Key Points
A recurring mutation in NDUFB11 causes congenital sideroblastic anemia.
The NDUFB11 p.93del mutation impairs erythroid proliferation, but not differentiation.
Abstract
The congenital sideroblastic anemias (CSAs) are a heterogeneous group of inherited blood disorders characterized by pathological mitochondrial iron deposition in erythroid precursors. Each known cause has been attributed to a mutation in a protein associated with heme biosynthesis, iron-sulfur cluster biogenesis, mitochondrial translation, or a component of the mitochondrial respiratory chain. Here, we describe a recurring mutation, c.276_278del, p.F93del, in NDUFB11, a mitochondrial respiratory complex I–associated protein encoded on the X chromosome, in 5 males with a variably syndromic, normocytic CSA. The p.F93del mutation results in respiratory insufficiency and loss of complex I stability and activity in patient-derived fibroblasts. Targeted introduction of this allele into K562 erythroleukemia cells results in a proliferation defect with minimal effect on erythroid differentiation potential, suggesting the mechanism of anemia in this disorder.
Introduction
The sideroblastic anemias (SAs) are a group of inherited and acquired bone marrow disorders characterized by pathological iron accumulation in the mitochondria of erythroid precursors.1-3 Abnormal, iron-laden mitochondria appear to encircle erythroblast nuclei, giving rise to the defining morphological feature of SAs, the ringed (or ring) sideroblast. All genetically defined congenital SAs (CSAs) can be attributed to mutations in 1 of 3 mitochondrial pathways: heme synthesis, iron-sulfur cluster biogenesis and protein synthesis, or, rarely, a mutation in a protein involved in mitochondrial respiration itself.2,4
Other than X-linked SA and ataxia, which is due to an iron-sulfur biogenesis defect, each of the genetically defined syndromic CSA phenotypes (thiamine-responsive megaloblastic anemia; Pearson marrow-pancreas syndrome; mitochondrial myopathy with lactic acidosis and ringed sideroblasts; and SA, immunodeficiency, fevers, and developmental delay5,6 ) is associated with a generalized defect in mitochondrial protein translation or synthesis of a specific mitochondrial protein.
Here, by whole-exome sequencing (WES), we identify a recurring mutation in NDUFB11, a nuclear-encoded mitochondrial complex I protein that is essential for mitochondrial oxidative phosphorylation. The mutation was identified in 5 individuals from 4 families with variably syndromic, normocytic CSA. In vitro experiments in an erythroid cell line demonstrate a proliferation, but not a differentiation, defect that can account for the anemic phenotype.
Study design
Patient samples were obtained with informed consent. WES was performed in 2 families using the SureSelect Target Enrichment kit (Agilent) on a HiSeq 2500 sequencer (Illumina) at a mean depth of ∼50X. Analysis was performed using a custom-built, rule-based algorithm that integrates single-nucleotide polymorphism genotypes, linkage, and next-generation sequencing analyses: “Variant Explorer” (K.S.-A. and K.M., unpublished computer program for analyzing genetic data). For gene validation experiments, zebrafish embryo morphants were prepared and analyzed as previously described with o-dianisidine staining and flow cytometry.7 Basal respiration, adenosine triphosphate production, and respiratory capacity of primary patient fibroblasts were assayed on a Seahorse XF96 Extracellular Flux Analyzer using the Mitochondrial Stress Test kit. Intact mitochondria were isolated from patient fibroblasts and K562 cell lines, and respiratory complex (RC) activity and assembly were determined using standard assays. For gene targeting, we transfected clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)/green fluorescent protein plasmid PX458 containing the NDUFB11-targeting guide sequence in combination with the single-stranded homologous recombination oligo donor sequence (supplemental Table 1, available on the Blood Web site) using an Amaxa nucleofector. Wells containing single clones were identified visually 2 weeks after transfection and correctly targeted clones were identified by HinfI digestion of a polymerase chain reaction product including the patient-specific knock-in variant (supplemental Table 1) and then confirmed by Sanger sequencing. Following differentiation, cells were stained with o-dianisidine to assess ability to hemoglobinize. A detailed description of the methods is contained in supplemental Methods.
