Key Points
OVOL2 is identified as a novel binding protein of ER71.
Interaction between ER71 and OVOL2 cooperatively regulates the generation of FLK1+ mesoderm, and endothelial and erythroid cells.
Abstract
In this study, we report that OVOL2, a C2H2 zinc finger protein, is a novel binding protein of ER71, which is a critical transcription factor for blood and vessel development. OVOL2 directly interacted with ER71, but not with ETS1 or ETS2, in the nucleus. ER71-mediated activation of the Flk1 promoter was further enhanced by OVOL2, although OVOL2 alone failed to activate it. Consistently, coexpression of ER71 and OVOL2 in differentiating embryonic stem cells led to a significant augmentation of FLK1+, endothelial, and hematopoietic cells. Such cooperative effects were impaired by the short hairpin RNA-mediated inhibition of Ovol2. Collectively, we show that ER71 directly interacts with OVOL2 and that such interaction is critical for FLK1+ cell generation and their differentiation into downstream cell lineages.
Introduction
It is widely accepted that blood cells are generated from 2 distinct cell populations during embryogenesis: hemangioblast and hemogenic endothelium.1,2 The blood islands (BIs) in the extraembryonic yolk sac as early as embryonic day (E) 7.5 is the first site of hematopoiesis. Both blood and endothelial cells in the BIs are generated from the hemangioblast. On the other hand, the hemogenic endothelium, a specialized endothelial cell population within the aorta-gonad-mesonephros, generates hematopoietic stem cells, which can populate hematopoietic organs including bone marrow. Despite the unsettled controversy regarding the hemangioblast and the hemogenic endothelium, it is evident that the generation of FLK1(VEGFR2)+ cells in the developing embryos is an essential step for both hematopoiesis and vessel development.3,4 In this regard, we and others have demonstrated that Er71/Etv2, a member of the ETS transcription factor family, is indispensable for specification of mesodermal precursors into endothelial and hematopoietic lineages during mouse embryogenesis.5-8 ER71 activates genes critical for both hematopoiesis and vessel development including Flk1, VECadherin (VE-Cad), Scl, and Lmo2 through direct binding to gene-associated promoters/enhancers.5,8-10 Studies in zebrafish and Xenopus also reported er71’s potent function,11,12 indicating an evolutionally conserved and essential role for ER71 in the establishment of the cardiovascular system.
The specificity and function of the majority of ETS-domain transcription factors are dependent upon their interacting proteins.13-15 However, very few studies have examined the role of ER71 interacting proteins.9,16 In this study, we show that ER71 interacts with OVOL2, a C2H2 zinc finger (ZF) transcription factor essential for early embryogenesis,17 and that this interaction plays an important role in the generation of FLK1+, as well as endothelial and erythroid cells.
Study design
Generation of inducible embryonic stem cells (ESCs)
Glutathione S-transferase (GST)-pull down assay and proteomics analysis
GST or GST-ER71 incubated overnight with 1 mg of whole cell lysate of day (D) 3.5 embryoid bodies (EBs) was washed, and the resulting beads were then subjected to liquid chromatography/mass spectrometry (MS) analysis. For complete details, see the supplemental Methods on the Blood Web site.
Results and discussion
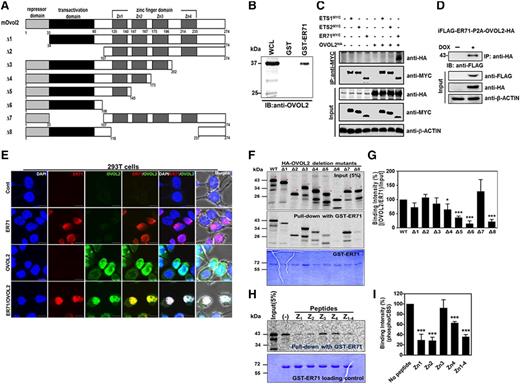
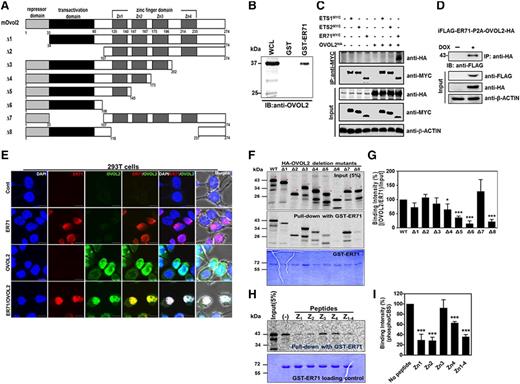
In this study we investigated whether ER71 could interact with other regulatory proteins to regulate FLK1+ cell generation in mouse ESC differentiation. To this end, a GST-ER71 fusion protein was incubated with lysates prepared from D3-3.5 EBs, a time at which the expression of ER71 reached its peak.5 Pull-down fractions were then subjected to liquid chromatography-MS/MS proteomic analysis. Among candidates, priority was given to OVOL2 (Figure 1A and supplemental Table 1), a member of the ZF transcription factor family, because Ovol2-deficient mouse embryos showed several developmental defects, including abnormal vessel formation.17,20 Subsequent pull-down assays between GST-ER71 and EB-cell lysates identified OVOL2 (Figure 1B). The interaction of ER71 and OVOL2 was further validated by coimmunoprecipitation in 293T cells (Figure 1C and supplemental Figure 1). Interestingly, OVOL2 did not interact with ETS1 or ETS2 (Figure 1C), which have been reported to activate the Flk1 promoter.21,22 This finding indicates that the binding of OVOL2 is specific to ER71. We also confirmed the colocalization of ER71 and OVOL2 in the nucleus of 293T cells by immunostaining (Figure 1E and supplemental Figure 2). To further characterize the interaction between ER71 and OVOL2, a series of deletion mutant forms of OVOL2 (Figure 1A) were subjected to in vitro pull-down with the GST-ER71 fusion protein. As shown in Figure 1F-G, in vitro translated wild-type (WT) OVOL2 (full-length, WT) were precipitated with GST-ER71, indicating direct interaction. Interestingly, OVOL2 mutants lacking ZF domains, especially Δ6 and Δ8, showed reduced binding to GST-ER71. The pull-down experiment with a GST protein control failed to precipitate OVOL2 (supplemental Figure 3). In agreement with these results, a binding inhibition assay showed that the peptides corresponding to each ZF domain of OVOL2 efficiently inhibited binding between ER71 and OVOL2 (Figure 1H-I). Collectively, these results suggest that ER71 can directly bind with OVOL2 partly through the ZF domains.
