Key Points
Integration of genome-wide copy number and whole transcriptome data identifies key mutational events in the pathogenesis of DLBCL.
Genomic deletions in RCOR1 are associated with a specific gene expression signature and with unfavorable clinical outcomes in DLBCL patients.
Abstract
Effective treatment of diffuse large B-cell lymphoma (DLBCL) is plagued by heterogeneous responses to standard therapy, and molecular mechanisms underlying unfavorable outcomes in lymphoma patients remain elusive. Here, we profiled 148 genomes with 91 matching transcriptomes in a DLBCL cohort treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisolone (R-CHOP) to uncover molecular subgroups linked to treatment failure. Systematic integration of high-resolution genotyping arrays and RNA sequencing data revealed novel deletions in RCOR1 to be associated with unfavorable progression-free survival (P = .001). Integration of expression data from the clinical samples with data from RCOR1 knockdowns in the lymphoma cell lines KM-H2 and Raji yielded an RCOR1 loss–associated gene signature comprising 233 genes. This signature identified a subgroup of patients with unfavorable overall survival (P = .023). The prognostic significance of the 233-gene signature for overall survival was reproduced in an independent cohort comprising 195 R-CHOP-treated patients (P = .039). Additionally, we discovered that within the International Prognostic Index low-risk group, the gene signature provides additional prognostic value that was independent of the cell-of-origin phenotype. We present a novel and reproducible molecular subgroup of DLBCL that impacts risk-stratification of R-CHOP-treated DLBCL patients and reveals a possible new avenue for therapeutic intervention strategies.
Introduction
Diffuse large B-cell lymphoma (DLBCL), the most common type of non-Hodgkin lymphoma, accounts for approximately 30% to 40% of all new lymphoma cases. DLBCL comprises at least 2 major molecular subtypes, reflecting the phenotype of the hypothetical cell of origin (COO): (1) activated B cell–like (ABC), derived from B cells exiting (or poised to exit) the germinal center; and (2) germinal center B cell–like (GCB), derived from B cells found in the germinal center.1 The advent of high-throughput sequencing has led to the discovery of various somatic genomic mutations linked to the pathogenesis of DLBCL. These include genomic copy number (CN) changes such as arm-length deletions of chromosome 6q, gains of chromosomes 3 and 18q,2,3 focal deletions of CDKN2A,4 and gains of REL,5 in addition to recurrent somatic point mutations in CREBBP, EP300, EZH2, and MLL26-9 that collectively implicate disruption of chromatin modification as a defining feature.
Despite the increasing knowledge base of genomic and transcriptomic abnormalities, a major clinical challenge persists: approximately 40% of DLBCL patients receiving standard therapy of rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisolone (R-CHOP) are not cured.10 The biological correlates of treatment failure are not well understood, and the variability in clinical outcomes cannot be explained by our current knowledge of the mutational landscape. Thus, additional biological heterogeneity and related associations to treatment outcomes remain to be discovered.
We set out to uncover prognostically significant molecular subgroups of DLBCL through simultaneous interrogation of the genomic and transcriptomic dimensions of tumors from a uniformly treated population of patients for whom comprehensive clinical follow-up data were available. We sought to identify somatic CN alterations with measureable impact on gene expression of transcriptional networks as primary candidates for functional and prognostic alterations.11 Although similar data sets have been generated by other groups,2,3,12-14 most studies have been restricted by small cohorts, low data resolution, and/or lack of association with clinical outcomes in the current era of R-CHOP therapy. The most recent integrative study15 presented the largest and highest resolution data set to date and focused on the synergy between aberrations in p53 and cell-cycle components. However, associations between gene-centric CN aberrations, clinical outcomes, and gene expression patterns were not described.
Here, we identify novel focal and recurrent CN changes in RCOR1 that are associated with a prognostically significant gene expression signature. This RCOR1 loss–associated gene signature identifies a subgroup of DLBCL patients with unfavorable overall survival (OS), both in our discovery cohort and in an independent cohort.16 Taken together, we have identified RCOR1 as a critically deregulated gene in DLBCL pathogenesis and identified a consequent gene expression signature as a novel risk-associated molecular profile in R-CHOP-treated patients.
Methods
Additional detailed methods are presented in supplemental Methods on the Blood Web site.