Results and discussion
WES was performed on a total of 67 unexplained CSA probands and 191 of their first-degree relatives. In 2 males (400-III-2 and 05-II-1) (Figure 1A; Table 1; supplemental Figure 1), there was an in-frame deletion of 3 nucleotides predicting loss of phenylalanine 93 (c.276_278del, p.F93del) in NDUFB11, an X-linked gene encoding a mitochondrial RC-I–associated protein with no specifically known function (supplemental Figure 2). This variant was not present in the Exome Aggregation Consortium (April 2016) Browser database (http://exac.broadinstitute.org). NDUFB11 is a 153-aa protein that is predicted to include a single transmembrane helical domain (residues 81-109).8 Residue F93 is predicted to be in the middle of this domain, contributing to its hydrophobicity. Deletion of F93 would lead to the shortening of the transmembrane domain and also cause a rotational rearrangement of all succeeding residues in the helix. Such an alteration could affect its localization on the mitochondrial membrane and/or the interaction of NDUFB11 with other mitochondrial membrane proteins.
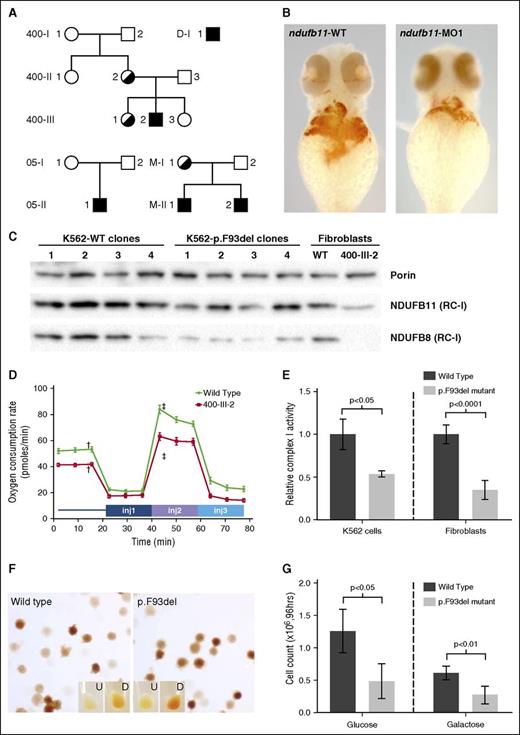
NDUFB11 mutations in CSA. (A) NDUFB11 CSA pedigrees. Black shading indicates the genotype. The genotype of M-I-1 is inferred on the basis of her 2 sons. (B) Zebrafish embryos stained for hemoglobin with o-dianisidine at 48 hours postfertilization (hpf) following microinjection with water control or antisense morpholino reagents targeting the fish ortholog of human NDUFB11. (C) Total mitochondrial protein was analyzed for the RC-I proteins NDUFB11 (middle) and NDUFB8 (bottom) in K562 cells and patient fibroblasts by western blot. Equivalent loading of mitochondrial lysates was confirmed by immunoblot analysis for porin (VDAC1) (top). (D) Mitochondrial function was measured using oxygen consumption rate (ocr) using the Seahorse Mitostress test. Patient and control fibroblasts were treated with 2 µM oligomycin injection 1 (inj1), 0.5 µM carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP) (inj2), and 1 µM rotenone/antimycin A (inj3). Error bars are ± standard error of the mean (SEM). Basal OCR was measured before adding treatments (†) and maximum OCR was measured after the addition of 0.5 µM FCCP (‡). The difference in basal and maximum OCR is plotted in supplemental Figure 6. (E) RC-I function was analyzed in K562 cell lines (n = 4, clones as in panel C) and control and patient fibroblasts (n = 6, technical replicates) and normalized to citrate synthase activity. Data are expressed relative to the wild-type (WT) value, whose mean was defined as 1.0. Ratios are ± standard deviation (SD). (F) Representative WT and p.F93del K562 cells (clones K562-WT3 and K562-p.F93del1) stained with o-dianisidine show normal hemoglobinization. Undifferentiated (U) and differentiated (D) K562 cells cultured in the presence of 6.0 mM sodium butyrate for differentiation. (G) Growth of undifferentiated K562 cells (n = 4 clones) in glucose or galactose after 96 hours. Cells were initially plated at 4 × 104 cells per mL and analyzed in triplicate.
