Abstract
Thiol isomerases are multifunctional enzymes that influence protein structure via their oxidoreductase, isomerase, and chaperone activities. These enzymes localize at high concentrations in the endoplasmic reticulum of all eukaryotic cells where they serve an essential function in folding nascent proteins. However, thiol isomerases can escape endoplasmic retention and be secreted and localized on plasma membranes. Several thiol isomerases including protein disulfide isomerase, ERp57, and ERp5 are secreted by and localize to the membranes of platelets and endothelial cells. These vascular thiol isomerases are released following vessel injury and participate in thrombus formation. Although most of the activities of vascular thiol isomerases that contribute to thrombus formation are yet to be defined at the molecular level, allosteric disulfide bonds that are modified by thiol isomerases have been described in substrates such as αIIbβ3, αvβ3, GPIbα, tissue factor, and thrombospondin. Vascular thiol isomerases also act as redox sensors. They respond to the local redox environment and influence S-nitrosylation of surface proteins on platelets and endothelial cells. Despite our rudimentary understanding of the mechanisms by which thiol isomerases control vascular function, the clinical utility of targeting them in thrombotic disorders is already being explored in clinical trials.
Introduction to thiol isomerases
In the early 1960s, Anfinsen et al described the ability of the inactive, denatured form of ribonuclease to refold into its native form and express full enzymatic activity.1 An enzyme from liver, called disulfide interchange enzyme, reshufflace, or rearrangease,2 was identified as facilitating the refolding of ribonuclease by forming the disulfide bonds to obtain the lowest energy structure.3 This enzyme came to be known as protein disulfide isomerase (PDI; EC.5.3.4.1) and was established as a critical component of the biosynthetic pathway for proteins.4 PDI, present in the endoplasmic reticulum, is the prototype thiol isomerase and has been extensively studied.5
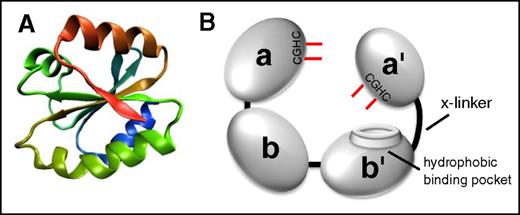
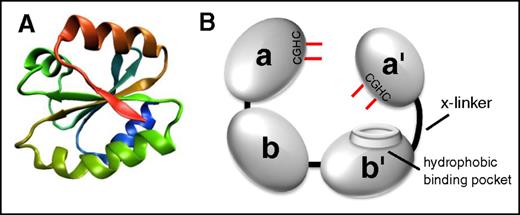
All thiol isomerases, of which now there are 21 in the PDI family, have evolved from an ancestral thioredoxin found in primitive unicellular organisms. PDI is the archetypical and most abundant thiol isomerase (Figure 1). These proteins contain a thioredoxin-like domain, which consists of a 4-stranded antiparallel β sheet surrounded by 3 α helices (Figure 1A).6,7 PDI family thiol isomerases have an active site motif Cys-X-X-Cys (CXXC) where X is any amino acid. Duplication and evolution of the thioredoxin-like fold8 resulted in multidomain thiol isomerases of the PDI family that contain both catalytic domains (termed a, a′, or a0 by convention), which contain the CXXC motif, and substrate-binding domains (termed b or b′ by convention). Catalytic domains contain the CXXC motif and perform oxidoreductase functions (Figure 2). Although the a and a′ domains are homologous, they are not functionally redundant. For example, only the a′ domain of PDI is critical for thrombus formation.9 The substrate binding domains have lost the catalytic motifs and developed features that facilitate protein–protein interactions such as the hydrophobic pocket on PDI (Figure 1B).10
The architecture of thiol isomerases. Thiol isomerases consist of tandem domains containing a thioredoxin-fold. (A) The structure of the thioredoxin-fold.7 (B) PDI is formed from 4 tandem domains containing thioredoxin-like folds that assume a U-shape. The 2 catalytic domains (a and a′) contain the CGHC motifs that contain the catalytic cysteines (shown in red). The catalytic cysteines face one another and are responsible for oxidoreductive activities. The substrate binding domains (b and b′) form the bottom of the U-shape. The b′ domain contains a hydrophobic binding pocket that is primarily responsible for substrate binding. The b′ and a′ domains are connected by a flexible 19-amino-acid peptide termed the x-linker.
The architecture of thiol isomerases. Thiol isomerases consist of tandem domains containing a thioredoxin-fold. (A) The structure of the thioredoxin-fold.7 (B) PDI is formed from 4 tandem domains containing thioredoxin-like folds that assume a U-shape. The 2 catalytic domains (a and a′) contain the CGHC motifs that contain the catalytic cysteines (shown in red). The catalytic cysteines face one another and are responsible for oxidoreductive activities. The substrate binding domains (b and b′) form the bottom of the U-shape. The b′ domain contains a hydrophobic binding pocket that is primarily responsible for substrate binding. The b′ and a′ domains are connected by a flexible 19-amino-acid peptide termed the x-linker.
