To the editor:
Primary cutaneous CD30+ lymphoproliferative disorders (CD30+ LPDs) are a disease spectrum including primary cutaneous anaplastic large cell lymphoma (C-ALCL) and lymphomatoid papulosis (LyP).1 C-ALCL presents as solitary, grouped, or, rarely, multifocal nodules and tumors that often ulcerate. Cutaneous relapses are common, but extracutaneous dissemination occurs in only 10% to 15% of patients and mainly involves regional lymph nodes.2-4 LyP is characterized by a chronic course of recurrent, self-healing papulonecrotic or nodular skin lesions. Extracutaneous disease rarely develops.3 The prognosis of both conditions is usually excellent with a 5-year disease-specific survival of approximately 90% for C-ALCL and almost 100% for LyP.2-4 However, about 10% of patients with primary cutaneous CD30+ LPD run a more aggressive clinical course. Apart from presentation with extensive skin lesions on legs or arms, risk factors for this unusually aggressive clinical course are currently unknown.2,3,5
Primary systemic ALCL is divided into ALK+ ALCL and ALK− ALCL, based on the presence or absence, respectively, of a rearrangement in the ALK gene that can reliably be detected by ALK immunohistochemistry.6 Systemic ALK+ ALCL has a better overall survival than systemic ALK− ALCL, with 5-year survival rates of approximately 80% for ALK+ ALCL and approximately 50% for ALK− ALCL.7,8
In systemic ALK− ALCL, recent studies detected 2 additional genes (DUSP22 and TP63) with recurrent chromosomal rearrangements that were shown to be mutually exclusive and absent in ALK+ ALCL. DUSP22 is rearranged in approximately 30% of the ALK− ALCL patients and is associated with a more favorable prognosis with a 5-year survival rate of 90%.8,9 Chromosomal rearrangements involving the TP63 gene, found in 8% of patients with ALK− ALCL, showed a poorer overall survival than ALK− ALCL without a DUSP22 or TP63 rearrangement with 5-year survival rates of 17% and 42%, respectively.8-10
In C-ALCL, ALK rearrangements are generally absent.11 However, unusual cases of ALK+ C-ALCL have been reported, many with an excellent prognosis.12-15 Rearrangements of DUSP22 have been detected in approximately 30% of C-ALCL patients and in a small subset of LyP.11,16,17 Clinical behavior and prognosis were similar to that of patients without DUSP22 rearrangements. TP63 rearrangements have only been detected in 2 of 41 patients with C-ALCL (5%), and both patients had an unusually aggressive disease course.10,18 TP63 rearrangements also have been detected in a single patient with transformed mycosis fungoides (MF) with an aggressive clinical course, but were not detected in 32 patients with LyP.18
The association of TP63 rearrangements with poor outcome, both in patients with systemic ALK− ALCL and 2 patients with C-ALCL, prompted us to study the presence of this rearrangement in a selected group of patients with C-ALCL and LyP with an aggressive clinical course.
From the 118 patients with C-ALCL included in the Cutaneous Lymphoma Registry of the Leiden University Medical Center (LUMC) between 1985 and 2015, those patients were particularly selected who had developed extracutaneous disease and/or died of ALCL (Table 1; patients 1-8 and 12) and patients who had presented with extensive skin lesions on the legs (patients 1-4, 8, 13, and 14). From the 185 patients with LyP registered in the same period, we selected the only 2 patients who died of systemic ALCL during follow-up (patients 15 and 16) and 1 patient who developed a large persistent tumor during follow-up (patient 17). The final study group contained 17 patients with CD30+ LPDs, including 14 patients with C-ALCL and 3 with LyP, type C. In all patients with C-ALCL, routine staging procedures, including physical examination, blood tests, computed tomography scans of neck, chest, and abdomen, and in most cases bone marrow biopsies, showed no signs of extracutaneous disease at the time of diagnosis.