Patient cohorts
We assembled a cohort of 151 diagnostic DLBCL samples (supplemental Figure 1) from the tissue repository of the Center for Lymphoid Cancer at the British Columbia Cancer Agency and Arizona Lymphoid Tissue Repository (“BCCA study cohort”). We generated 151 high-resolution CN microarrays (Affymetrix SNP 6.0) from the BCCA study cohort samples, of which 148 passed quality control (supplemental Methods). A subset of patients corresponding to 139 of these samples was treated with combined immunochemotherapy (R-CHOP/R-CHOP-like therapy [“R-CHOP cohort”]) and used in outcome correlation analysis. Analysis of clinical characteristics (age, stage, performance status, extranodal sites, lactate dehydrogenase, and the derived International Prognostic Index [IPI] score) indicated that the R-CHOP cohort (n = 139) and the BCCA study cohort (n = 148) were representative of the DLBCL R-CHOP-treated population at the BCCA (Table 1 and supplemental Table 1). Previously published gene expression profiles were matched to 91 samples by RNA sequencing (RNA-seq) analysis6 ; of these samples, 85 were treated with R-CHOP/CHOP-like therapy. Two rediscovery cohorts were used from the studies of Lenz et al16 and Monti et al.15 The research was approved by the local ethics board and was conducted in accordance with the Declaration of Helsinki.
CN analysis
Affymetrix SNP 6.0 microarrays were used to profile the CN architecture. The SNP 6.0 microarrays were preprocessed using the PennCNV-Affy17 protocol. OncoSNP18 was then used to simultaneously segment and predict CN data. The CN states and logR were subsequently projected onto gene locations (Ensembl version 72), followed by filtering of CN polymorphisms. To evaluate for enrichment for CN aberrations in either the ABC or the GCB subtype, a Fisher’s exact test was performed. The Genomic Identification of Significant Targets in Cancer (GISTIC) algorithm19 was run to find commonly CN-aberrated regions.
FISH experiments
Fluorescence in situ hybridization (FISH) analysis was performed on fixed cell suspensions (methanol/acetic acid) or nuclei extracted from formalin-fixed paraffin-embedded DLBCL samples according to standard protocols as described elsewhere using in-house bacterial artificial chromosome clones.20
Gene expression analysis in clinical samples
RNA-seq libraries6 matching the CN cohort were available to quantify the gene expression levels in 91 samples. These libraries were aligned using the Genomic Short-Read Nucleotide Alignment Program (GSNAP) split-read aware aligner.21 Gene expression values were generated using the metric reads per kilobase of transcript per million reads mapped and then combined across all samples to form a gene expression matrix followed by log2 transformation and quantile normalization. Coexpression was predicted using the Spearman rank correlation test.
Integrative CN and gene expression analysis
Cis/trans correlations of CN and gene expression data were performed at the gene-centric level using the Spearman rank correlation test and the Kruskal-Wallis test.
Prediction of driver CN-aberrated genes was performed using DriverNet.11 In brief, a genomic aberration is correlated with the expression of genes it interacts with through highly curated transcriptional networks published in literature. The greater the impact a genomic alteration has on the genes it interacts with, the higher the probability that the genomic aberration has a functional role in the disease.
IHC on primary lymphoma and reactive tonsil samples
Immunohistochemistry (IHC) was performed on formalin-fixed paraffin-embedded tissue samples of 68 DLBCL cases with matching CN data, of which 40 had matching gene expression data. A reactive tonsil specimen was used to assess the staining pattern in the germinal center. Four-micrometer sections of tissue microarrays or whole tissue sections were stained with an anti-RCOR1 antibody (clone S72-8, LSBio, Seattle, WA; dilution 1:500) using routine protocols for automated procedures on the Ventana Benchmark XT (Ventana Medical Systems, Tucson, AZ).
Virus production, transduction and transcript expression
Stable RCOR1 knockdowns were generated in the classical Hodgkin lymphoma–derived cell line KM-H2 (KM-H2 Cl2 KD, KM-H2 Cl5 KD) and the Burkitt lymphoma–derived cell line Raji (Raji OB KD) using lentiviral transduction of a vector expressing a small hairpin RNA that, after being processed to mature small interfering RNA, interferes with RCOR1 messenger (m)RNA (Open Biosystems pGIPZ system). For comparison, nonsilencing lentiviral control plasmids were used (KM-H2 NS, Raji OB NS). Additionally, we used Sigma’s MISSION small hairpin RNA system on the Raji cell line (Raji M KD) and the respective nonsilencing controls (Raji M NS). Transductions were carried out following standard protocols using a multiplicity of infection of 10 and predetermined puromycin selection. One week after transduction, cells from time-matched samples were harvested for RNA and protein extraction. Knockdown was evaluated by measuring residual expression of the transcript by quantitative reverse transcriptase–polymerase chain reaction. Protein levels were assessed by western blot analysis using an RCOR1 antibody (Abcam ab183711), including a β-actin loading control.