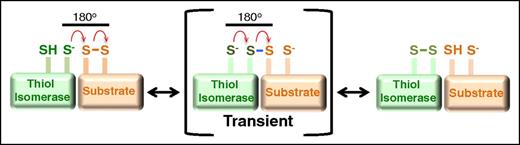
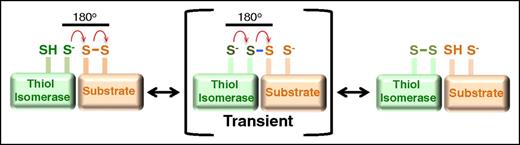
Mechanism of oxidoreductive cleavage by thiol isomerases. The reduced thiol isomerase aligns with a substrate that contains a disulfide bond such that the free thiolate forms an 180° angle with the vector of the disulfide. A second-order nucleophilic substitution SN2-type reaction then occurs in which the active site sulfur ion nucleophile of the thiol isomerase attacks the adjacent sulfur atoms of the disulfide bond.74 This nucleophilic substitution results in a transient mixed disulfide (blue). The mixed disulfide spontaneously decomposes, resulting in the formation of a disulfide bond in the thiol isomerase and reduction of the disulfide bond in the substrate.
Mechanism of oxidoreductive cleavage by thiol isomerases. The reduced thiol isomerase aligns with a substrate that contains a disulfide bond such that the free thiolate forms an 180° angle with the vector of the disulfide. A second-order nucleophilic substitution SN2-type reaction then occurs in which the active site sulfur ion nucleophile of the thiol isomerase attacks the adjacent sulfur atoms of the disulfide bond.74 This nucleophilic substitution results in a transient mixed disulfide (blue). The mixed disulfide spontaneously decomposes, resulting in the formation of a disulfide bond in the thiol isomerase and reduction of the disulfide bond in the substrate.
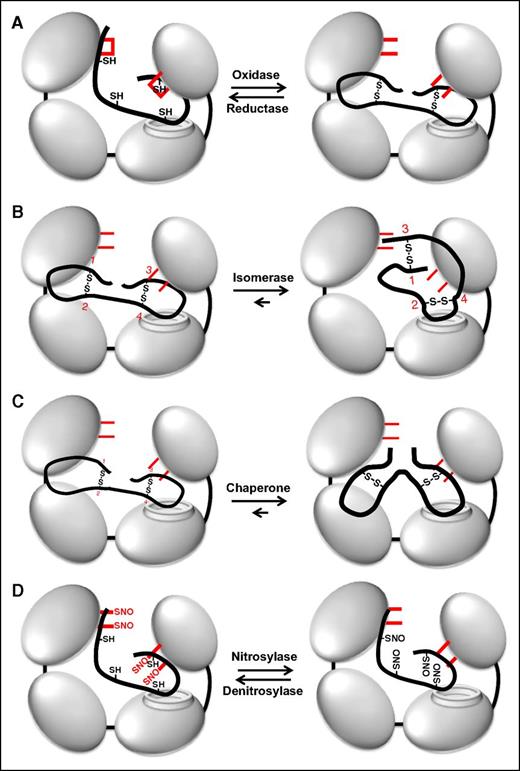
Depending on the context, thiol isomerases function as reductases, oxidases, or isomerases (Figure 3). Thiol isomerases can act as oxidases when engaging a thiol-containing substrate with a lower redox potential, forming disulfide bonds within the substrate (Figure 3A, reaction from left to right). Acting as a reductase, thiol isomerases cleave disulfide bonds within substrate proteins (Figure 3A, reaction from right to left). They can also act as an isomerase, changing protein structure by modified disulfide bond formation without a net change in free thiol content (Figure 3B). By facilitating protein folding in the absence of catalytic activity, they can act as chaperones (Figure 3C).11,12 Modifications of the catalytic cysteines enable thiol isomerases to act as nitrosylases/denitrosylases, transferring nitric oxide (NO) to or from substrates, respectively (Figure 3D).13-15 Variations in vascular NO levels may drive these S-nitrosylation reactions. The catalytic cysteines of thiol isomerases can also react with glutathione and other redox active plasma species.16,17 Thus, thiol isomerases are mutlidomain, multifunctional proteins.
Multiple functions of PDI. (A) PDI is an oxidoreductase that can both form (oxidase) and cleave (reductase) disulfide bonds. Active site cysteines are shown in red. Note that the catalytic cysteines are either reduced or oxidized during these processes. (B) Through its oxidoreductase activity, PDI acts as an isomerase changing disulfide bond patterns within a protein. In this example, PDI modifies disulfides formed between cysteines 1-2 and 3-4 into disulfides formed between 1-3 and 2-4 (numbering in red). Note that there is no net change in the redox state of the catalytic cysteines on PDI. (C) PDI can act as a chaperone, modifying the conformation of a protein independent of its oxidoreductase and chaperone activities. The redox state of the catalytic cysteines is unchanged. (D) The active site cysteines within PDI can also become S-nitrosylated. PDI can then transfer NO to substrates, thus acting as a nitrosylase. PDI can also act as a denitrosylase, removing NO from substrates. S-nitrosylation typically inactivates substrates, whereas denitrosylation activates substrates.
Multiple functions of PDI. (A) PDI is an oxidoreductase that can both form (oxidase) and cleave (reductase) disulfide bonds. Active site cysteines are shown in red. Note that the catalytic cysteines are either reduced or oxidized during these processes. (B) Through its oxidoreductase activity, PDI acts as an isomerase changing disulfide bond patterns within a protein. In this example, PDI modifies disulfides formed between cysteines 1-2 and 3-4 into disulfides formed between 1-3 and 2-4 (numbering in red). Note that there is no net change in the redox state of the catalytic cysteines on PDI. (C) PDI can act as a chaperone, modifying the conformation of a protein independent of its oxidoreductase and chaperone activities. The redox state of the catalytic cysteines is unchanged. (D) The active site cysteines within PDI can also become S-nitrosylated. PDI can then transfer NO to substrates, thus acting as a nitrosylase. PDI can also act as a denitrosylase, removing NO from substrates. S-nitrosylation typically inactivates substrates, whereas denitrosylation activates substrates.
