Key Points
Deletion of Hoxa genes reduces the engraftment potential of adult hematopoietic stem cells.
Ectopic overexpression of Hoxa9 partially restores Hoxa−/− hematopoietic stem cell activity.
Abstract
Determination of defined roles for endogenous homeobox (Hox) genes in adult hematopoietic stem and progenitor cell (HSPC) activity has been hampered by a combination of embryonic defects and functional redundancy. Here we show that conditional homozygous deletion of the Hoxa cluster (Hoxa−/−) results in a marked reduction of adult HSPC activity, both in vitro and in vivo. Specifically, proliferation of Hoxa−/− HSPCs is reduced compared with wild-type (WT) cells in vitro and they are less competitive in vivo. Notably, the loss of Hoxa genes had little impact on HSPC differentiation. Comparative RNA sequencing analyses of Hoxa−/− and WT hematopoietic stem cells (CD150+/CD48−/Lineage−/c-kit+/Sca-1+) identified a large number of differentially expressed genes, three of which (Nr4a3, Col1a1, and Hnf4a) showed >10-fold reduced levels. Engineered overexpression of Hoxa9 in Hoxa−/− HSPCs resulted in partial phenotypic rescue in vivo with associated recovery in expression of a large proportion of deregulated genes. Together, these results provide definitive evidence linking Hoxa gene expression to proliferation of adult HSPCs.
Introduction
Specific developmental phenotypes are associated with loss of individual Hoxa cluster genes in the mouse.1-4 Interestingly, most of these Hoxa genes are expressed in hematopoietic cells, with the highest levels in primitive hematopoietic stem and progenitor cells (HSPC) and much lower levels in mature cells.5-8
Evaluation of individual Hoxa gene mutant mice has provided only limited insight into their role in adult hematopoiesis, indicative of a level of functional redundancy of cluster genes in this tissue.1,9,10 The Hoxa9 homozygous null mice display the most overt hematopoietic phenotype characterized by mild leucopenia resulting from bone marrow (BM) hypocellularity, particularly of myeloid and B cells. This phenotype is associated with impairment in activity, but not the number of the long-term (LT) hematopoietic stem cells (HSCs).11
Further supporting the importance of Hoxa genes in hematopoiesis, we previously showed that heterozygous Hoxa cluster (Hoxa+/−) adult LT-HSCs are less competitive than wild-type (WT) cells in transplantation assays.5 Moreover, Di Poï et al previously reported that recipients of fetal liver-derived LT-HSCs in which Hoxa gene expression is reduced to 20% (HoxAc/− cells)12 have impaired ability to repopulate adult mice, particularly in the output of early erythrocytes. This phenotype was much less obvious in HoxAc/− newborn mice, possibly indicating that Hoxa cluster genes are more important in activity of adult vs fetal HSPCs.
To address this point, we now assess the impact of the complete ablation of Hoxa cluster genes in adult HSPCs and explore the consequence of this deletion on the transcriptome using RNA sequencing (RNASeq). Results clearly establish an essential function for Hoxa genes in proliferation, but not differentiation, of adult HSPCs.
Study design
Mice and Hoxa gene deletion
To delete the Hoxa locus Hoxaflox/flox/ MxCre mice13 (referred to as Hoxa−/− after deletion), MxCre controls received 7 injections of 10 µg poly I/poly C (pIpC) (GE Healthcare Life Sciences, Baie-d’Urfé, QC, Canada) per gram body weight for a maximum of 250 µg per mice intraperitoneally every 2 days. Hoxa cluster deletion was confirmed by polymerase chain reaction. All our animal studies were approved by the local Animal Care Committee under legislation of the Canadian Council on Animal Care.
Infection of primary BM cells
Lineage (Lin)− BM cells were purified from Hoxa−/− and control mice 4 weeks post-pIpC, and transfected with murine stem cell virus-pgk green fluorescent protein (GFP) or murine stem cell virus-HOXA9-pgk GFP retroviral supernatants by spinoculation (2250 rpm) for 90 minutes. After 48 hours, 0.25 × 106 sorted GFP+ were IV injected without helper cells, in irradiated (8 Gy) adult B6SJL congenic recipient mice.