Gene expression analysis of in-vitro knockdown cells
RNA-seq libraries were generated for each of the in-vitro RCOR1 knockdown clones (4 biological replicates: KM-H2 Cl2 KD, KM-H2 Cl5 KD, Raji OB KD, Raji M KD) and their matching nonsilencing control (KM-H2 NS, Raji OB NS, Raji M NS). The RNA-seq libraries were generated as per previous protocols6 and preprocessed using the same methodology as stated in the “Gene expression analysis in clinical samples” section. Differential expression was calculated using fold-change difference using a threshold of 0.3 for RCOR1 knockdowns vs nonsilencing controls.
RCOR1 loss–associated gene signature analysis
Genes classified as being differentially expressed in the in-vitro RCOR1 knockdown experiments and genes coexpressed with RCOR1 were intersected, followed by filtering to include only genes that were concordant in their up and down directionality, resulting in the RCOR1 loss–associated gene signature (supplemental Table 8). Next, hierarchical unsupervised clustering was performed on the BCCA RNA-seq cohort using the RCOR1 loss–associated gene signature. The Ward criteria were used for linking the clusters. The sample cluster dendrogram was cut to form 3 distinct clusters (supplemental Figure 15). The cluster with the lowest average RCOR1 expression was defined as the RCOR1-low cluster. The cluster with the highest average RCOR1 expression was defined as the RCOR1-high cluster. The remaining cluster was defined as the unclassified cluster.
For the analysis of external cohorts, only the genes in the RCOR1 loss–associated signature were carried over; thus, this strategy was defined as “rediscovery” rather than validation. These genes were used to perform a de novo clustering in these cohorts, followed by defining of the clusters using the same methodology as the study cohort (ie, cluster with the lowest average RCOR1 expression was defined as the RCOR1-low cluster).
Survival analysis
Survival analyses were performed at the gene-centric CN level by dichotomizing samples into deletions vs CN neutral. CN neutral loss-of-heterozygosity samples were excluded from outcome correlation analyses. The log-rank test was used to test whether outcomes were different between groups using OS, DSS, and PFS as end points. OS was defined as death from any cause. DSS was defined as death specifically from lymphoma. PFS was defined as the time from initial diagnosis to disease progression, lymphoma relapse, or death from any cause.
Similarly, for the RCOR1 loss–associated gene signature, we used the log-rank test comparing OS of patients in the RCOR1-low vs RCOR1-high expression clusters. To test for the prognostic independence of the RCOR1 aberrations and gene signature from the IPI and COO, we performed pairwise multivariate Cox regression.
Pathway enrichment analysis
Pathway enrichment analysis was performed using the Reactome FI Cytoscape Plugin (version 2013).22
Data availability
Affymetrix SNP 6.0 data have been deposited in the European Genome-Phenome Archive database (accession number EGAS00001001000).
Results
High-resolution CN analysis of DLBCL
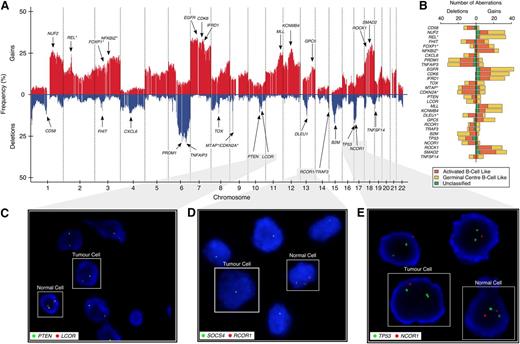
Genomic gains and losses were profiled in the 148-sample BCCA study cohort (Figure 1A). On average, 16.8% of the genome—affecting 3106 (15.3%) protein coding genes—was aberrant per sample (the full distributions are shown in supplemental Figure 2). We confirmed previously reported, highly recurrent, large-scale chromosome alterations, including gains of the entire chromosome 7, COO-specific deletions of 6q, and gains of chromosome 3 and 18q (enriched in ABC-DLBCL)2,3 (supplemental Figure 3). Gains in REL (n = 34; 22.9% of patients)5 were enriched in GBC-DLBCL (P < .001; Figure 1B), whereas gains in FOXP1 (n = 27; 18.2%, P = .03) and NFKBIZ (n = 31; 21%, P < .001)13 were enriched in ABC-DLBCL samples. We also observed deletions in CDKN2A/MTAP (n = 33/29; 22.3%/19.6%, P = .002/P = .003 enriched in ABC-DLBCL),4 TNFAIP3 (n = 43; 29.1%),7 CD58 (n = 18; 12.2%), B2M (n = 26; 17.6%),15 FHIT (n = 15; 10.1%),23 and PRDM1 (n = 43; 29.1%),24 corroborating previous reports. Commonly affected regions can be found in supplemental Figure 4 and supplemental Tables 2 and 3.