Thiol isomerases as redox sensors
Effect of redox environment on thiol isomerases
The activity of cell surface thiol isomerases is controlled by the redox environment. In a reducing environment, the catalytic cysteines within the active site motif are reduced and thiol isomerases act as reductases. Under oxidizing conditions, the catalytic cysteines will form a disulfide bond and thiol isomerases act as oxidases. The main redox buffers in the vasculature are the glutathione system (GSH/GSSG) and the cysteine system (Cys/CySS) and these determine the redox potential of plasma.18 Plasma redox potentials (Eh) are −130.9 ± 22.9 mV for GSH/GSSG and −72.4 ± 12.8 mV for Cys/CySS.19 The active site cysteines in PDI have a redox potential of −190 ± 10 mV.20 Based on these considerations, one would expect vascular PDI to be primarily oxidized in the extracellular environment. However, redox conditions can vary locally. In addition, protein disulfide oxidases such as ERO1α, which undergoes activation-dependent cell surface expression in platelets,21 and protein disulfide reductases can enzymatically modify catalytic cysteines within active site motifs.22 Little is known about the function of enzymatic control of redox states of extracellular vascular thiol isomerases.
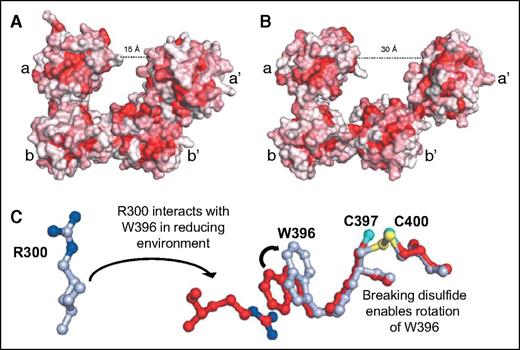
Thiol isomerases are sensitive to changes in redox potential, undergoing substantial conformational changes in response to modifications the redox environment. In reducing conditions, PDI assumes a compact structure (Figure 4A-B).23 The conformational change is mediated by reduction of the catalytic cysteines in the CGHC motif, which triggers rotation of Trp-396 on the a′ domain, enabling it to interact with Arg-300 on the b′ domain (Figure 4C). This switch initiates a series of interactions at the b′–a′ interface that positions the a′ domain over the hydrophobic binding site on the b′ domain.23,24 This constrained conformation is thought to be a means to limit substrate binding under reducing conditions.24 Under oxidizing conditions, the constraint of disulfide bond formation between Cys-397 and Cys-400 prevents Trp-396 from interacting with Arg-300 and the hydrophobic binding site on b′ remains exposed and facilitating interactions with substrates. PDI is thus a more effective chaperone under oxidizing conditions,24 but can also engage substrates under reducing conditions. Recent studies show that on binding a substrate, PDI assumes a more compact conformation.25 Substrate binding at the hydrophobic pocket activates an allosteric switch involving the x-linker, which results in enhanced reductase activity in the a and a′ domains and appears to be a cooperative mechanism whereby substrate engagement activates enzymatic activity.
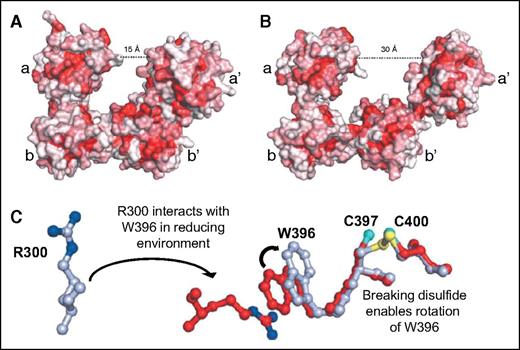
Redox sensitivity of PDI. (A) In a reducing environment, PDI assumes a compact structure characterized by a 15-Å distance between the a and a′ domains. (B) In an oxidizing environment, the distance between the a and a′ domain increases to 30 Å. The increase in distance results from loss of interactions between the b′ and a′ domains that allows movement of the x-linker and a′ domains. Adapted from Wang et al.23 (C) In a reducing environment, the disulfide bond between C397 and C400 is broken, enabling the rotation of W396. This rotation positions W396 so that it can interact with R300. The interaction between W396 and R300 brings the a′ domain in closer proximity to the b′ domain, facilitating additional interactions that lead to a more compact structure. Adapted from Wang et al.23
Redox sensitivity of PDI. (A) In a reducing environment, PDI assumes a compact structure characterized by a 15-Å distance between the a and a′ domains. (B) In an oxidizing environment, the distance between the a and a′ domain increases to 30 Å. The increase in distance results from loss of interactions between the b′ and a′ domains that allows movement of the x-linker and a′ domains. Adapted from Wang et al.23 (C) In a reducing environment, the disulfide bond between C397 and C400 is broken, enabling the rotation of W396. This rotation positions W396 so that it can interact with R300. The interaction between W396 and R300 brings the a′ domain in closer proximity to the b′ domain, facilitating additional interactions that lead to a more compact structure. Adapted from Wang et al.23
Generating surface free thiols
Activation of platelets results in a greater than fourfold increase in surface protein free thiols.26 Although the percentage of active sites that are dithiols is only 26% in resting platelets, the number increases to 81% in activated platelets.26 PDI is prominent among the sources of activation-dependent free thiols. Free thiols within catalytic domains have several potential functions in addition to the oxidoreductive functions discussed above. They could react with reactive oxygen species, thereby buffering platelets from oxidant damage. Free thiols within the catalytic domains of PDI could also act as denitrosylases, activating S-nitrosylated proteins, although this function has not been well characterized.