For the purpose of this study, p63 immunohistochemistry and fluorescence in situ hybridization (FISH) with break-apart probes for TP63 were performed on the formalin-fixed and paraffin-embedded (FFPE) skin biopsies of 16 patients and an involved lymph node of 1 patient (patient 15); these were collected from the archives of the LUMC Department of Pathology in accordance with the Dutch Code for Proper Secondary Use of Human Tissue and approved by the medical ethics committee of the LUMC. In all FFPE skin biopsies, CD30+ tumor cells made up at least 70% of the total infiltrate. As a positive control, the FFPE skin biopsy of 1 patient with tumor stage MF with a TP63 rearrangement (t(3,6)(q28p22.3)) detected by whole genome sequencing (unpublished data) was included. Immunohistochemistry was automatically performed using Dako Autostainer Link 48 with monoclonal mouse anti-human p63 antibodies from Dako (DAK-P63, M7317) in a dilution of 1:50. For positive staining, a cutoff value of 30% was used.9 For the FISH probes, DNA was isolated from bacterial artificial chromosome clones: RP11-718B1, RP11-179O12, and RP11-24F1 proximal and RP11-791J2, RP11-10K11, and RP11-204624 distal of the TP63 gene, purchased from the BACPAC resources center, Children’s Hospital Oakland Research Institute, California, and based on the probe design as used by Vasmatzis et al.10 The isolated DNA was labeled using nick translation either with digoxigenin- or biotin-coupled dideoxynucleotides as haptens. FFPE whole-tissue sections were deparaffinized in xylene, pretreated in 10 mM citric acid buffer, digested in 0.4% pepsin in 0.02M HCL, and codenatured and hybridized with the designed probes. The hapten-labeled probes were subsequently incubated with the mouse anti-digoxigenin antibody conjugated with fluorescein isothiocyanate (FITC) and streptavidin conjugated with Cy3 followed by a second detection using RabbitαMouse anti-biotin antibody conjugated with FITC, followed by counterstaining with 4,6 diamidino-2-phenylindole. In some cases, a probe for the centromere of chromosome 3 was added. For the analysis, 100 to 250 tumor cells per section were manually scored.
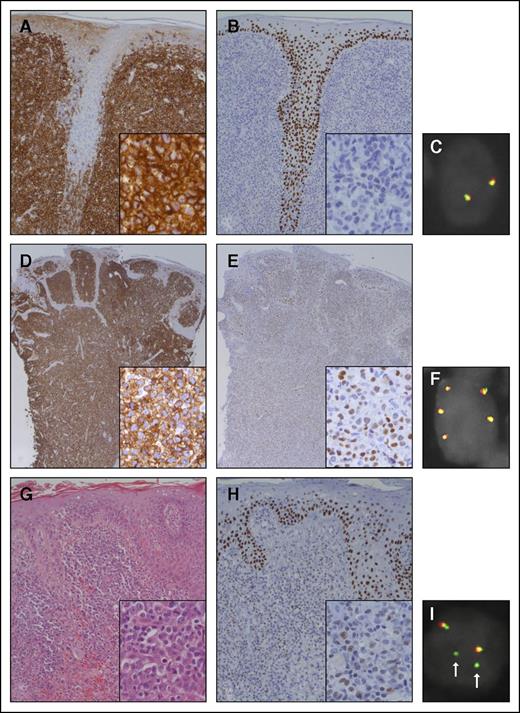
In all CD30+ LPD patients and the MF-positive control, both p63 immunohistochemistry and FISH for TP63 were successfully performed (Table 1; Figure 1). Expression of p63 in more than 30% of the tumor cells was present in 6/17 patients (35%), of which 3 showed expression in more than 75% of the tumor cells. In 7 patients, staining was completely negative. The MF-positive control expressed p63 in more than 30% of the neoplastic T cells. There was no relationship between p63 expression and the clinical course of the patients. A TP63 rearrangement was detected by FISH analysis in none of the 17 CD30+ LPD patients, suggesting that expression of p63 protein is transcriptionally regulated by the p63 promotor and/or influenced by epigenetic factors (eg, chromatin modifications, mitochondrial RNAs). In 3 patients, 1 to 3 extra fusion signals were seen in more than 5% of the scored cells: 6%, 12%, and 43%, respectively. In these cases, the probe for chromosome 3 also showed extra signals, indicating gain of chromosome 3 or polyploid tumor cells. It is unknown whether this has any implications on the regulation of the TP63 gene, though no clear association with p63 expression or with the clinical course of these patients was noted. The MF-positive control showed an unbalanced translocation with gain of the green signal (distal to TP63) (Figure 1).