Genome-wide CN architecture of 148 DLBCL patients. (A) A genome-wide linear representation of the CN profile, summarized at the gene level, across all 148 samples, with gains in red, and deletions in blue. Selected genes with recurrent somatic genomic aberrations are annotated with arrows. Germline CN polymorphisms have been subtracted from this plot. (B) Stacked horizontal-bar plot indicating the absolute number of gains (on right) and deletions (on left) of the selected genes from panel A. Different colors represent the aberration distribution according to COO classification, with asterisks indicating whether there is an enrichment of the aberration in a particular subtype. (C-E) Two-color FISH assays, with red probe interrogating a target gene of interest (LCOR, RCOR1, and NCOR1) and green probe interrogating an established tumor suppressor on the same chromosomal arm (PTEN, SOCS4, and TP53) as a reference.
Genome-wide CN architecture of 148 DLBCL patients. (A) A genome-wide linear representation of the CN profile, summarized at the gene level, across all 148 samples, with gains in red, and deletions in blue. Selected genes with recurrent somatic genomic aberrations are annotated with arrows. Germline CN polymorphisms have been subtracted from this plot. (B) Stacked horizontal-bar plot indicating the absolute number of gains (on right) and deletions (on left) of the selected genes from panel A. Different colors represent the aberration distribution according to COO classification, with asterisks indicating whether there is an enrichment of the aberration in a particular subtype. (C-E) Two-color FISH assays, with red probe interrogating a target gene of interest (LCOR, RCOR1, and NCOR1) and green probe interrogating an established tumor suppressor on the same chromosomal arm (PTEN, SOCS4, and TP53) as a reference.
Dysregulated transcriptional networks identified by integration of CN and gene expression data
We integrated the gene expression profiles of 91 matching RNA-seq libraries to investigate CN alterations impacting gene expression profiles. We estimated 22.1% of protein-coding genes to be cis-correlated (Spearman rank correlation test and Kruskal-Wallis test; false discovery rate <0.1) (supplemental Tables 4 and 5). These cis-correlated genes were enriched for the biological processes neurotrophin signaling pathway, B-cell receptor signaling pathway, signaling events mediated by histone deacetylase (HDAC) class I, and class I major histocompatibility complex–mediated antigen processing and presentation (supplemental Table 6).
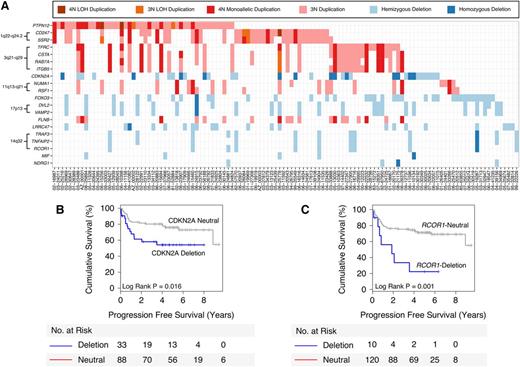
To pinpoint candidate functional aberrations, we performed a DriverNet analysis,11 which links CN and gene expression data through known transcriptional networks (trans correlations). The top 10 candidate deleted genes and the top 10 candidate gained genes that were predicted to significantly impact the expression of their cognate genes are listed in Figure 2A. These genes included the known tumor suppressor CDKN2A, as well as genomic loci that harbored multiple candidate genes 1q22-q24.2 (CD247, SSR2), 3q21-q29 (TFRC, CSTA, RAB7A, ITGB5), 11q13-q21 (NUMA1, RSF1), 14q32 (RCOR1, TRAF3, TNFAIP2), and 17p13 (DVL2, VAMP2).
Top 20 candidate genes selected by DriverNet. (A) Matrix showing the top 20 DriverNet genes (in rows) and samples containing genomic aberrations in any one of these genes (in columns). To simplify the visualization, any gains affecting deleted genes and any deletions affecting gained genes have been removed. (B-C) Kaplan-Meier analyses for CDKN2A and RCOR1 deletions, respectively, demonstrate an association with poor PFS.
Top 20 candidate genes selected by DriverNet. (A) Matrix showing the top 20 DriverNet genes (in rows) and samples containing genomic aberrations in any one of these genes (in columns). To simplify the visualization, any gains affecting deleted genes and any deletions affecting gained genes have been removed. (B-C) Kaplan-Meier analyses for CDKN2A and RCOR1 deletions, respectively, demonstrate an association with poor PFS.