In endothelium, reaction of free thiols with oxidants and free radicals is thought to protect endothelium from oxidant damage. Several thiol isomerases including PDI, ERp46, and ERp5 are oxidized on incubation of endothelial cells with the neutrophil-derived oxidant HOCl.27 Similarly, thiol isomerases, particularly thioredoxin, are capable of regenerating protein inactivated by oxidative stress.28,29 Thiol isomerases on endothelium expose a free thiol (Cys326) on the surface of β2GPI and β2GPI reduced by endothelial thiol isomerases protects endothelium from cell death induced by oxidative stress.30
S-nitrosylation of thiol isomerases
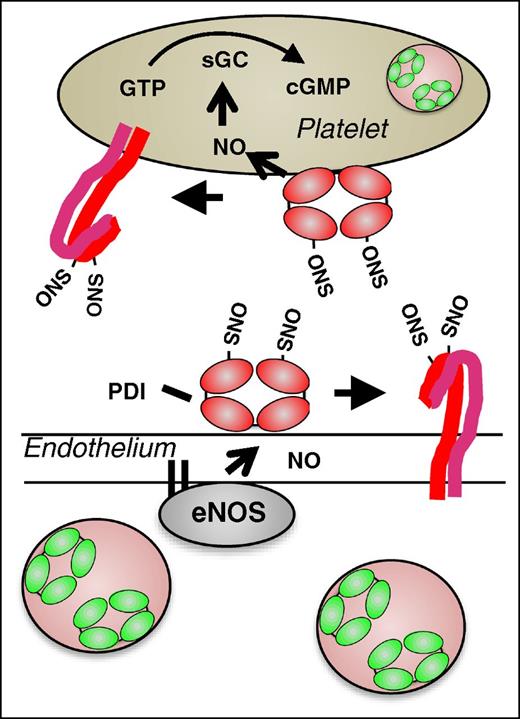
Generation of NO by endothelial cells is an essential mechanism for maintaining vascular quiescence. NO produced by endothelium acts on platelets to maintain them in a resting state. Exposure to NO results in a decreased number of free thiols on megakaryocyte and platelet surfaces31 and S-nitrosylation of multiple platelet receptors including αIIbβ3 is observed following exposure to NO.32 PDI undergoes S-nitrosylation following exposure to NO donors.33 In contrast to S-nitrosylation at regulatory cysteines within platelet receptors such as αIIbβ3,34 S-nitrosylation of catalytic cysteines within PDI enables transfer of NO to other proteins and into cells. Several lines of evidence demonstrate this transnitrosylase activity of PDI. Delivery of NO from an S-NO donor into cytosol is inhibited following knockdown of PDI in HEL cells.13 Inhibition of PDI blocks the delivery of NO from the physiologic NO donor, S-nitrosoglutathione, into both megakaryocytes31 and into platelets.35 PDI-mediated transnitrosylation is proposed to occur via the delivery of NO into cell membranes in the form of N2O3.14 Intracellular substrates are S-nitrosylated as they interact at the membrane–cytosol interface.15 NO can be transferred from PDI to the heme moiety in guanylyl cyclase, as evidenced by its activation and marked increased in guanosine 3′,5′-cyclic monophosphate (cGMP) levels.13,35 PDI could also transfer NO to surface proteins14 including platelet receptors, maintaining them in an inactivated conformation. Thiol isomerase–mediated S-nitrosylation of surface proteins could therefore function in maintaining vascular quiescence (Figure 5). However, the significance of this mechanism in vivo has not yet been tested.
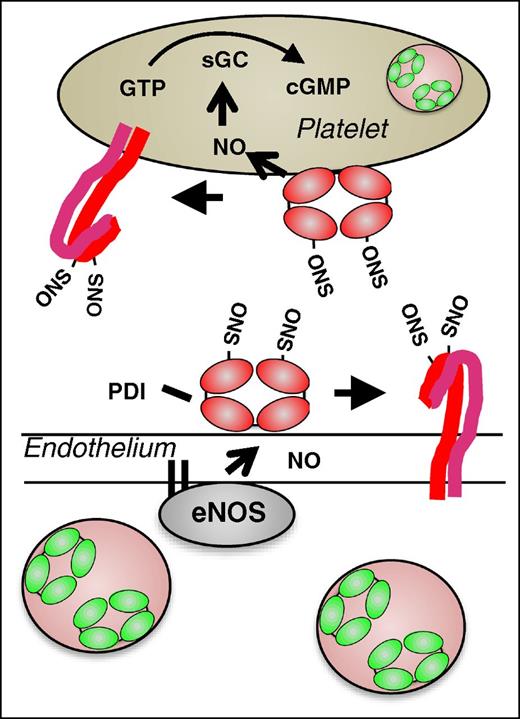
Proposed model for the role of PDI in regulating vascular NO activity. In the quiescent vasculature, when NO levels are high, PDI may be S-nitrosylated. Endothelial SNO-PDI could transfer NO to surface proteins, thereby inhibiting their activity, or transfer NO intracellularly. Intracellular NO could bind to guanylyl cyclase, stimulating the generation of cGMP and contributing the platelet quiescence.
