Key Points
GWAS can identify allele mismatch associated with aGVHD development.
Three novel candidate loci for minor histocompatibility antigens significantly associate with aGVHD.
Abstract
Acute graft-versus-host disease (aGVHD) represents one of the major complications in allogeneic stem cell transplantation and is primarily caused by genetic disparity between the donor and recipient. In HLA-matched transplants, the disparity is thought to be determined by loci encoding minor histocompatibility antigens (minor H antigens), which are presented by specific HLA molecules. We performed a genome-wide association study (GWAS) to identify minor H antigen loci associated with aGVHD. A total of 500 568 single nucleotide polymorphisms (SNPs) were genotyped for donors and recipients from 1589 unrelated bone marrow transplants matched for HLA-A, -B, -C, -DRB1, and -DQB1, followed by the imputation of unobserved SNPs. We interrogated SNPs whose disparity between the donor and recipient was significantly associated with aGVHD development. Without assuming HLA unrestriction, we successfully captured a known association between HLA-DPB1 disparity (P = 4.50 × 10−9) and grade II-IV aGVHD development, providing proof of concept for the GWAS design aimed at discovering genetic disparity associated with aGVHD. In HLA-restricted analyses, whereby association tests were confined to major subgroups sharing common HLA alleles to identify putative minor H antigen loci, we identified 3 novel loci significantly associated with grade III-IV aGVHD. Among these, rs17473423 (P = 1.20 × 10−11) at 12p12.1 within the KRAS locus showed the most significant association in the subgroup, sharing HLA-DQB1*06:01. Our result suggested that a GWAS can be successfully applied to identify allele mismatch associated with aGVHD development, contributing to the understanding of the genetic basis of aGVHD.
Introduction
Allogeneic (allo) hematopoietic stem cell transplantation (HSCT) has been established as the standard choice of potentially curative therapy for many high-risk leukemias and other intractable hematologic neoplasms1,2 in which the major therapeutic benefit is primarily obtained from alloimmune reactions directed against the recipient’s leukemic cells mediated by engrafted donor T cells (graft-versus-leukemia [GVL] effect).3 However, the same kind of allo-reaction can also be induced against normal host tissues, giving rise to a severe complication known as graft-versus-host disease (GVHD).4-6 GVHD represents one of the major causes of mortality and morbidity after allo-HSCT. In particular, the early onset of GVHD before day 100 after transplant (acute GVHD [aGVHD]) has been consistently associated with poor overall survival, whereas chronic GVHD, which takes place after day 100, may correlate with low relapse rates in some leukemia types.7,8 Thus, preventing life-threatening GVHD while harnessing GVL effects is a key to successful transplantation for leukemia.9 The importance of circumventing severe aGVHD is further underscored in transplantation for benign disorders such as aplastic anemia and severe combined immunodeficiency.10,11
In allo-HSCT, the immunologic targets for aGVHD are genetically defined alloantigens that are not shared by the donor; thus, they could be recognized by engrafted donor T cells to elicit destructive immune reactions.6,12 Among these, the most important in allo-HSCT are HLAs, particularly HLA-A, -B, -C, -DR, and -DQ, which are requested to be strictly matched between the donor and recipient in standard transplant procedures to prevent life-threatening GVHD and graft rejection.2,13-17 However, even in HLA-matched transplantation, severe aGVHD does occur in 6% to 17% of related and 11% to 42% of unrelated transplant recipients.2,15,18-20
In HLA-matched transplantation, the antigens responsible for GVHD are considered minor histocompatibility antigens (minor H antigens), which are typically defined by single-nucleotide polymorphisms (SNPs) in the recipient or by other polymorphic alleles not shared by the donor and presented on the recipient’s tissues in the context of particular HLA types.21,22 Donor T cells can recognize these minor H antigens to cause graft-versus-host (GVH)/GVL reactions, but only when the minor H antigens are presented by the same HLAs that are shared by the recipient (Figure 1).5,22,23 These allo-reactions may be further modified by other genetic factors in the donor and/or recipient and by environmental factors such as polymorphisms in immune regulators (eg, tumor necrosis factor α) and tissue damage caused by conditioning regimens before transplant.24,25 Therefore, for better control of severe aGVHD in HLA-matched HSCT, it is important to identify the relevant minor H antigens and other genetic factors for GVHD. These are plausible targets for GWASs.