RCOR1 deletions define a subgroup of DLBCL patients with unfavorable survival in a homogenously R-CHOP-treated cohort
We investigated the prognostic ability of the DriverNet-identified aberrations using clinical outcome data available for the 139 R-CHOP-treated DLBCL patients. For genes with ≥5% aberration frequency, CDKN2A deletions were associated with unfavorable 5-year PFS (54.5% deleted vs 76.3% neutral, P = .016; Figure 2B), DSS (P = .006; supplemental Figure 5E), and OS (P = .016; supplemental Figure 5I) as previously reported,4 and the set of genes located in the 14q32 locus were associated with 5-year PFS: RCOR1 (22.5% deleted vs 71.1% neutral, P = .001; Figure 2C), TRAF3 (18.3% deleted vs 72% neutral, P < .001; supplemental Figure 5C), and TNFAIP2 (18.3% deleted vs 72% neutral, P < .001; supplemental Figure 5D). Similar statistical trends were found using the end points DSS and OS (supplemental Figure 5): RCOR1 (DSS = 0.037; OS = 0.093), TRAF3 (DSS = 0.003; OS = 0.01), and TNFAIP2 (DSS = 0.003; OS = 0.01).
Given the known role of RCOR1 in chromatin modification, we pursued this gene in our study. Further analysis revealed an association between deletion of the transcriptional corepressor and PFS that was independent of standard prognostic risk factors included in the IPI (P = .005) and COO phenotyping (P = .005) using pairwise multivariate Cox regression (supplemental Table 7). RCOR1 deletions were significantly correlated with gene expression (supplemental Figure 6) and protein expression by IHC (supplemental Figure 7C-D), with representative RCOR1 IHC staining cases shown in supplemental Figure 8. Additionally, protein expression was correlated with gene expression (supplemental Figure 7E), and low protein expression was associated with unfavorable survival (supplemental Figure 9). Lastly, IHC on a benign reactive tonsil revealed strong nuclear staining of the germinal center cells compared with the mantle zone cells that were negative (supplemental Figure 8D), suggesting that the RCOR1 deletions would have a pathogenic consequence because their normal counterparts express RCOR1.
Recurrent deletions in members of the corepressor family
In addition to deletions in RCOR1 (n = 11, 7.5%), we also observed recurrent deletions in members of the corepressor gene family: LCOR (n = 13, 8.8%) and NCOR1 (n = 21, 14.2%). We also analyzed CN data from an independent DLBCL cohort (n = 77),7 identifying recurrent deletions in these 3 genes (RCOR1: n = 6, 7.8%; LCOR: n = 2, 2.6%; and NCOR1: n = 15, 19.5%). Focal examples are shown in Figure 3 and supplemental Figure 10.
Focal view of raw CN values. Each panel contains raw CN values from 2 samples. The top sample is from the BCCA study cohort and the bottom sample is from the Pasqualucci et al7 cohort. (A) Focal view of chr14:102,058,998-104,377,837 demonstrating predicted RCOR1 deletions in samples 05-19287 and 00003883_ Columbia_GW6.0. (B) Focal view of chr10:97,592,017-99,740,800 demonstrating predicted LCOR deletions in samples 99-25549 and 00003861_Columbia_GW6.0. (C) Focal view of chr17:14,934,718-17,119,010 demonstrating predicted NCOR1 deletions in samples 03-26969 and 00003861_Columbia_GW6.0.
Focal view of raw CN values. Each panel contains raw CN values from 2 samples. The top sample is from the BCCA study cohort and the bottom sample is from the Pasqualucci et al7 cohort. (A) Focal view of chr14:102,058,998-104,377,837 demonstrating predicted RCOR1 deletions in samples 05-19287 and 00003883_ Columbia_GW6.0. (B) Focal view of chr10:97,592,017-99,740,800 demonstrating predicted LCOR deletions in samples 99-25549 and 00003861_Columbia_GW6.0. (C) Focal view of chr17:14,934,718-17,119,010 demonstrating predicted NCOR1 deletions in samples 03-26969 and 00003861_Columbia_GW6.0.
In selected index cases, deletions in LCOR (n = 6) and NCOR1 (n = 7), as well as in all RCOR1 (n = 11) were validated using FISH with a reference probe interrogating a known tumor suppressor on the same chromosomal arm (PTEN, TP53, SOCS4). Figure 1 shows 3 specific examples illustrating the focal nature of the observed deletions: a homozygous LCOR deletion (10q24.1; 13 kb; case 99-25549), PTEN neutral (10q23.31) (Figure 1C); a homozygous RCOR1 deletion (14q32.31; 143 kb; case 05-19287), hemizygous SOCS4 deletion (14q22.3) (Figure 1D); and a hemizygous NCOR1 deletion (17p12; 159 kb; case 01-19969), TP53 neutral (17p13.1) (Figure 1E). These examples suggest that the genomic deletions may have been selected for, independent of known proximal tumor suppressors in at least a subset of cases.