Proposed model for the role of PDI in regulating vascular NO activity. In the quiescent vasculature, when NO levels are high, PDI may be S-nitrosylated. Endothelial SNO-PDI could transfer NO to surface proteins, thereby inhibiting their activity, or transfer NO intracellularly. Intracellular NO could bind to guanylyl cyclase, stimulating the generation of cGMP and contributing the platelet quiescence.
Vascular thiol isomerases
A subset of thiol isomerases is found in the vasculature (Table 1)36,37 and some of these, including PDI, ERp57, and ERp5, have known functional importance on the surfaces of platelets and endothelial cells (Figure 6). The first report of a thiol isomerase being released from platelets was in the early 1990s, when Chen et al showed that PDI is released by activated platelets.38 The Essex laboratory subsequently showed that PDI localizes to the platelet membrane39,40 and functions in platelet activation.41 Several other groups subsequently demonstrated that thiol isomerases traffic to secretory granules/membrane systems and are released from platelets in an activation-dependent manner.36,42-45 Kim et al showed that platelets lacking PDI have reduced activation-dependent aggregation.46 Platelet surface PDI has been postulated to modulate αIIbβ3 function,47-49 collagen binding to α2β1,50 GP1bα,26 thrombospondin 1,51-53 and vitronectin.54 ERp555 and ERp5756,57 also localize to the platelet surface and their inhibition blocks platelet activation. The precise mechanisms by which thiol isomerases function in the activation of αIIbβ3 or other platelet receptors, however, are not well understood.
Domain structures of vascular thiol isomerases. (A) The domain structures of thiol isomerases with known activity in thrombus formation (PDI, ERp57, and ERp5) are shown. Catalytic domains (tan), binding domains (blue), and c domains (green) are indicated. The location of the CGHC motif is indicated in red. Numbers indicate the amino acid position in the mature protein.
Domain structures of vascular thiol isomerases. (A) The domain structures of thiol isomerases with known activity in thrombus formation (PDI, ERp57, and ERp5) are shown. Catalytic domains (tan), binding domains (blue), and c domains (green) are indicated. The location of the CGHC motif is indicated in red. Numbers indicate the amino acid position in the mature protein.
Thiol isomerases have also been found on the surface of the endothelium. Cell-impermeable probes capable of reacting with closely spaced thiols on cell surfaces have been used to identify PDI on the surface of endothelial cells.15,58 Subsequent studies showed that PDI forms a complex with αVβ3 on the endothelial cell membranes, providing a mechanism by which it is retained on these membranes.59 This endothelial cell surface PDI is capable of activating both β1 and β3 integrins.59,60 Cell surface PDI has also been shown to regulate phosphatidylserine exposure,61 as well as nitric oxide delivery on endothelial cells.15,62 Multiple endothelial cell agonists including thrombin, histamine, and phorbol 12-myristate 13-acetate have been shown to stimulate PDI release from endothelial cells, and PDI is released from endothelium in vivo following laser injury.63 However, it was not until the function of thiol isomerases was evaluated in the context of thrombus formation that the in vivo importance was appreciated.
Thiol isomerases in thrombus formation
The discovery that thiol isomerases participate in thrombus formation was accidental and facilitated by the intersection of the development and application of high-speed multicolor digital intravital microscopy to the study of thrombus development in a live mouse,64 the hypothesis of Hogg et al that the mechanism of tissue factor deencryption involved oxidation of 2 free sulphydryls and formation of a disulfide bond,65 and the suggestion that PDI might be the enzyme that catalyzes the formation of that disulfide bond.66 Although subsequent efforts to examine the oxidation of tissue factor following tissue injury in in vivo systems neither proved nor disproved the Hogg hypothesis, several studies showed that platelets and endothelium secrete PDI on tissue injury in a live animal.63,67 That PDI, important for disulfide bond formation during protein synthesis, is released from vascular cells and plays an important extracellular role was in itself a surprising finding. However, the observation that inhibition of PDI with blocking antibodies demonstrated inhibition of thrombus formation provided direct evidence that PDI is required for thrombus formation after tissue injury.67 The importance of PDI in thrombus formation has now been demonstrated in several models of thrombus formation including a laser injury model,67 a ligation model,68 and a ferric chloride model (Table 2).69,70
Since the discovery of the importance of PDI in thrombus formation, similar observations have been extended to other thiol isomerases, including ERp57 and ERp5 (Table 2).45,56,57 ERp57, with a domain organization parallel to PDI, is also secreted by platelets and endothelial cells on cell activation. Three groups have related ERp57 to thrombus formation using (1) blocking antibodies to ERp5756 ; (2) genetic alterations of megakaryocyte/platelet PDIA3,71 the gene that encodes ERp57; and (3) in vivo morpholinos in mice to decrease expression of ERp57.70
ERp5 is yet another thiol isomerase whose inhibition blocks thrombus formation. Using polyclonal monospecific, immunoaffinity purified antibodies to ERp5 antibodies in vivo in the laser injury model in mice, Passam et al showed that inhibition of ERp5 blocked thrombus formation including both platelet aggregation and fibrin generation.45 ERp5 is derived from both platelets and endothelial cells on cell activation with vascular injury. ERp5 binds to αIIbβ3 but does not bind through the active site domain.