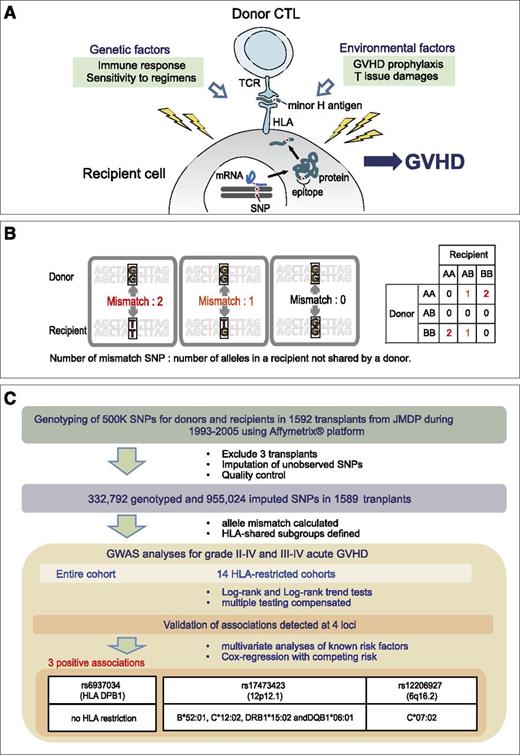
GVHD development in HSCT. (A) GVHD is an alloimmune response that develops after allogeneic transplantation, in which donor T cells recognize the host-derived alloantigens presented on their targets. In HLA-matched transplantation, these minor H antigens are typically defined by the host SNPs or by other polymorphisms that are not shared with the donor. GVHD development is also affected by other genetic and environmental factors. (B) The number of allele mismatches is defined as the number of alleles (0, 1, or 2) that are not shared by the donor. (C) Schematic diagram of the study design. For each SNP showing >5% of minor allele frequency in the donor cohort, association was tested between the presence/absence of grade II-IV aGVHD or grade III-IV aGVHD and the number of allele mismatches for each donor-recipient pair using log-rank and/or log-rank trend tests (GWAS), with corrections for multiple testing. GWAS was performed including the entire cohort, or to identify minor H antigen–related loci confined to those subsets sharing 1 of the 14 HLA alleles observed in >20% of the current transplant cohort. CTL, cytotoxic T lymphocyte; GWAS, genome-wide association study; JMDP, Japan Marrow Donor Program; mRNA, messenger RNA; TCR, T-cell receptor.
GVHD development in HSCT. (A) GVHD is an alloimmune response that develops after allogeneic transplantation, in which donor T cells recognize the host-derived alloantigens presented on their targets. In HLA-matched transplantation, these minor H antigens are typically defined by the host SNPs or by other polymorphisms that are not shared with the donor. GVHD development is also affected by other genetic and environmental factors. (B) The number of allele mismatches is defined as the number of alleles (0, 1, or 2) that are not shared by the donor. (C) Schematic diagram of the study design. For each SNP showing >5% of minor allele frequency in the donor cohort, association was tested between the presence/absence of grade II-IV aGVHD or grade III-IV aGVHD and the number of allele mismatches for each donor-recipient pair using log-rank and/or log-rank trend tests (GWAS), with corrections for multiple testing. GWAS was performed including the entire cohort, or to identify minor H antigen–related loci confined to those subsets sharing 1 of the 14 HLA alleles observed in >20% of the current transplant cohort. CTL, cytotoxic T lymphocyte; GWAS, genome-wide association study; JMDP, Japan Marrow Donor Program; mRNA, messenger RNA; TCR, T-cell receptor.
In the present study, we performed a series of GWAS analyses involving 1589 matched HLA-A, -B, -C, -DRB1, and -DQB1 unrelated bone marrow transplants from the JMDP, in an attempt to identify the genetic loci relevant to the development of severe aGVHD.
Methods
Subjects and genotyping
A total of 1589 unrelated bone marrow transplants performed through the JMDP from 1993-2005 were included. Bone marrow was the exclusive source of stem cells. All the transplants were completely matched for HLA-A, -B, -C, -DRB1, and -DQB1 loci on the basis of high-resolution DNA typing,26 although 995 transplants (62.6%) were mismatched for HLA-DPB1 in the GVH direction. T-cell depletion was not performed in any transplants. For prophylaxis of aGVHD, calcineurin inhibitors (cyclosporine A or tacrolimus) in combination with methotrexate were used in all transplants. Anti-thymocyte globulin was used in 104 (6.5%) transplants (Table 1). In total, 622 and 229 recipients developed grade II-IV and III-IV aGVHD, respectively, according to the criteria proposed by Glucksberg et al.27
Including 42 individuals who performed multiple marrow donations, 1547 donors and 1589 recipients were genotyped for 500 568 SNPs using the Affymetrix Human Mapping 500K Array Set (Table 1). We excluded from further analyses 3 sets of transplants in which call rates of SNP typing in either donors or recipients were below 90%. Genotypes of unobserved SNPs were imputed on the basis of the published HapMap data (see supplemental Methods, available on the Blood Web site).28,29 A total of 332 792 genotyped and 955 024 imputed SNPs passed quality control.