RNA isolation and RNASeq library preparation
Total RNA was extracted from 60 000 to 100 000 Hoxa−/− and control LT-HSC (CD150+/CD48−/Lin−/c-kit+/Sca-1+ [LKS]) and used to generate transcriptome libraries. Paired end (2 × 100 bp) sequencing was performed using an Illumina HiSeq2000 (Illumina, San Diego, CA). RNASeq data were analyzed with Cuffdiff or the DeSeq R package.14
Results and discussion
MxCre-induced conditional deletion of Hoxa genes in adult HSPCs resulted in a significant reduction of white blood cells, red blood cells, and platelets in the peripheral blood (PB), and moderate reductions in cellularity in all hematopoietic organs, primarily due to a severe decrease in B cells (see supplemental Figure 1A-D, available on the Blood Web site). Flow cytometry and clonogenic assays also demonstrated reduced HSPCs (Figure 1A and supplemental Figure 1E-G, and confirmation of deletion in LKS cells in Figure 1B), consistent with observations made in fetal liver-derived HoxAc/− HSCs.12 However, unlike the fetal-derived equivalents, adult Hoxa−/− HSCs demonstrated reduced proliferation potential in vitro (supplemental Figure 1H), but were more in cycle in vivo (supplemental Figure 2A-C). In accordance with the in vitro results, Hoxa−/− BM cells demonstrated a reduced ability in reconstituting irradiated congenic hosts compared with WT controls (Figure 1C and supplemental Figure 1I-J) that is partly due to an increase in apoptosis (supplemental Figure 2D-E), but not due to homing defects or non–Hoxa-deleted escapees (supplemental Figure 2F-H). Repopulation ability of Hoxa−/− cells was completely absent in secondary recipients, in contrast to control cells, suggesting an exhaustion of the HSC pool (supplemental Figure 1K). In this in vivo model, B cells were also underrepresented, whereas Hoxa−/− cells reconstituted most other cell lineages proportionally (Figure 1D and supplemental Figure 1L). These data confirm our earlier observation that adult HSC-derived B cells have an increased sensitivity for Hoxa gene levels5 , compared with fetal HSC-derived B cells.12
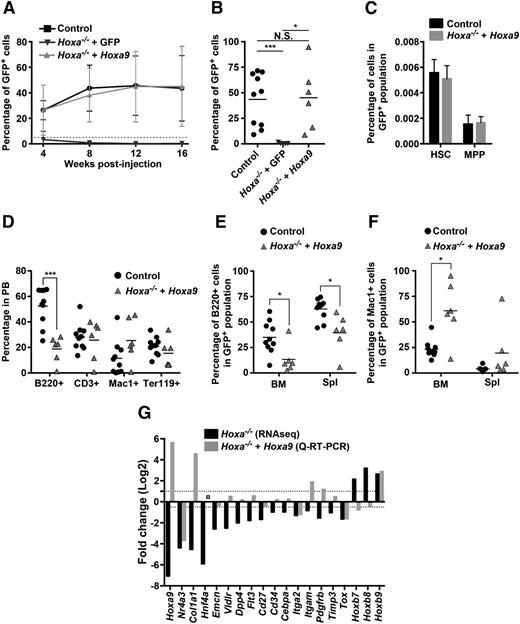
Deletion of Hoxa cluster genes in adult mice and transcriptome analysis of Hoxa−/− HSCs. (A) Bar graphs depicting absolute numbers of CD150+/CD48−/CD244−/LKS HSCs and CD150+/CD48−/CD244+/LKS MPPs in BM of Hoxa−/− and control mice (n = 4). BM cells isolated from femur and tibia 4 weeks after pIpC injection. (B) Polymerase chain reaction analysis showing the deletion of the Hoxa cluster in purified LKS cells at 4 weeks following the last pIpC injection (n = 5). (C) Engraftment of Hoxa−/− (n = 14) and control (n = 15) cells in the PB, BM, spleen, and thymus of recipient mice at ∼24 weeks after transplantation. Each dot represents a CD45.1 recipient mouse transplanted with 106 congenic CD45.2 BM cells. (D) Proportions of B (B220+), myeloid (Mac1+), T (CD3+), and erythroid (Ter119+) cells in PB of reconstituted mice shown in (C). (E) Scatterplot showing transcriptome analysis of genes that were up- (above axis) or downregulated (below axis) in Hoxa−/− vs WT control CD150+/CD48−/LKS cells sorted 1 month after the last pIpC injection. Values are expressed as average (log10 [(average RPKM)*1000 + 1]). Cut-off limit for showing gene identify was set at 3, representing a RPKM value of ∼0.1. Highlighted genes show 10-fold differences between the 2 groups. (F-G) Comparative expression of Hoxa (F) and Hoxb (G) genes in pairs of Hoxa−/− and control HSC samples (n = 3). *P < .05; **P < .01; ***P < .001. Cre, causes recombination; CT, cycle time; HPRT, hypoxanthine-guanine phosphoribosyltransferase; MPP, multipotent progenitors; RQ, relative quantification; Spl, spleen; Thy, thymus.