In summary, a series of thiol isomerase has been identified in the vessel wall and in platelets that, on cell activation, stimulates activation of extracellular thiol isomerase activity72 and accompanies thrombus formation in vivo.45,67 Each of these thiol isomerases is either absolutely required or partially required for thrombus formation. In addition to these in vivo murine models, a role for thiol isomerases in platelet activation and thrombus formation has also been shown in vitro using human samples.72 Several surprises can be noted. First, these thiol isomerases reside in the endoplasmic reticulum and, in the case of PDI, also the T-granule43 and yet have extracellular function; second, they have important function with regard to thrombus formation. An initiation pathway requiring thiol isomerases for thrombus formation has never been previously described.
The master switch hypothesis
As bits and pieces of this new system were discovered, it was difficult to synthesize these observations to understand their biological function. What is the pathway in which inhibition of PDI or ERp57 or ERp5 individually block both platelet activation and fibrin generation in vivo? Are these thiol isomerases acting in series in the pathway? That is, is there an electron transport chain in which thiol isomerases act in a linear fashion as enzyme and substrate in a redox-active manner? Or do they act in parallel, with each of the vascular thiol isomerases acting on a unique set of substrates? Are the actions of thiol isomerases on substrates only about changes in the covalent structure of disulfide bonds or are the other functions of thiol isomerases involved?
With only fragments of information available about this new pathway(s), we considered that the intravascular release of these thiol isomerases primed the triggers for initiating thrombus formation. We posit that (1) vascular thiol isomerases are intracellular under normal conditions; (2) thiol isomerases are released on tissue damage or cellular activation; (3) only on release of thiol isomerases can thrombus formation be initiated; and (4) extracelluar thiol isomerases catalyze changes in vascular surface receptors and plasma protein structure. In essence, this master switch hypothesis postulates that encoded into many vascular proteins are functional disulfide bonds that lock the proteins in an inactive state. Such allosteric disulfides have previously been described in vascular proteins and proposed to function in thrombus formation.73,74 These allosteric disulfides could form an embedded network of clamps that prevents the untoward initiation of thrombus formation. Thiol isomerases, released from activated or injured cells, are required to deactivate this embedded network of safeguards so as to enable blood coagulation and platelet activation. An essential component of this network is that its activation is required for coagulation and platelet activation, but it is not itself capable of eliciting either.
The specific substrates of thiol isomerases and identification of cysteines involved in clamping vascular proteins in an inactive state are largely undefined. Yet some examples are illustrative. PDI binds to platelet αIIbβ348 and platelets deficient in PDI demonstrate impaired αIIbβ3 activation.46 Although the disulfides on αIIbβ3 that are targeted by PDI have not been identified, regulatory disulfides in the epidermal growth factor domains have been implicated.47,75 PDI interacts with αVβ3 on endothelial cells59 and αMβ2 on neutrophils.76 ERp545,55 and ERp5756,71 also bind to and modify αIIbβ3 activity. Thrombospondin 1 is another thiol isomerase substrate. PDI has previously been found to form disulfide linked complexes with thrombospondin 1 from platelets51 and expose a cryptic RGD sequence that facilitates the association of thrombospondin 1 with endothelial cell αVβ3.53 PDI has been proposed to activate tissue factor by forming and allosteric bond between Cys186 and Cys209.65,77 Although intriguing, this activation mechanism remains controversial.61,78 Overall, the importance of these thiol isomerase–substrate interactions in thrombus formation remains to be proven in an in vivo model. However, the hypothesis that distinct thiol isomerases act on an overlapping but distinct set of substrates to prime the vasculature for thrombus formation following vessel injury or activation is consistent with current observations.
Clinical implications of thiol isomerases in hemostasis and thrombosis
The modification of functional disulfide bonds by thiol isomerases represents a previously unrecognized layer of control of vascular function in general and specifically thrombosis. Modification of allosteric disulfide bonds by thiol isomerases is akin to cleavage of amino acid bonds by serine proteases of the coagulation cascade. Yet fundamental distinctions exist between these 2 mechanisms that have important clinical implications. Unlike proteases of the coagulation pathway, active thiol isomerases do not directly cause blood to coagulate. Thus, thiol isomerases could support hemostasis in coagulopathies without the thrombotic risks associated with infusion of coagulation proteins (eg, FEIBA, FVIIa). Conversely, because thiol isomerases control thrombus formation, but are not the final effectors of clotting, inhibition of thiol isomerases could achieve control of thrombosis without concurrent bleeding risks. In fact, genetic deletion of megakaryocyte (MK)/platelet PDI impairs thrombosis without prolonging tail bleeding times.46 The extremely high abundance of intracellular thiol isomerases (200 μM in endoplasmic reticulum)79 renders likely the possibility that diseases characterized by increased cell proliferation and/or cell death will show elevated thiol isomerase levels. Chronic exposure to elevated thiol isomerase levels may predispose to thrombosis. These distinguishing characteristics of thiol isomerases compared with serine proteases have important implications with regard to the clinical utility of thiol isomerase-targeted therapeutics and diagnostics.