Data analysis and association tests for GVHD
Our primary interest was to identify polymorphic histocompatibility loci for which disparity between the donor and recipient is associated with severe aGVHD. Accordingly, our GWAS analyses involved 2 genotypes of both the donors and recipients: for each SNP locus in each donor-recipient pair, we determined the presence or absence of GVH disparity and enumerated the recipient alleles (0, 1, or 2) that were not shared by the donor. The association of the disparity and the number of mismatched alleles with the development of aGVHD was tested for each SNP locus across the entire genome. We employed 2 discrete aGVHD end points: grade II-IV and grade III-IV aGVHD, whereby those with grade 0-I and 0-II aGVHD, respectively, were treated as having no GVHD (control), assuming that the severity of aGVHD was differentially affected depending on the genetic loci. For each of the 2 end points (II-IV and III-IV aGVHD), log-rank statistics were calculated at each SNP locus for the presence or absence of mismatched SNPs. The trends of a higher aGVHD grade with an increasing number of mismatched alleles were also tested using trend log-rank statistics, given that the underlying genetic/immunologic model for GVHD was unclear (supplemental Methods). In the latter test, we hypothesized that GVH reactions were enhanced with an increasing number of mismatched alleles (Figure 1B). Competing risk, defined as death without aGVHD, and potential confounding risk factors for aGVHD, including age, sex, methods of GVHD prophylaxis, and use of total body irradiation or anti-thymocyte globulin, were not considered for the genome-wide screening but were included in the calculation to confirm the positive associations (supplemental Methods).30 Donor or recipient SNPs relevant to the development of aGVHD were also interrogated by GWAS analyses employing simple genotypes of either donors or recipients using log-rank and log-rank trend statistics (also described in supplemental Methods).
Results
Identification of HLA-DPB1 mismatch for the risk of grade II-IV aGVHD
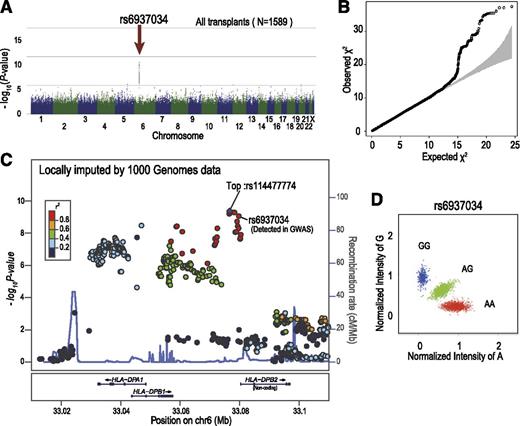
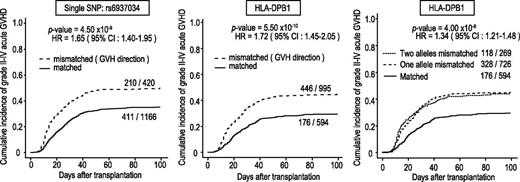
In total, 3142 DNA specimens from the donors and recipients of 1592 transplants were genotyped. After the exclusion of 3 transplants with a donor or recipient showing a call rate <90% in either the donor or the recipient, a total of 1589 transplants were included in the analyses, with a mean call rate of 99.2%. The genomic control inflation factors were lower than 1.05 for all analyses, indicating a low possibility of false-positive associations resulting from population stratification or genotype misclassification.31,32 We first tested the association of simple genotypes in the donors and recipients with the development of aGVHD, finding no significant associations with regard to grade II-IV or grade III-IV aGVHD. Thus, we next tested the association of allele mismatch between donor and recipient with the development of grade II-IV and grade III-IV aGVHD using the entire cohort, under the assumption of no HLA restriction. As shown in Figure 2, a significant peak that was associated with grade II-IV aGVHD, but not with grade III-IV aGVHD, was detected at rs6937034 in 6p21, in the vicinity of the HLA-DPA1/DPB1 loci, with a minimum P value of 9.06 × 10−10 (log-rank test). The test remained significant (P = 4.50 × 10−9), even when competing risk was taken into consideration (Figure 3; Table 2). The association remained significant (P = 7.90 × 10−10) after known risk factors and competing risk were incorporated in a multivariable analysis (Table 3). When the association was directly tested for HLA-DPB1 mismatch, a stronger association was observed, with an HR of 1.72 (95% CI, 1.45-2.05; P = 5.50 × 10−10) in the competing-risk regression analysis (Figure 3, left panel). The significant peak disappeared completely when the analysis was stratified for HLA-DPB1 mismatch before the association test, suggesting that the HLA-DPB1 mismatch was causative for the association, recapitulating our recent observation in the extended JMDP cohort.33 This was also an example in which allele mismatch between highly polymorphic alleles could be successfully captured by dichotomous SNP alleles.