Histologic features of 2 representative patients with primary C-ALCL and a patient with tumor stage MF included as a positive control. The dermis of the first patient with C-ALCL (patient 10) shows a dense dermal infiltrate of CD30+ tumor cells with infiltration of the hair shaft (A). The tumor cells are predominantly negative for p63 (B). FISH with break-apart probes for TP63 (TP63 FISH) shows 2 normal fusion signals (C). The second patient with C-ALCL (patient 4) also shows a dense dermal infiltrate of CD30+ tumor cells (D). The tumor cells variably express p63 (E). TP63 FISH shows 3 extra copies of the fusion signal, indicating gain of chromosome 3 or polyploid tumor cells (F). The hematoxylin and eosin staining of the patient with MF shows some epidermotropism and diffuse infiltration of the dermis by blastic tumor cells (G). The tumor cells variably express p63 (H). TP63 FISH shows 2 fusion signals and 2 separate green signals (I; arrows), suggesting gain of chromosome 3 with an unbalanced translocation of TP63. Note that the epidermis shows expression of p63 in the basal and suprabasal layers (B,E,H). Original magnification ×200 for panels A-B,G-H, ×100 for panels D-E, and ×400 for insets.
Histologic features of 2 representative patients with primary C-ALCL and a patient with tumor stage MF included as a positive control. The dermis of the first patient with C-ALCL (patient 10) shows a dense dermal infiltrate of CD30+ tumor cells with infiltration of the hair shaft (A). The tumor cells are predominantly negative for p63 (B). FISH with break-apart probes for TP63 (TP63 FISH) shows 2 normal fusion signals (C). The second patient with C-ALCL (patient 4) also shows a dense dermal infiltrate of CD30+ tumor cells (D). The tumor cells variably express p63 (E). TP63 FISH shows 3 extra copies of the fusion signal, indicating gain of chromosome 3 or polyploid tumor cells (F). The hematoxylin and eosin staining of the patient with MF shows some epidermotropism and diffuse infiltration of the dermis by blastic tumor cells (G). The tumor cells variably express p63 (H). TP63 FISH shows 2 fusion signals and 2 separate green signals (I; arrows), suggesting gain of chromosome 3 with an unbalanced translocation of TP63. Note that the epidermis shows expression of p63 in the basal and suprabasal layers (B,E,H). Original magnification ×200 for panels A-B,G-H, ×100 for panels D-E, and ×400 for insets.
In conclusion, this study suggests that in patients with CD30+ LPD, an aggressive clinical course cannot be defined by the presence of TP63 rearrangements, as was recently shown in systemic ALK− ALCL. In addition, immunohistochemical expression of p63 is variable in CD30+ LPD and is not correlated with overall survival.
Authorship
Acknowledgments: The authors thank D. de Jong, Department of Molecular Cell Biology, Leiden University Medical Center, The Netherlands, for providing valuable assistance in performing FISH.
This work was supported by funding from the Dutch Cancer Society (grant UL2013-6104).
Contribution: A.M.R.S. performed the research, analyzed the data, and wrote the paper; Y.-Y.C. performed the research, contributed critical reagents, and analyzed the data; P.M.J. analyzed the data; A.N.B.T. contributed critical reagents; K.S. contributed analytical tools and analyzed the data; C.P.T. designed the research and contributed analytical tools; and R.W. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interest.
Correspondence: Anne M. R. Schrader, Leiden University Medical Center, Department of Pathology, P.O. Box 9600, 2300 RC Leiden, The Netherlands; e-mail: a.m.r.schrader@gmail.com.