing of band in panel A, red residues were identified by mass spectrometry analysis of band in panel A, and underlined residues were identified by mass spectrometry analysis of band in panel C. (C) Fibrinogen was partially digested with plasmin, incubated with biotinylated Aβ42, and Aβ42 was pulled down with streptavidin (SA)-coated beads. A fibrinogen fragment that bound to Aβ was observed (top arrow).
To confirm that blockage of a plasmin cleavage site on the fibrin(ogen) Aα chain by Aβ42 is involved in increased PRFF formation in the presence of Aβ42, we investigated whether Aβ42 can bind to this region of fibrinogen. Fibrinogen was partially digested with plasmin to generate large fibrinogen degradation products. Pulldown of the fibrinogen degradation products with biotinylated Aβ42 identified a fragment similar in size to the PRFF (arrow in Figure 6C). Mass spectrometry analysis demonstrated that this fragment maps to the same region of the α-chain as identified for the PRFF (Figure 6B). Together, these results suggest that Aβ42 binds to the α-chain of fibrin(ogen) and blocks its cleavage by plasmin.
Discussion
We examined the fibrinogen-binding region within Aβ42 using 4 distinct biochemical approaches. The results from all 4 experimental approaches (summarized in supplemental Figure 4) are closely aligned, but there is a slight difference between the results obtained using antibodies blocking specific regions of Aβ42 (Figure 3) and the approaches using Aβ fragments. Competitive inhibition of the Aβ-fibrinogen interaction using unlabeled Aβ subpeptides (Figure 2) showed that Aβ17-42 (IC50, 1.03 µM) had 10-fold higher inhibitory efficacy than Aβ17-40 (IC50, 13.4 µM). This supports our previous results showing that Aβ42 has a higher binding affinity to fibrinogen than Aβ40,2 and suggests that the C terminus of Aβ42 is important for its binding to fibrinogen. However, antibodies against the C terminus of Aβ42 (G2-11; Figure 3) failed to block Aβ42-fibrinogen binding. One possible explanation for this difference is that although the C-terminus of Aβ42 may not be directly involved in the interaction between Aβ42 and fibrinogen, the 2 C-terminal residues of Aβ42 (Ile41 and Ala42) increase the stability of Aβ’s tertiary structure and promote its oligomerization,23,24 which may enhance Aβ-fibrinogen binding due to the fact that Aβ oligomers have stronger binding affinity to fibrinogen than monomers.2 It is therefore possible that once the tertiary structure and/or oligomeric state of Aβ42 are stabilized by its C-terminal residues, blocking the C terminus using antibodies does not inhibit the Aβ42-fibrinogen interaction.
Overall, our results suggest that fibrinogen interacts with the central region of tertiary structured Aβ42, which is stabilized by its C-terminal residues. This binding model is similar to the interaction between ApoE3 and Aβ42, where binding is mediated by the central region of Aβ42 and further enhanced by its C-terminal residues.25 By defining the binding regions on Aβ and fibrinogen involved in their interaction, our results provide a basis for structure-based rational design of small molecule or antibody inhibitors targeting this interaction. Our results also suggest that targeting the tertiary stability of Aβ42 via its 2 C-terminal residues may be an alternative strategy for inhibiting Aβ42-fibrinogen binding.
We found that naturally occurring p3 peptides (Aβ17-40 and Aβ17-42) inhibit the Aβ42-fibrinogen interaction. The role of p3 peptides in AD pathogenesis is controversial. Some studies indicate that Aβ17-42 exhibits enhanced aggregation relative to full-length Aβ4226 and that p3 peptides are prevalent in diffuse deposits and in a subset of dystrophic neurites in AD patients.27 However, other studies suggest that p3 peptides might be a benign form of amyloid because they lack domains associated with microglial activation,28 do not form oligomers, the most toxic species of Aβ,18 and do not have negative effects on neuronal synaptic function.29 Our present study demonstrates that p3 peptides compete with Aβ42 for binding to fibrinogen, suggesting that they may attenuate AD pathology. Therefore, the inhibition of the Aβ42-fibrinogen interaction by p3 peptides may be another beneficial consequence of enhancing α-secretase activity as a therapeutic strategy for AD.30
Prolonged incubation of Aβ42 with fibrinogen resulted in SDS-stable complex formation. Previously, Aβ42 has been shown to form SDS-stable complexes with ApoE2 and 3.25 Because SDS-stable complex formation between Aβ and its binding partners may impair protein function, studies aimed at identifying the nature of the SDS-stable complexes involving Aβ may shed light on new mechanisms by which Aβ may contribute to pathological processes.
We have previously shown that Aβ42 binding to fibrin(ogen) delays fibrinolysis by interfering with the binding of plasminogen and plasmin to fibrin8 and that Aβ42 binds to the β-chain of fibrinogen fragment D.7 We also confirmed Aβ42 binding to fibrin D-dimer (supplemental Figure 5). The current study identifies an additional binding site for Aβ42 on the fibrin(ogen) αC region, which is known to bind plasminogen and include several plasmin cleavage sites. Aβ42 binding to this αC region results in the formation of a PRFF, likely via Aβ42-mediated interference with plasmin(ogen) binding to fibrin and with plasmin-mediated cleavage of fibrin at this site. However, it is possible that Aβ42 binding to this region may inhibit other molecules that bind the αC region of fibrin, such as tPA and α2-antiplasmin.31-33 Investigating whether Aβ binding to the fibrin(ogen) αC region affects tPA and α2-antiplasmin binding to fibrin and whether there are functional consequences of these inhibitions would clarify the mechanism of PRFF formation. Furthermore, studies measuring the levels of PRFFs in the blood and brain parenchyma of individuals at various stages of disease would shed light on whether the PRFF could be useful as a biomarker for AD.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank The Rockefeller University High-Throughput and Spectroscopy Resource Center for access to experimental equipment and technical support. The authors thank N. Trikannad for experimental assistance, and M. Foley, E. Norris, and Z. Chen for helpful discussion.
This work was supported by the Alzheimer’s Drug Discovery Foundation and Robertson Therapeutic Development Fund (H.J.A.); National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke grant NS50537; the Tri-Institutional Therapeutics Discovery Institute; the Alzheimer’s Drug Discovery, Thome Memorial Medical, Litwin, Rudin Family, Blanchette Hooker Rockefeller, and Mellam Family Foundations; and John A. Herrmann (S.S.). The Rockefeller University Structural Biology Resource Center was supported by grants 1S10RR022321-01 and 1S10RR027037-01 from the NIH National Center for Research Resources.
Authorship
Contribution: D.Z. and H.J.A. designed the study, performed experiments, analyzed data, and wrote the manuscript; H.E.B.-R., P.K.S. and D.S.S. performed experiments and analyzed data; D.A.O. designed the structural study and assisted in experiments and data analysis; S.S. designed the study and participated in data analysis and manuscript preparation; M.K. and K.A. designed the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hyung Jin Ahn, Laboratory of Neurobiology and Genetics, The Rockefeller University, 1230 York Ave, New York, NY 10065; e-mail: hahn@rockefeller.edu.