SNPs around the HLA-DPB1 locus showed strong association with the development of grade II-IV aGVHD. Manhattan plot (A) and QQ plot (B) of GWAS analysis, in which the log-rank test statistic was calculated for each SNP according to the number of mismatching alleles in the GVH direction between the donor and patient. In the QQ plot, 95% CIs are depicted in gray. (C) Unobserved SNPs were imputed using the data from the 1000 Genomes Project. The P values from the log-rank test for each SNP around the HLA-DPB1 locus were plotted as circles, the color of which indicates the r2 value between each SNP and the SNP showing the minimum P value (rs114477774; “Top”). In the diagram, genetic recombination rates are overlaid with a light blue line. (D) Cluster plot for rs6937034. Specimens genotyped as AA, AG, and GG are shown in red, green, and blue, respectively. CI, confidence interval; QQ, quantile-quantile.
SNPs around the HLA-DPB1 locus showed strong association with the development of grade II-IV aGVHD. Manhattan plot (A) and QQ plot (B) of GWAS analysis, in which the log-rank test statistic was calculated for each SNP according to the number of mismatching alleles in the GVH direction between the donor and patient. In the QQ plot, 95% CIs are depicted in gray. (C) Unobserved SNPs were imputed using the data from the 1000 Genomes Project. The P values from the log-rank test for each SNP around the HLA-DPB1 locus were plotted as circles, the color of which indicates the r2 value between each SNP and the SNP showing the minimum P value (rs114477774; “Top”). In the diagram, genetic recombination rates are overlaid with a light blue line. (D) Cluster plot for rs6937034. Specimens genotyped as AA, AG, and GG are shown in red, green, and blue, respectively. CI, confidence interval; QQ, quantile-quantile.
Association of grade II-IV aGVHD with mismatches of HLA-DPB1 and rs6937034. Cumulative incidence of grade II-IV aGVHD. HRs and P values were calculated by competing-risk regression analysis, in which death before aGHVD was considered a competing risk. Association was tested according to the mismatch for the single SNP (rs6937034) (left), whereas HLA-DPB1 mismatch (middle) in the GVH direction was used to draw Kaplan-Meier curves. This trend in association was not obvious in increased numbers of HLA-DPB1 mismatches (right). HR, hazard ratio.
Association of grade II-IV aGVHD with mismatches of HLA-DPB1 and rs6937034. Cumulative incidence of grade II-IV aGVHD. HRs and P values were calculated by competing-risk regression analysis, in which death before aGHVD was considered a competing risk. Association was tested according to the mismatch for the single SNP (rs6937034) (left), whereas HLA-DPB1 mismatch (middle) in the GVH direction was used to draw Kaplan-Meier curves. This trend in association was not obvious in increased numbers of HLA-DPB1 mismatches (right). HR, hazard ratio.
No conspicuous trend of association with development of aGVHD was observed with an increasing number of mismatched HLA-DPB1 alleles (Figure 3, right panel), although the log-rank trend statistics showed a detectable peak corresponding to the HLA-DBP1 locus. No other loci, including HLA loci, were significantly associated with aGVHD.
Association tests under the assumption of HLA restriction for minor H antigen recognition
In HLA-matched transplants, the antigen relevant to the development of GVHD is thought to be presented by a particular HLA subtype (HLA restriction in minor H antigen presentation/recognition). For this reason, in an attempt to interrogate aGVHD-related minor H antigen loci, we next performed a series of GWASs under the assumption of HLA restriction, whereby each GWAS was confined to a subset of all the transplants that shared a given HLA. In these analyses, to prevent unacceptable loss of statistical power due to a reduced number of mismatched transplants in individual analyses, we limited the HLA subtypes to be tested to 14 common alleles that account for >20% of the present cohort, instead of exhaustively repeating multiple underpowered tests for many minor HLA alleles. Due to the presence of common haplotypes among the Japanese population, only 7 of the 14 alleles were thought to be independent (supplemental Table 1; supplemental Figure 1).