Deletion of Hoxa cluster genes in adult mice and transcriptome analysis of Hoxa−/− HSCs. (A) Bar graphs depicting absolute numbers of CD150+/CD48−/CD244−/LKS HSCs and CD150+/CD48−/CD244+/LKS MPPs in BM of Hoxa−/− and control mice (n = 4). BM cells isolated from femur and tibia 4 weeks after pIpC injection. (B) Polymerase chain reaction analysis showing the deletion of the Hoxa cluster in purified LKS cells at 4 weeks following the last pIpC injection (n = 5). (C) Engraftment of Hoxa−/− (n = 14) and control (n = 15) cells in the PB, BM, spleen, and thymus of recipient mice at ∼24 weeks after transplantation. Each dot represents a CD45.1 recipient mouse transplanted with 106 congenic CD45.2 BM cells. (D) Proportions of B (B220+), myeloid (Mac1+), T (CD3+), and erythroid (Ter119+) cells in PB of reconstituted mice shown in (C). (E) Scatterplot showing transcriptome analysis of genes that were up- (above axis) or downregulated (below axis) in Hoxa−/− vs WT control CD150+/CD48−/LKS cells sorted 1 month after the last pIpC injection. Values are expressed as average (log10 [(average RPKM)*1000 + 1]). Cut-off limit for showing gene identify was set at 3, representing a RPKM value of ∼0.1. Highlighted genes show 10-fold differences between the 2 groups. (F-G) Comparative expression of Hoxa (F) and Hoxb (G) genes in pairs of Hoxa−/− and control HSC samples (n = 3). *P < .05; **P < .01; ***P < .001. Cre, causes recombination; CT, cycle time; HPRT, hypoxanthine-guanine phosphoribosyltransferase; MPP, multipotent progenitors; RQ, relative quantification; Spl, spleen; Thy, thymus.
Comparative RNASeq transcriptome data from Hoxa−/− and control HSCs identified 881 significantly differentially expressed genes, the vast majority of which (614) were downregulated (supplemental Table 1). Functional annotation clustering using Gene Ontology terms revealed that differentially expressed genes were associated with cell proliferation and differentiation, cell activation, signaling, regulation of gene expression, hematopoiesis, migration, and apoptosis (supplemental Table 2). Moreover, differentially expressed genes were associated with several pathways according to the Kyoto Encyclopedia of Genes and Genomes, which included hematopoietic cell lineage and cancer pathways, supporting a known role for Hoxa genes in hematopoiesis and leukemia (supplemental Table 3).