Although RCOR1, LCOR, and NCOR were all found to be the focal target of heterozygous or homozygous deletion events, none of these 3 genes were affected by single-point mutations in the RNA-seq cohort as previously reported.6 When analyzing mutational patterns including RCOR1, LCOR, and NCOR1 deletions (supplemental Figure 11A), we found that LCOR and NCOR1 deletions significantly co-occurred with other somatic mutations and CN aberrations, such as TP53 mutations (correlated with NCOR1) and FAS mutations (correlated with LCOR) (supplemental Figure 11B). However, RCOR1 deletions did not significantly co-occur with any other somatic mutations.
An RCOR1 loss–associated gene expression signature derived by in-vitro knockdown
Having established an association between outcomes and RCOR1 deletions, we sought to validate the effects of RCOR1 loss at the transcriptional level using in-vitro knockdown in the two B-cell lymphoma lines KM-H2 and Raji. Quantitative reverse transcriptase–polymerase chain reaction confirmed the reduction of RCOR1 in KM-H2 cells to 21.5% ± 0.1% and in Raji cells to 25.5% ± 0.02% compared with the nonsilencing controls (supplemental Figure 12A). Western blot analysis also confirmed the reduction of RCOR1 at the protein level (supplemental Figure 12B). Differential expression analyses between KM-H2 and Raji revealed a strong correlation in fold-change directionality (P < .001), as well as consistency in the dysregulated pathways (supplemental Figure 13), providing the confidence to combine the results of the 2 in-vitro knockdown B-cell lines to produce a single list of differentially expressed genes (n = 1588). This set of genes was significantly overlapping (P < .001) with the genes coexpressed with RCOR1 in the RNA-seq cohort (n = 1639) (supplemental Figure 14). We defined the list of the overlapping genes (n = 233) as the RCOR1 loss–associated gene signature (supplemental Table 8). This gene signature was enriched for biological processes that included upregulation of the proteasome, processing of capped intron-containing pre-mRNA, and downregulation of signaling events mediated by HDAC class II (supplemental Table 9).
The RCOR1 loss–associated gene expression signature is associated with unfavorable outcome
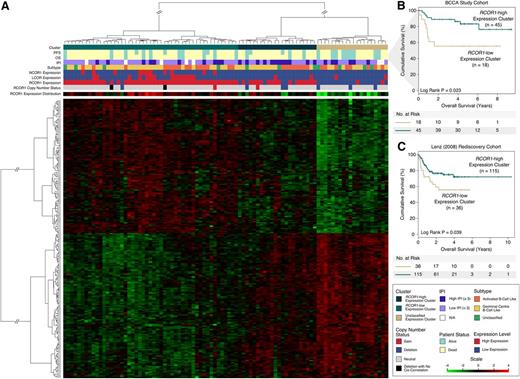
We next investigated whether the RCOR1 loss–associated gene signature correlated with outcomes following R-CHOP chemotherapy in our BCCA study cohort. Using the RNA-seq-derived expression measurements (from 91 patients) of the RCOR1 loss genes as features, we performed hierarchical clustering and found 3 distinct subgroups (Figure 4A and supplemental Figure 15), including a group of patients (n = 20, 22%) exhibiting low RCOR1 expression (defined as the RCOR1-low group). This subgroup demonstrated a differential gene expression profile distinct from another group of patients (n = 49, 53.8%) exhibiting high RCOR1 expression (defined as the RCOR1-high group; P < .001; supplemental Figure 16A), as well as high LCOR and NCOR1 expression (supplemental Figure 16B-C). A third group of patients (n = 22, 24.2%) demonstrated a mixture of the expression profile from both the RCOR1-low and RCOR1-high groups, and we defined this as the unclassified group. When placing the RCOR1 deletions in the context of the gene signature groups, we found that RCOR1 deletions trended toward clustering in the RCOR1-low group (P = .079). When considering also the unclassified group, the RCOR1 deletions clustered into either the RCOR1-low or unclassified group (P = .039). In the subgroup of R-CHOP-treated patients (n = 63), the RCOR1-low expression cluster showed unfavorable OS (Figure 4B) relative to the RCOR1-high expression clusters (5-year OS: 55.6% RCOR1-low vs 83.4% RCOR1-high, P = .023).