Inhibition of thiol isomerases
Thiol isomerases function in coagulation primarily to modify substrates on endothelial cells, platelets, and in plasma to create conditions conducive for coagulation. Inhibition of thiol isomerases interferes with this priming mechanism and blocks thrombus formation in animal models. As described above, antibodies directed against PDI,67 ERp57,56 or ERp580 inhibit thrombus formation. Small molecules targeting ERp57 such as ADTM81 or compounds targeting PDI such as the flavonoid quercetins (eg, quercetin-3-rutinoside and isoquecetin) also inhibit thrombus formation.69 When tested in mice, quercetin-3-rutinoside inhibited thrombus formation at doses as low as 0.5 mg/kg. Infusion of recombinant PDI reversed the inhibitory effect of quercetin-3-rutinoside in mice, indicating that PDI was the relevant target of quercetin-3-rutinoside in this thrombosis model. Orally administered quercetin-3-rutinoside was also antithrombotic.69 Importantly, quercetin-3-rutinoside blocked injury-induced thrombus formation at concentrations commonly used by individuals who consume quercetin-3-rutinoside supplements. Epidemiologic studies indicate that consumption of flavonoids is associated with reduced risk of cardiovascular death and decreased incidence of nonfatal and fatal stroke.82,83
Isoquercetin, a flavonoid quercetin that is more readily absorbed than quercetin-3-rutinoside, is currently being evaluated as an antithrombotic in the Cancer Associated Thrombosis and Isoquercetin (CAT-IQ) trial. The CAT-IQ study is a 600-patient, randomized, placebo-controlled, phase 2/3 clinical trial (#NCT02195232) designed to determine the efficacy and safety of isoquercetin in preventing cancer-related thrombosis.84 The efficacy outcome measurements include thrombosis at 60 days and d-dimer levels. Bleeding will be monitored to assess safety. As with any antithrombotic, it will be valuable to identify assays capable of monitoring the anticoagulant effect of PDI-targeted therapies. Ancillary studies will assess the PDI inhibitory activity in plasma to determine whether adequate plasma concentrations of isoquercetin or its metabolites are reached. Although inhibition of PDI does not affect standard coagulation studies, it does have some effect on platelet aggregation41,85 and platelet-dependent thrombin generation.86 Successful completion of the CAT-IQ trial will provide proof-of-principle for inhibition of PDI in primary prophylaxis of thrombosis and allow us to develop methods to monitor PDI-targeted therapies.
Thiol isomerases in coagulopathy
If thiol isomerases do not initiate thrombosis themselves, but rather prime the vasculature for clot formation, then it is possible that they will be effective in promoting hemostasis in the setting of coagulopathy. Similarly, it is possible that lack of thiol isomerase release contributes to coagulopathy. The latter premise is supported by studies of Hermansky-Pudlak syndrome (HPS). HPS is characterized by occulocutaneous albinism and a deficiency in platelet dense granules.87 Mice with HPS have prolonged bleeding times and impaired thrombus formation, including impaired fibrin formation, on vascular injury. Platelets and endothelial cells from HPS mice are deficient in the secretion of PDI.44 Furthermore, infusion of recombinant PDI reverses the thrombus formation defect in HPS mice (S. H. Kim and B. Furie,unpublished observations, 2013). Whether recombinant PDI shortens bleeding times in mice has not yet been determined. Nor is it known whether infusion of recombinant thiol isomerases can improve bleeding in other coagulopathies.
Selecting candidate diseases for thiol isomerase-targeted therapy
Our immature understanding of the mechanisms by which thiol isomerases control thrombosis complicates identification of optimal clinical settings for thiol isomerase-targeted therapy. If our underlying premise is that thiol isomerases work on multiple substrates to create a prothrombotic state, then thiol isomerase-targeted therapies could be antithrombotic in almost any hypercoagulable condition. Such therapies could be most effective in disease states in which baseline thiol isomerase levels are elevated. Currently, there are very little data on circulating levels of vascular thiol isomerases other than thioredoxin. Normal ranges for any of the vascular thiol isomerases shown to function in thrombosis (PDI, ERp5, ERp57) have not been established. Autoantibodies directed against these enzymes, however, indicates that these thiol isomerases circulate at increased levels in certain disease states. Antibodies to PDI have been identified in cancer and SLE,88 as well as in infection.89 ERp57 autoantibodies have also been found in cancer,90,91 and infection.92 Thiol isomerases that bind to cell surfaces could be detected by the immune system and generate autoantibodies even if circulating levels are undetectable.
The possibility that thiol isomerase inhibition does not substantially enhance bleeding risk suggests additional indications. Currently, primary prophylaxis is not offered for certain thrombotic risk factors because the risk of bleeding from the antithrombotic outweighs the protective benefit (eg, heterozygous factor V Leiden with single thrombosis, antiphospholipid antibody without thrombosis). Thiol isomerase-targeted therapies could provide protection from thrombosis for these conditions. Because thiol isomerase inhibition acts by a mechanism other than direct inhibition of coagulation factor synthesis or activity, it is possible that the activity of the two approaches will be synergistic. The addition of thiol isomerase inhibition to existing anticoagulation may be useful in the case of refractory thrombotic syndromes. However, the finding that inhibition or deletion of thiol isomerases such as PDI is not associated with bleeding is not universally observed and requires additional testing in more reliable hemostasis models. In general, more basic understanding and clinical experience with thiol isomerase inhibition will be required to establish the optimal use of agents targeting these enzymes.