NDUFB11 mutations in CSA. (A) NDUFB11 CSA pedigrees. Black shading indicates the genotype. The genotype of M-I-1 is inferred on the basis of her 2 sons. (B) Zebrafish embryos stained for hemoglobin with o-dianisidine at 48 hours postfertilization (hpf) following microinjection with water control or antisense morpholino reagents targeting the fish ortholog of human NDUFB11. (C) Total mitochondrial protein was analyzed for the RC-I proteins NDUFB11 (middle) and NDUFB8 (bottom) in K562 cells and patient fibroblasts by western blot. Equivalent loading of mitochondrial lysates was confirmed by immunoblot analysis for porin (VDAC1) (top). (D) Mitochondrial function was measured using oxygen consumption rate (ocr) using the Seahorse Mitostress test. Patient and control fibroblasts were treated with 2 µM oligomycin injection 1 (inj1), 0.5 µM carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP) (inj2), and 1 µM rotenone/antimycin A (inj3). Error bars are ± standard error of the mean (SEM). Basal OCR was measured before adding treatments (†) and maximum OCR was measured after the addition of 0.5 µM FCCP (‡). The difference in basal and maximum OCR is plotted in supplemental Figure 6. (E) RC-I function was analyzed in K562 cell lines (n = 4, clones as in panel C) and control and patient fibroblasts (n = 6, technical replicates) and normalized to citrate synthase activity. Data are expressed relative to the wild-type (WT) value, whose mean was defined as 1.0. Ratios are ± standard deviation (SD). (F) Representative WT and p.F93del K562 cells (clones K562-WT3 and K562-p.F93del1) stained with o-dianisidine show normal hemoglobinization. Undifferentiated (U) and differentiated (D) K562 cells cultured in the presence of 6.0 mM sodium butyrate for differentiation. (G) Growth of undifferentiated K562 cells (n = 4 clones) in glucose or galactose after 96 hours. Cells were initially plated at 4 × 104 cells per mL and analyzed in triplicate.
Sequencing amplified exons of other members of the index families showed that the mutation occurred de novo in proband 05-II-1 and in the mother (400-II-2) of proband 400-III-2; the variant was also present in 1 nonanemic sister (400-III-3) of 400-III-2. X-inactivation studies in peripheral blood leukocytes demonstrated skewing toward the unaffected X chromosome in 2 heterozygous females (400-II-2 and 400-III-3; Table 1). The de novo nature of the variant and X-inactivation pattern in carrier females also supports an X-linked–recessive phenotype and the likelihood that the NDUFB11 variant is causative.
To further assess the causality of the NDUFB11 variant, we performed Sanger sequencing of the complete coding sequence of the gene in 24 additional unrelated patients with CSA who were negative for mutations in all other known CSA disease-associated genes. Two additional male probands and 1 clinically affected male sibling were identified with the identical c.276_278del (p.F93del) variant (Table 1). The recurrent nature of the allele is likely a consequence of polymerase slipping in a repeat sequence (TTCTTCTTT) encoding a series of 3 phenylalanines (p.F91_F93). No other rare NDUFB11 coding variants were found in any other CSA proband, suggesting that this is an allele-specific phenotype.
Loss of Ndufb11 in Chinese hamster fibroblasts results in mild mitochondrial respiratory defects,9 whereas germ line heterozygous null alleles in female humans result in 2 distinct phenotypes: histiocytoid cardiomyopathy10 or microphthalmia with linear skin defects,11 depending upon the pattern of somatic X inactivation. An NDUFB11 null allele is embryonically lethal in males.11 Thus, it is likely that the pF93del allele is a partial loss-of-function variant. Furthermore, despite the genotypic homogeneity, there was a great deal of phenotypic heterogeneity in affected individuals. All were ascertained by having CSA, and several had coexisting myopathy, lactic acidosis, or short stature; other neurodevelopmental or organ dysgenesis phenotypes were present in individual families (Table 1).
To determine a potential role for NDUFB11 in erythropoiesis, we examined the phenotype of zebrafish ndufb11 morphant embryos. The splice-blocking morpholino (MO1) resulted in normal ndufb11 RNA processing, but an overall decrease in steady-state messenger RNA (supplemental Figure 3A). We found that this partial knockdown of ndufb11 resulted in a decrease in the number of erythroid cells by flow cytometry using a Tg(globinLCR:eGFP) line (supplemental Figure 3B), and diminished hemoglobin by o-dianisidine staining (Figure 1B). This finding supports the hypothesis that NDUFB11 could have a phylogenetically conserved role in erythropoiesis.