RCOR1 loss–associated signature is associated with unfavorable outcome. (A) The heat map produced from unsupervised clustering on the BCCA cohort using the RCOR1 loss–associated gene expression signature (n = 233). (B) Kaplan-Meier analyses performed on the RCOR1-low vs RCOR1-high expression clusters in the BCCA study cohort. (C) Kaplan-Meier analyses performed on the RCOR1-low vs RCOR1-high expression clusters in the Lenz rediscovery cohort. N/A, not available.
RCOR1 loss–associated signature is associated with unfavorable outcome. (A) The heat map produced from unsupervised clustering on the BCCA cohort using the RCOR1 loss–associated gene expression signature (n = 233). (B) Kaplan-Meier analyses performed on the RCOR1-low vs RCOR1-high expression clusters in the BCCA study cohort. (C) Kaplan-Meier analyses performed on the RCOR1-low vs RCOR1-high expression clusters in the Lenz rediscovery cohort. N/A, not available.
The prognostic association of the RCOR1 loss–associated gene signature was reproducible in an independent cohort of R-CHOP-treated patients from Lenz et al16 (Lenz cohort). This cohort included 233 samples (rediscovery cohort) with microarray-derived gene expression data and clinical data including OS but not PFS. The set of genes from the gene signature was carried over to this rediscovery cohort and used to perform de novo clustering. As per the BCCA study cohort, this rediscovery cohort was stratified into RCOR1-low (n = 53), RCOR1-high (n = 128), and unclassified (n = 52) gene expression clusters (supplemental Figure 17). After removing 38 overlapping samples from our study cohort and rediscovery cohort for outcome analysis, the RCOR1-low expression patients had unfavorable OS (Figure 4C) relative to patients in the RCOR1-high and unclassified expression clusters (5-year OS: 55.8% RCOR1-low vs 72.2% RCOR1-high, P = .039). To further test the prognostic value of the gene signature, it was tested in a second independent rediscovery cohort from Monti et al15 (n = 90; supplemental Figure 18A) that produced a statistical trend for prognostic significance (5-year OS: 47.1% RCOR1-low vs 72.6% RCOR1-high, P = .187; supplemental Figure 18B).
We next tested the prognostic value of the gene signature with respect to known prognostic markers such as COO and IPI using a multivariate Cox regression analysis. The prognostic value was independent of COO in our study cohort but was linked to IPI (supplemental Table 7). We investigated this further and observed that the gene signature adds prognostic value in the IPI low-risk group (P = .043; supplemental Figure 19A). In the Lenz cohort, we again observed an enhancement of the prognostic value within the IPI low-risk group (P < .001; supplemental Figure 19B) that was independent of COO (supplemental Table 7).
Taken together, the gene expression data from an orthogonal platform derived from 2 nonoverlapping cohorts of similarly treated patients confirm the prognostic value of the RCOR1 loss–associated gene expression signature.
Discussion
Using integrative analysis of high-resolution CN and RNA-seq data in a large cohort of DLBCL patients, we identified novel focal and recurrent deletions in the transcriptional regulator RCOR1 and established a prognostic signature of 233 genes that stratified patients into a distinct subgroup associated with reduced RCOR1 expression. Our methodology focused on identification of CN alterations that: (1) affected the mRNA expression of genes harbored within the regions of chromosomal imbalance; (2) led to genome-wide changes in transcriptional networks; and (3) were associated with clinical outcomes. Although this study focused primarily on CN alterations, additional candidate functional alterations could be revealed through integration with somatic point mutations and will be an important aspect in future studies.
From the inventory of CN alterations affecting gene expression, RCOR1 deletions stood out from other identified gene loci because these deletions were associated with a pronounced effect on key cellular pathways (DriverNet analysis) and unfavorable survival. Moreover, the RCOR1 loss–associated gene signature proved to be a prognostic indicator of survival that added prognostic value within the IPI low-risk group independent of COO.
Based on our validation work using FISH, we demonstrated the specificity of selected deletions that were independent of other known tumor suppressor gene loci in close proximity. The concept of synergistic tumorigenic effects of codeleted or coamplified genes on the same or different chromosomes has been widely assessed in lymphoma and other cancers.25-27 Indeed, RCOR1 deletions were associated with deletions of TRAF3 (located in close vicinity). TRAF3 is a key molecule in tumor necrosis factor α and Toll-like receptor signaling, acting as a negative regulator of nuclear factor–κB-inducing kinase.5,28 TRAF3 has also been identified as a target of somatic mutations and deletions in a number of cancers.7,29-31 We propose that when RCOR1 and TRAF3 are codeleted, the combination of transcriptional pattern changes mediated by RCOR1 loss and the downstream effects on alternative nuclear factor–κB signaling may cooperate and contribute to the malignant phenotype.