Summary
Vascular thiol isomerases serve an essential role in thrombus formation and there is growing evidence that they have additional functions in regulating the vascular redox environment. Nonetheless, investigators trying to unravel the mechanisms of how vascular thiol isomerases function in thrombus formation face several challenges. While investigators have demonstrated the role of PDI, ERp5, and ERp57 in thrombus formation in several thrombosis models, the full repertoire of thiol isomerases that participate in thrombosis remains to be identified and how extracellular thiol isomerases interact with one another in the context of thrombosis is currently unknown. Understanding how and under what conditions thiol isomerases are released is central to understanding their function and to developing the possibility of using plasma thiol isomerase levels diagnostically. Identifying substrates of thiol isomerases will be critical to understanding activation pathways that rely on disulfide bond rearrangements (Figure 7). How thiol isomerases are regulated within the vasculature remains to be determined. As these facets of vascular thiol isomerase biology are elucidated, investigators need to form a compelling general model of how thiol isomerases prime the vasculature for thrombus formation. Even as mechanistic studies are underway, clinical studies evaluating how to best use thiol isomerase-targeted therapies are taking place and will help pave the way for new approaches to disorders of hemostasis and thrombosis.
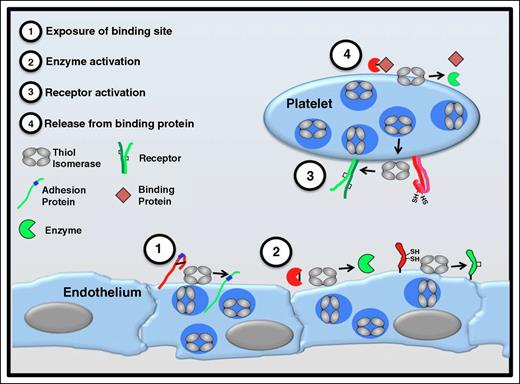
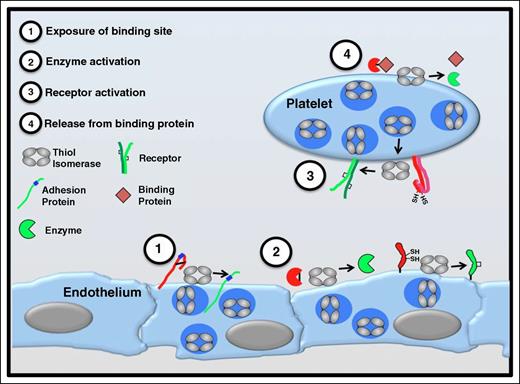
Functional consequences of thiol isomerase-mediated disulfide bond modifications. Thiol isomerases are secreted from platelets and endothelium following cell activation and mediate modifications of functional disulfide bonds in multiple substrates. Several categories of modifications are observed. (1) Exposure of binding sites in adhesion proteins. Cleavage of disulfide bonds can reveal cryptic binding sites such as RGD sequences (shown in blue) enabling a nonbinding adhesion protein (red) to adhere to binding partners on the cell surface (green). Thrombospondin is an adhesion protein that is modified in this manner.53 (2) Activation of enzyme function. Functional disulfide bond cleavage could also expose an encrypted active site within an enzyme or modify its conformation, converting the enzyme to a more active conformation. Conversely, formation of a disulfide bond could activate an enzyme or coenzyme (eg, tissue factor).65,66,93 (3) Receptor activation. Disulfide bond shuffling could contribute to the formation or stabilization of the active conformation of cell surface receptors (eg, αIIbβ3).47,75 (4) Release from binding protein. Cleavage of a disulfide bond could release a protein from its binding partner, thereby activating the protein (eg, TGF-β).94
Functional consequences of thiol isomerase-mediated disulfide bond modifications. Thiol isomerases are secreted from platelets and endothelium following cell activation and mediate modifications of functional disulfide bonds in multiple substrates. Several categories of modifications are observed. (1) Exposure of binding sites in adhesion proteins. Cleavage of disulfide bonds can reveal cryptic binding sites such as RGD sequences (shown in blue) enabling a nonbinding adhesion protein (red) to adhere to binding partners on the cell surface (green). Thrombospondin is an adhesion protein that is modified in this manner.53 (2) Activation of enzyme function. Functional disulfide bond cleavage could also expose an encrypted active site within an enzyme or modify its conformation, converting the enzyme to a more active conformation. Conversely, formation of a disulfide bond could activate an enzyme or coenzyme (eg, tissue factor).65,66,93 (3) Receptor activation. Disulfide bond shuffling could contribute to the formation or stabilization of the active conformation of cell surface receptors (eg, αIIbβ3).47,75 (4) Release from binding protein. Cleavage of a disulfide bond could release a protein from its binding partner, thereby activating the protein (eg, TGF-β).94
Acknowledgments
This work was supported by National Institutes of Health, National Heart, Lung, and Blood Institute grants U54 HL112302, HL125275, HL112809, and T32 HL007917, and National Institute on Drug Abuse grant DA032476.
Authorship
Contribution: R.F. and B.F. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert Flaumenhaft, Division of Hemostasis and Thrombosis, Department of Medicine, Beth Israel Deaconess Medical Center, 330 Brookline Ave, Boston, MA 02215; e-mail: rflaumen@bidmc.harvard.edu.