The subgroup analyses were performed under stratification for HLA-DPB1 mismatch to avoid false positives arising from the heterogeneity of the population. We initially identified 3 discrete positive loci (P < 10−8) for 7 HLA subgroups (Table 2) and validated the observed or imputed genotypes of SNPs within the selected positive loci across the entire cohort using the MassARRAY system (Agena Bioscience, San Diego, CA), in which we obtained an excellent concordance (99.4%) with SNP array-based genotyping for all SNPs examined (supplemental Table 2).
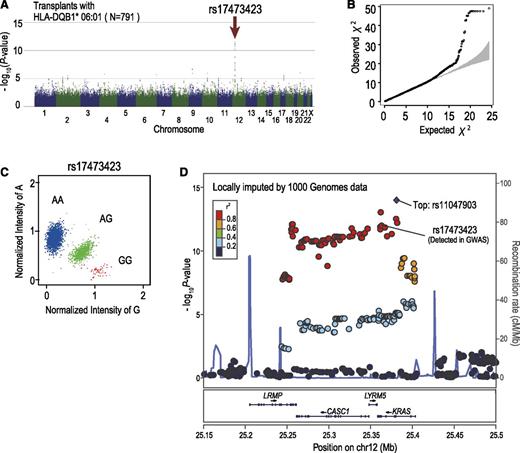
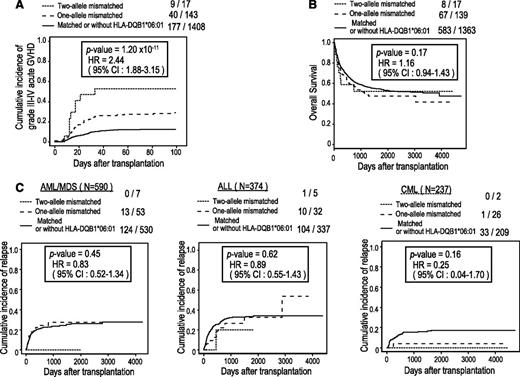
Among these 3 loci, the strongest association was observed at rs17473423 on 12p12.1 for the subgroup sharing the DQB1*06:01 allele, whereby the allele mismatch was significantly associated with grade III-IV aGVHD, showing an HR of 2.44 (95% CI, 1.88-3.15; P = 1.20 × 10−11) after competing risk was taken into consideration. The association remained significant (P = 5.9 × 10−11), even after adjustment for the effects of known risk factors (Table 3). Significant associations at the same locus were also identified for 3 other HLA alleles with similar HRs, including C*12:02, B*52:01, and DRB1*15:02. This result was expected because these HLA alleles compose the most prevalent haplotype in the Japanese population (A*24:02-C*12:02-B*52:01-DRB1*15:02-DQB1*06:01), and except for A*24:02, which is also shared by other HLA haplotypes, they show near-complete linkage with DQB1*06:01. Because of the strong linkage among the 4 highly correlated HLA alleles, it was difficult to determine which of these HLA alleles would be responsible for the presentation of the antigen relevant to aGVHD development. As for the putative minor H antigen epitope, the LD block (∼150 kb) contained 4 genes, including CASC1, KRAS, LRMP, and LYRM5, in which no candidate protein-coding SNP for the minor H antigen epitope was identified in the HapMap data. We further interrogated the putative epitope by imputing unobserved SNPs using the data from the 1000 Genomes Project (Figure 4D).34 We found 6 SNPs within the 3′ untranslated region or intronic sequence of KRAS or LRMP. However, showing significantly lower P values than those of rs17473423, these SNPs were not likely to directly regulate the expression of the relevant epitope. Based on the estimate for rs17473423, 20.5% of the transplants with the relevant HLA subtypes, or 10.2% of the entire cohort, carried the allele mismatch at the relevant locus. Despite the significant association with grade III-IV aGVHD, the allele mismatch at this locus did not substantially affect the overall survival or leukemic relapse (Figure 5).