RCOR1 encodes a corepressor of the RE1-silencing transcription factor REST that binds to RE1 neuron-restrictive silencer elements to repress gene expression in nonneuronal cells.32,33 RCOR1 is part of the BRAF35–histone deacetylase complex, where it associates with the C-terminal domain of REST, the histone deacetylases 1 and 2 (HDAC1/2), and KDM1A, and regulates gene expression through chromatin remodeling.34-36 Further, NCOR1, a paralog of RCOR1, that we found deleted and cis-correlated in our cohort, binds to the N-terminus of REST, further recruiting HDAC1/2 to RE1/NRSE DNA-binding sites.37 When intersecting the in-vitro RCOR1 knockdown signature with RCOR1 coregulated genes in clinical samples to define the gene expression signature, we revealed gene enrichment in pathways associated with HDAC class II signaling events and processing of capped intron-containing pre-mRNA. Thus, global deregulation of gene expression is a likely consequence of RCOR1 loss.
Taken together and in 2 separate, independent cohorts, the identified outcome correlations of genomic RCOR1 deletions and the RCOR1 loss–associated gene signature suggest that these findings may be valuable as novel prognostic biomarkers in DLBCL patients. We suggest that the RCOR1 loss–associated gene signature as a biomarker is the best representation of the initial finding of RCOR1 deletions because it is stable (based on multiple gene features), reproducible (2 independent cohorts), and biologically meaningful (defined by RCOR1 in-vitro knockdown). This biologically defined RCOR1 loss–associated gene expression signature identified an RCOR1-low cluster that had unfavorable OS in both the BCCA and the rediscovery cohorts. Although the deletions tended to cluster with the RCOR1-low cluster, several cases in this cluster had low expression but had no RCOR1 deletion. Alternative molecular mechanisms such as promoter methylation along with other epigenetic modifications are possible explanations for the lack of expression in these cases and would need to be explored in future studies.
The predictive capacity of the RCOR1 loss–associated gene signature had added prognostic value in the IPI low-risk group in our study and in the Lenz cohort. Additionally, the added prognostic value in the IPI low-risk group was prognostically independent of COO classification. Thus, RCOR1 loss–related biology is likely to add prognostic value to COO identification in DLBCL patients. In conjunction with other emerging biomarkers such as COO subtype, a signature of RCOR1 loss could be combined in gene expression analyses to improve risk stratification. Lastly, we suggest that targeting the biology associated with RCOR1 loss provides a road map for improving therapeutic intervention in a poor-outcome subgroup of DLBCL patients.
Presented in part at the 54th annual meeting of the American Society of Hematology, Atlanta, GA, December 8-11, 2012.
The data reported in this article have been deposited in the European Genome-Phenome Archive database (accession number EGAS00001001000).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sarah Mullaly for comments on earlier versions of the manuscript.
This work was supported by Career Investigator Awards by the Michael Smith Foundation for Health Research (S.P.S. and C.S.) and by a Terry Fox Research Institute team grant (grant 1023) (C.S. and R.D.G.).
Authorship
S.P.S. and C.S. oversaw the project, designed the research, and wrote the manuscript; F.C.C. designed and performed the research, analyzed and interpreted the data, and wrote the manuscript; A.T. and S. Healy performed in-vitro knockdown experiments and analyzed the results; S.B.-N. performed FISH experiments and analyzed the results; A.M. performed IHC experiments and analyzed the results; R.L. and R.D.M. analyzed RNA-seq data; M.D. and S. Hu performed the experiments and analyzed the results; J.D. performed and analyzed the DriverNet results; G.H. provided CN analysis expertise; D.W.S. provided and interpreted the clinical data; R.K. performed the experiments; A.B. provided cis/trans analysis expertise; S.R. analyzed SNP 6.0 data; N.J. designed the research and generated SNP 6.0 data; L.M.R., L.S., and J.M.C. oversaw the collection of data; M.A.M. participated in the design of the original project and reviewed the manuscript; and R.D.G. designed the research, oversaw the collection of data, and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Steidl, Department of Lymphoid Cancer Research, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC, Canada V5Z 1L3; e-mail: csteidl@bccancer.bc.ca; and Sohrab P. Shah, Department of Molecular Oncology, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC, Canada V5Z 1L3; e-mail: sshah@bccrc.ca.
References
Author notes
S.P.S. and C.S. contributed equally to this study.