In order to assess the functional consequences of the NDUFB11p.F93del allele, we examined protein levels and mitochondrial phenotypes in patient-derived fibroblasts. Western blotting of mitochondrial extracts showed that the amount of NDUFB11 protein was decreased in patient (400-III-2) cells (Figure 1C). Furthermore, commensurate with the previously reported association of NDUFB11 with RC-I, and typical of many respiratory protein mutations, including YARS2 CSA,12 we found that there was a deficiency of fully assembled RC-I (Figure 1C; supplemental Figure 4), but not other RCs (supplemental Figure 5). Using the Seahorse XF96 Extracellular Flux Analyzer, we measured basal respiration, adenosine triphosphate production, and respiratory capacity and found that the basal and maximum oxygen consumption rates were decreased by approximately one-quarter in patient fibroblasts (Figure 1D; supplemental Figure 6). Each of these findings was corroborated by a specific loss of RC-I, but not RC-IV, activity measured in vitro (Figure 1E; supplemental Figure 7). This respiratory insufficiency phenotype is entirely consistent with the lactic acidosis, myopathy, and other phenotypes observed in some of the patients.
To further validate the functional significance of the NDUFB11p.F93del allele, we targeted the variant into K562 erythroleukemia cells using CRISPR/Cas9 gene-editing technology. Four biallelically targeted clones and 4 nontargeted clones that had been subject to the same transfection and selection procedures were studied (supplemental Figure 2). We did not observe ring sideroblasts in differentiated cells stained with Prussian blue or by electron microscopy (not shown), but these abnormalities have never been observed in vitro or in vivo in nucleated erythroid cells in any CSA model, save Alas2-deficient mice bearing a human ALAS2 transgene.13 However, we found that NDUFB11p.F93del recombinants uniformly had a proliferation defect (Figure 1G), but that they did not exhibit abnormalities in hemoglobinization when induced to differentiate with sodium butyrate (Figure 1F). This observation is consistent with the normochromic anemia characterized by reduced cell numbers in affected patients.
In total, the weight of evidence indicates that NDUFB11p.F93del causes a novel form of X-linked SA, almost certainly due to an incomplete loss of function of RC-I activity. In addition, in the course of preparing these studies for publication, a single patient with the same mutation in NDUFB11 was described with SA and lactic acidosis, further supporting our conclusions.14 Like other mitochondrial translation or RC deficiencies associated with SA, the phenotype is syndromically associated with mitochondrial respiratory phenotypes such as lactic acidosis and myopathy, which will distinguish this disorder from certain other CSAs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and families as well as their referring physicians, without whom this work would not be possible. Sarah Ducamp is acknowledged for providing a critical review of the manuscript. The authors thank the laboratory of Feng Zheng for their generous donation of CRISPR/Cas9 plasmid PX458, the laboratory of Rosalyn Adams at Boston Children’s Hospital for providing the Amaxa nucleofector 2b, Dan Bauer and colleagues for advice on CRISPR/Cas9 gene editing, the Flow Cytometry Core at Boston Children’s Hospital for their cell-sorting services, and Rebecca King for the images of the bone marrow sample.
This work was supported by National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases DK087992 (M.D.F.) and R01 DK070838 and National Heart, Lung, and Blood Institute P01 HL03226 2 (B.H.P.).
Authorship
Contribution: D.A.L. designed and performed CRISPR/Cas9 experiments and A.W.C. designed and performed mitoblot and mitochondrial functional assays, supervised by A.K.S. and P.J.S.; D.R.C. performed and coordinated sequence analysis; C.M.S. performed Seahorse experiments and was supervised by A.K.; K.S.-A. designed and implemented the informatics pipeline and was supervised by K.M.; M.D.K. performed phenotypic analyses in zebrafish and was supervised by B.H.P.; C.M.N., J.P., M.M.P., A.M., M.M.H., and S.S.B. ascertained and phenotyped patients; and M.D.F. coordinated and oversaw all genomic and functional analyses.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark D. Fleming, Department of Pathology, Boston Children’s Hospital, Bader 124.1, BCH 3027, 300 Longwood Ave, Boston, MA 02115; e-mail: mark.fleming@childrens.harvard.edu.