Significant association with grade III-IV aGVHD detected in 12p12.1. Manhattan plot (A) and QQ plot (B) of GWAS analysis. A log-rank trend test statistic was calculated to compare the cumulative rates of grade III-IV aGVHD, in which analyses were performed based on the number of allele mismatches between donor and recipient. Under the assumption that the minor H antigen was presented by HLA-DQB1*06:01, association was tested within the subgroup of transplants sharing HLA-DQB1*06:01. In the QQ plot, 95% CIs are depicted in gray. (C) Cluster plot of rs17473423 showing the highest significance. Specimens genotyped as AA, AG, and GG are shown in blue, green, and red, respectively. (D) Regional association plots are depicted in which unobserved SNPs were imputed using the data from the 1000 Genomes Project. Genetic recombination rates estimated using genotyping data from the 1000 Genomes Project are depicted by a light blue line; r2 values between SNPs and the most significantly associated SNP (rs11047903; “Top”) are color-coded.
Significant association with grade III-IV aGVHD detected in 12p12.1. Manhattan plot (A) and QQ plot (B) of GWAS analysis. A log-rank trend test statistic was calculated to compare the cumulative rates of grade III-IV aGVHD, in which analyses were performed based on the number of allele mismatches between donor and recipient. Under the assumption that the minor H antigen was presented by HLA-DQB1*06:01, association was tested within the subgroup of transplants sharing HLA-DQB1*06:01. In the QQ plot, 95% CIs are depicted in gray. (C) Cluster plot of rs17473423 showing the highest significance. Specimens genotyped as AA, AG, and GG are shown in blue, green, and red, respectively. (D) Regional association plots are depicted in which unobserved SNPs were imputed using the data from the 1000 Genomes Project. Genetic recombination rates estimated using genotyping data from the 1000 Genomes Project are depicted by a light blue line; r2 values between SNPs and the most significantly associated SNP (rs11047903; “Top”) are color-coded.
Association of SNP rs17473423 with aGVHD, survival, and relapse. (A) Cumulative incidence of grade III-IV aGVHD. The P value and HR were calculated in a competing risk regression analysis in which death before aGVHD was considered a competing risk. (B) Kaplan-Meier curves for overall survival according to the number of mismatching alleles. The P value and HR were calculated by Cox regression analysis. (C) Cumulative incidence curves of relapse of AML/MDS (left), ALL (middle), and CML (right) in which death before relapse was considered a competing risk.
Association of SNP rs17473423 with aGVHD, survival, and relapse. (A) Cumulative incidence of grade III-IV aGVHD. The P value and HR were calculated in a competing risk regression analysis in which death before aGVHD was considered a competing risk. (B) Kaplan-Meier curves for overall survival according to the number of mismatching alleles. The P value and HR were calculated by Cox regression analysis. (C) Cumulative incidence curves of relapse of AML/MDS (left), ALL (middle), and CML (right) in which death before relapse was considered a competing risk.
Other significant associations were identified for grade III-IV aGVHD: at rs9657655 in 9q31.3 with HLA B*44:03-C*14:03 (P = 7.00 × 10−10) and at rs12206927 in 6q16.2 with C*07:02 (P = 6.97 × 10−9) (Table 2; supplemental Figure 2), although no significant peaks were detected for grade III-IV aGVHD. The HR values for grade III-IV aGVHD were 3.31 (95% CI, 2.02-5.42) and 3.68 (95% CI, 2.40-5.64) for mismatch at rs9657655 and rs12206927, respectively. However, when multivariate analysis was performed for these loci, the significance at rs9657655 was substantially reduced (P = 9.3 × 10−3), although rs12206927 remained significant (P = 4.0 × 10−9) (Table 3). No known genes have been mapped within the corresponding LD blocks (supplemental Figure 2). The mismatches at rs9657655 and rs12206927 were expected to occur in 12.3% and 11.9% of transplants sharing the corresponding HLA alleles or in 2.5% and 2.5% of all transplants, respectively. However, again, the high rate of GVHD did not appear to affect overall survival or disease relapse (supplemental Figure 2). No significant association was detected when the development of grade II-IV aGVHD was used as an end point.