Only 13 genes expressed at reads per kilobase of transcript per million mapped reads (RPKM) values >1 showed a >10-fold differential expression between the 2 conditions (Figure 1E; supplemental Table 1). Genes downregulated in Hoxa−/− cells were mostly from the Hoxa cluster, but also included Nr4a3, Col1a1, and Hnf4a (Figure 1E-F). Nr4a3, also known as Nor-1, codes for an orphan nuclear receptor transcription factor highly homologous to Nr4a1 (Nur77) and Nr4a2 (Nurr1). Co-deficiency of Nor-1 and Nur77 is associated with an aggressive acute myeloid leukemia,15 whereas individual mutants have only weak phenotypes both related to proliferation and apoptosis.16,17 The latter phenotype corresponds with those for the Hoxa−/−, indicating that Nr4a3 may have contributed to the Hoxa−/− phenotype. Two genes, Hoxb8 and microRNA 196a-1, were significantly upregulated (>10-fold) in Hoxa−/− HSCs (Figure 1E-G), possibly pointing to cross-regulation between Hoxa and Hoxb cluster genes and their inter-cluster–located microRNAs.18,19
We next investigated if overexpression of a single Hox gene, Hoxa9, could at least in part, rescue the HSPC phenotype found in recipients of Hoxa−/− cells. Hoxa−/− HSPCs were transduced with Hoxa9 or control GFP virus and transplanted into congenic animals. We found that Hoxa9 overexpression restored HSPC engraftment to control WT levels (Figure 2A-C and supplemental Figure 3A). However, and in line with our previous observations,5 the B and myeloid lineages are skewed and therefore do not recapitulate the normal distribution observed in control mice. Specifically, B cell numbers remained persistently lower and a substantial increase of Mac1+ cells and myeloid progenitors (LKS−) was observed in mice overexpressing Hoxa9 (Figure 2D-F and supplemental Figure 3B-C). Notably, overexpression of Hoxa9 did provide partial recovery at the molecular level of over 70% (14 of 19) of the deregulated genes found in the Hoxa−/− mutant (Figure 2G). Despite this, Hoxa9 overexpression did not correct the decrease in B cells, and instead favored production of myeloid cells, indicating that qualitative contributions of individual Hoxa genes are vital for balanced hematopoiesis.
Rescue of Hoxa−/− HSC engraftment by overexpression of Hoxa9. (A) Average kinetics of engraftment in mice receiving 0.2 × 106 of either control Hoxa+/+ + GFP (n = 13), Hoxa−/− + GFP (n = 5), and Hoxa−/− + Hoxa9-GFP (n = 6) BM cells. Percentage of donor cells are measured by flow cytometry for GFP+ in the PB at indicated times. (B) LT engraftment of HSCs with indicated genotype in PB of individual mice 16 weeks posttransplantation. (C) Bar graphs depicting percentage of Hoxa+/+ + GFP and Hoxa−/− + Hoxa9 CD150+/CD48−/LKS HSCs (n = 3) and CD150−/CD48−/LKS MPPs (n = 3) in GFP+ cell population. (D) Percentage of Hoxa+/+ + GFP and Hoxa−/− + Hoxa9-derived B cells (B220+), T cells (CD3+), myeloid cells (Mac1+), and erythroid cells (Ter119+) in the PB of individual mice 16 weeks posttransplantation. (E-F) Percentage of B cells (B220+) (E) and myeloid cells (Mac1+) (F) of GFP+ cells in BM and spleen. (G) Gene expression analysis in RNASeq HSC and Lin− cells overexpressing Hoxa9. ○ indicates quantity not sufficient defined as corrected CT value >36. *P < .05; ***P < .001. N.S., non-significant; MPP, multipotent progenitors; Q-RT-PCR, quantitative-reverse transcription-polymerase chain reaction; Spl, spleen.
Rescue of Hoxa−/− HSC engraftment by overexpression of Hoxa9. (A) Average kinetics of engraftment in mice receiving 0.2 × 106 of either control Hoxa+/+ + GFP (n = 13), Hoxa−/− + GFP (n = 5), and Hoxa−/− + Hoxa9-GFP (n = 6) BM cells. Percentage of donor cells are measured by flow cytometry for GFP+ in the PB at indicated times. (B) LT engraftment of HSCs with indicated genotype in PB of individual mice 16 weeks posttransplantation. (C) Bar graphs depicting percentage of Hoxa+/+ + GFP and Hoxa−/− + Hoxa9 CD150+/CD48−/LKS HSCs (n = 3) and CD150−/CD48−/LKS MPPs (n = 3) in GFP+ cell population. (D) Percentage of Hoxa+/+ + GFP and Hoxa−/− + Hoxa9-derived B cells (B220+), T cells (CD3+), myeloid cells (Mac1+), and erythroid cells (Ter119+) in the PB of individual mice 16 weeks posttransplantation. (E-F) Percentage of B cells (B220+) (E) and myeloid cells (Mac1+) (F) of GFP+ cells in BM and spleen. (G) Gene expression analysis in RNASeq HSC and Lin− cells overexpressing Hoxa9. ○ indicates quantity not sufficient defined as corrected CT value >36. *P < .05; ***P < .001. N.S., non-significant; MPP, multipotent progenitors; Q-RT-PCR, quantitative-reverse transcription-polymerase chain reaction; Spl, spleen.