Tissue-specific effects of mismatched alleles
Detection of positive association was critically affected by the grade of aGVHD used as an end point. To examine this result in more detail, we evaluated the correlations between organ-specific aGVHD stages and the number of mismatched alleles at the detected positive loci. The disparity at rs6937034 was associated with stage 2-3 lesions in the skin but was not significantly associated with stage 4 skin lesions or with aGVHD in the liver and the intestine (supplemental Figure 3, left panel). Thus, the association was captured only in the analysis using grade II-IV aGVHD as an end point. In the case of the disparity at rs17473423, there was no significant increase in stage 2-3 skin lesions or stage 1 lesions in the liver and the intestine, but the disparity was significantly associated with stage 4 skin and stage 2-4 intestinal lesions (Figure 6). Similarly, the disparity at rs12206927 was associated with stage 4 skin, stage 2-4 liver, and stage 2-4 intestine lesions, whereas stage 2-3 skin and stage 1 liver and intestine lesions were not substantially affected (supplemental Figure 3, right panel). Thus, the association was seen only with grade III-IV aGVHD and not with grade II-IV aGVHD.
Distinct effects of rs17473423 allele mismatch on different target organs. Relative frequency of transplants showing the indicated organ stages of GVHD having an increasing number of allele mismatches of rs17473423 for targeted SNPs for corresponding HLA subtypes.
Distinct effects of rs17473423 allele mismatch on different target organs. Relative frequency of transplants showing the indicated organ stages of GVHD having an increasing number of allele mismatches of rs17473423 for targeted SNPs for corresponding HLA subtypes.
Discussion
aGVHD remains one of the major complications negatively affecting the outcome of allo-HSCT.6 However, current knowledge about the molecular pathogenesis of aGVHD is still incomplete, especially with respect to the responsible minor H antigens in HLA-matched transplants. Most minor H antigens reported thus far have been isolated as the targets of recipient-derived cytotoxic T cells that recognized recipients’ leukemic cells. They were, accordingly, more likely to be associated with GVL than with GVHD.35,36 Alternatively, several antigens have been identified by testing the genetic association of candidate polymorphic alleles with GVHD.3 Genetic association has also been used to establish links between candidate donor/recipient variants and the development of GVHD.37-42 However, the lack of reliable criteria for the selection of plausible candidates has largely limited its application to unbiased detection of causative variants.23,43
Recently, GWAS has been established as a method for unbiased detection of genetic variants associated with a phenotype of interest and has been successfully applied to the identification of disease-associated loci.44 In the present study, we demonstrated that GWAS could also be applied to the detection of histocompatibility loci at which allele mismatches conferred a risk of severe aGVHD. We first performed GWAS analysis under the assumption of no HLA restriction and successfully detected the known allele mismatch for HLA-DPB1 as a risk of grade II-IV aGVHD, providing proof of concept that GWASs involving donor and recipient genotypes can successfully capture clinically relevant allele mismatches for histocompatibility. Moreover, using common HLA subtypes in the assumption of HLA restriction for antigen presentation, we identified previously unreported associations at 12p12.1 and 6q16.2. The results were affected by the end points used for GWAS because the target organs and the severity of GVHD in each organ were different depending on the mismatched loci, most likely reflecting tissue-specific expression of the relevant minor H antigens and different antigenicity.
Our results indicate that we should, whenever possible, avoid transplantation when donors show disparities at the loci we have just identified in the corresponding HLA context. In general, GWAS is expensive and time consuming. Moreover, GWAS in a population having higher ethnic heterogeneity with respect to allo-HSCT may lead to more statistical noise than expected for more homogeneous populations like the Japanese. However, when a large GWAS is conducted to identify allele mismatches associated with major HLA restrictions, the result could be widely used in many transplantation centers for better donor selection from relatives or through a nationwide donor program to minimize the risk of severe GVHD. However, this strategy would impose some limitations on donor selection, whereby otherwise eligible donors are excluded owing to disparity at certain high-risk loci, or could cause a dilemma if we are forced to choose a donor showing disparity at relevant loci, especially when no other HLA-matched donors are available. Nevertheless, the present result is of potential clinical significance, in that we could evaluate the risk of otherwise unpredictable life-threatening grade III-IV aGVHD by genotyping these SNPs in the donor and recipient. In addition, the limitation could be partly mitigated, given that the putative minor H locus is associated with the most common HLA haplotypes found in the Japanese population,45 allowing the identification of more potential candidate donors, and given that the disparity is expected to be observed for <20.5% of such candidates.