Together, the data identifies a critical role for Hoxa cluster genes in adult HSPC function, and furthermore, a Hoxa gene-dependent gene signature that underlies this function.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge Martine Dupuis and Nathalie Henley for cell sorting, and in particular, Dr Denis Duboule (University of Geneva, Geneva) and Dr Marie Kmita (Institut De Recherches Cliniques De Montreal) for providing the mice. The authors also thank the staff at the animal facility for taking care of the animals; Mona Hassawi, Gratianne Vaisson, and Gloria Giono for their help with the isolation of BM cells for the RNASeq; and Dr Julie Lessard and Dr Martin Guimond for critically reading the manuscript.
This work was supported by a grant from the Canadian Cancer Society (#20399). C.-E.L.-G. and M.F. are recipients of a department scholarship and award from the Cole foundation (M.F.).
Authorship
Contribution: C.-E.L.-G. performed experiments; C.-E.L.-G., M.F., and J.J.B. analyzed data and mounted the figures; C.-E.L.-G., A.T., G.S., and J.J.B. designed the research and wrote the paper; and L.K. was involved in the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Janet Bijl, Maisonneuve-Rosemont Hospital Research Center, 5415 Blvd De l’Assomption, Montreal, QC, Canada H1T 2M4; e-mail: janettabijl@yahoo.ca.

![Figure 1. Deletion of Hoxa cluster genes in adult mice and transcriptome analysis of Hoxa−/− HSCs. (A) Bar graphs depicting absolute numbers of CD150+/CD48−/CD244−/LKS HSCs and CD150+/CD48−/CD244+/LKS MPPs in BM of Hoxa−/− and control mice (n = 4). BM cells isolated from femur and tibia 4 weeks after pIpC injection. (B) Polymerase chain reaction analysis showing the deletion of the Hoxa cluster in purified LKS cells at 4 weeks following the last pIpC injection (n = 5). (C) Engraftment of Hoxa−/− (n = 14) and control (n = 15) cells in the PB, BM, spleen, and thymus of recipient mice at ∼24 weeks after transplantation. Each dot represents a CD45.1 recipient mouse transplanted with 106 congenic CD45.2 BM cells. (D) Proportions of B (B220+), myeloid (Mac1+), T (CD3+), and erythroid (Ter119+) cells in PB of reconstituted mice shown in (C). (E) Scatterplot showing transcriptome analysis of genes that were up- (above axis) or downregulated (below axis) in Hoxa−/− vs WT control CD150+/CD48−/LKS cells sorted 1 month after the last pIpC injection. Values are expressed as average (log10 [(average RPKM)*1000 + 1]). Cut-off limit for showing gene identify was set at 3, representing a RPKM value of ∼0.1. Highlighted genes show 10-fold differences between the 2 groups. (F-G) Comparative expression of Hoxa (F) and Hoxb (G) genes in pairs of Hoxa−/− and control HSC samples (n = 3). *P < .05; **P < .01; ***P < .001. Cre, causes recombination; CT, cycle time; HPRT, hypoxanthine-guanine phosphoribosyltransferase; MPP, multipotent progenitors; RQ, relative quantification; Spl, spleen; Thy, thymus.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/1/10.1182_blood-2015-02-626390/5/m_87f1.jpeg?Expires=1769166423&Signature=zERAtuXsrfc2EunTwAyiDyQAKfQ1EBg~FdiQg5DT9-6btVjqFpUHfzAcHC8FC5cxwsmfmb0lrWAsRQ6M-9AD2llDcSdEiN0ZietRQOFujP--5n3n7jN3lcw4y~13agyZv3AaydYzHaqkG4J23GdjyTOD87MwgkLJAnDiZUQl-S4vxQlC4tD8RBot40Q8nCxthl4StTRyFKUqABGaMFb6Y7XlBFNoyfoVEfIGnu68D39wcl9HZGAFL3T1D3lf-LPx00iLYJH3eObEZW3tMP46C3G~CuaX~qhyrKjWZHqBu~lSNPvWuXEXPWkMnBm7jezKR2RWZvoqOAyFfxIVqWVxZw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)