Although we supposed that the loci detected in this study would correspond to the relevant minor H antigens involved in severe aGVHD, we could not identify any protein-coding units predicted to undergo amino acid alterations in consequence of the SNPs identified by the GWAS analysis or other SNPs tightly linked to these SNPs within the corresponding LD blocks. Thus, the precise polymorphic epitopes for the putative minor H antigens that correspond to these positive loci remain to be elucidated. Several mechanisms have been implicated in altered antigenicity or expression of minor H antigens caused by an SNP, such as cryptic start codons, the presence of unknown genes using different reading frames, and other SNPs/polymorphisms within regulatory regions affecting gene expression. Thus, we still cannot exclude the possibility that the detected SNPs were responsible for the presentation of some minor H antigens to cause severe GVHD.22 In addition, the strong LD associated with common HLA haplotypes in the Japanese population45 largely prevented the determination of the HLA types that are responsible for the presentation of the putative minor H antigens implicated in the detected association. Future studies are warranted to determine the precise epitopes of the relevant minor H antigens and their tissue distribution, and the corresponding HLA types with which they are presented.
There are several potential caveats to be considered when applying GWAS to the identification of GVHD-associated histocompatibility antigens. First, it is not a simple genotype in donors or recipients but the disparity of histocompatibility defined by 2 independent genotypes for the 2 individuals that is tested for association with aGVHD, which could potentially compromise the statistical power to capture the responsible loci, as compared to typical GWASs. For a simple genotype, the power of capturing the causative SNP using another marker SNP in a GWAS is attenuated by a factor of r2, where r is a coefficient of correlation between the 2 SNPs (0 ≤ r ≤ 1).46 However, when the association is tested for a combination of 2 independent genotypes, the factor will be further decreased by a factor of ∼r,4 especially in poorly covered genomic regions. Second, under the assumption of HLA restriction for antigen presentation, the number of mismatched transplants will be substantially reduced, further compromising power.40,41 Despite this drawback, the presence of major HLA subtypes found in the Japanese population at relatively high frequencies enabled us to interrogate relevant mismatched loci with acceptable power, even when HLA restriction was assumed.
Some minor H antigens such as HA-122,43 are recognized by donor T cells only when the relevant alleles are owned by the recipient. The presence of these minor H antigens showing unidirectional antigenicity, which we did not assume in the current study, could also compromise the power, although the reduction of power would not be prominent, so long as the minor allele frequencies of those SNPs remain low, whereas they could become substantial with increasing minor allele frequencies. Another caveat comes from the fact that we performed trend tests based on the numbers of allele mismatches, assuming that the expression level of mismatched minor H antigens would affect the severity of aGVHD. However, even in the case of the presence of biallelic mismatch, GVHD may be eased off if the donor T cells are suppressed by the recipients’ residual T cells in the direction of rejection. Such a response would not be induced in the case of only 1 allele mismatch because the recipient carries a donor allele in the GVH direction (Figure 1B).
Lastly, because of the very large number of SNP loci tested, there is a chance of detecting those loci whose mismatch distribution within the study cohort coincidentally conformed to that of 1 or more known risk factors, generating a false-positive association. The probability is particularly increased for a smaller cohort size, with a higher frequency of cases having the confounding risk. In fact, 1 of the 3 loci initially detected in the GWAS is likely to be coincidentally confounded by the use of a TBI-containing regimen. To avoid such false-positive findings, the initial findings of the GWAS should be validated, taking known risks into consideration. Unfortunately, it may not be realistic to take into account all possible confounding factors, again suggesting the importance of the biological confirmation of the relevance of our GWAS findings.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all the medical staff who participated in transplants through JMDP or worked at the data center.
This work was supported by the Core Research for Evolutional Science and Technology, Japan Science and Technology Agency; a Grant-in-Aid from the Ministry of Health, Labor, and Welfare, Japan (H23-Immunology-010) and the Ministry of Education, Culture, Sports, Science, and Technology, Japan (3224-22133002, 22133003, 22133009, 22133011); and the Practical Research Project for Allergic Disease and Immunology (Research on Technology of Medical Transplantation), supported by the Japan Agency for Medical Research and Development.
Authorship
Contribution: A.S.-O. performed large-scale genotyping and GWAS analysis and wrote the manuscript; Y.O., M. Sanada, G.C.K., and Y.N. performed large-scale genotyping and data analysis; K.K. and M. Satake performed high-resolution HLA typing; M.O., K.K., F.A., H.I., M. Satake, and Y.M. assisted with sample preparation, data control, and DNA banking; A.S.-O., Y.A., S.C., H.S., S.M., K.Y., Y.K., Y.M., and T.S. designed the study and discussed the data; and S.O. led the entire project and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Seishi Ogawa, Pathology and Tumor Biology, Graduate School of Medicine, Kyoto University, Yoshida-Konoe-cho, Sakyo-ku, Kyoto 606-8501, Japan; e-mail: sogawa-tky@umin.ac.jp.