ER71 directly interacts with OVOL2. (A) A schematic diagram of OVOL2 and its deletion mutants. (B) GST-ER71 interacts with OVOL2. Binding between recombinant GST-ER71 and OVOL2 from D3.5 EB was determined by immunoblotting by anti-OVOL2 antibody. (C) Coimmunoprecipitation between ER71 and OVOL2. Cell lysates of 293T cells that were transfected with the control or expression plasmid of ER71-MYC, ETS1-MYC, ETS2-MYC, and/or HA-OVOL2 were immunoprecipitated (IP) with anti-MYC antibodies and immunoblotted with anti-MYC and anti-HA antibodies, respectively. β-ACTIN was used for loading control. (D) iFLAG-ER71-HA-OVOL2 ESCs in which the expression of ER71 and OVOL2 is dependent upon doxycycline (DOX) were differentiated in a serum-containing media, treated with DOX at D1 and harvested at D3.5 of differentiation. Subsequently, cell lysate was subjected to immunoprecipitation with anti-HA antibody, followed by immunoblotting with anti-FLAG antibodies. β-ACTIN was used for loading control. (E) Colocalization of ER71 and OVOL2; 293T cells transiently transfected with ER71-MYC and/or HA-OVOL2 were immunostained with antibodies against MYC or HA, and then analyzed by confocal microscopy. Red and green denote ER71 and OVOL2, respectively. DAPI was used for visualization of nucleus (blue), and all fluorescent images were overlaid with phase contrast images (Merge, last column of panels). All images are magnified views of the original images in supplemental Figure 2. Scale bars, 10 μm. (F) GST-ER71 pull-down with in vitro translated OVOL2. Purified GST-ER71 was incubated with each 35S-labeled WT or mutants OVOL2. Subsequently, pull-down products were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by analysis of radioactive signals by phosphor imager. The phosphor image of input amount of mutant OVOL2 given for binding (top); the phosphor image of mutant OVOL2 that was precipitated by GST-ER71 (middle), and Coomassie blue staining for GST-mER71 that was used for each binding (bottom). (G) Quantification of radioactive signals from in vitro pull-down assays. Binding affinity between GST-ER71 and 35S-labeled OVOL2 mutants were analyzed and presented in the graph. The density of 35S-labeled OVOL2 was normalized against that of GST-ER71, after which it was renormalized against OVOL2 input signal. Value from WT OVOL2 was given at 100%. Results are mean ± standard error of the mean (SEM) from 3 replications. (*P < .05 and ***P < .001 compared with WT). (H-I) Peptide competition assay. (H) The phosphor image of 35S-labeled OVOL2 precipitated with GST-ER71 in the absence or presence of ZF peptide. (I) Quantification of radioactive signals from peptide competition assays. Binding affinity between GST-ER71 and 35S-labeled OVOL2 were analyzed. The density of OVOL2 was normalized against that of GST-ER71. Results are mean ± SEM from 3 replications (***P < .001 compared with no peptide control).
ER71 directly interacts with OVOL2. (A) A schematic diagram of OVOL2 and its deletion mutants. (B) GST-ER71 interacts with OVOL2. Binding between recombinant GST-ER71 and OVOL2 from D3.5 EB was determined by immunoblotting by anti-OVOL2 antibody. (C) Coimmunoprecipitation between ER71 and OVOL2. Cell lysates of 293T cells that were transfected with the control or expression plasmid of ER71-MYC, ETS1-MYC, ETS2-MYC, and/or HA-OVOL2 were immunoprecipitated (IP) with anti-MYC antibodies and immunoblotted with anti-MYC and anti-HA antibodies, respectively. β-ACTIN was used for loading control. (D) iFLAG-ER71-HA-OVOL2 ESCs in which the expression of ER71 and OVOL2 is dependent upon doxycycline (DOX) were differentiated in a serum-containing media, treated with DOX at D1 and harvested at D3.5 of differentiation. Subsequently, cell lysate was subjected to immunoprecipitation with anti-HA antibody, followed by immunoblotting with anti-FLAG antibodies. β-ACTIN was used for loading control. (E) Colocalization of ER71 and OVOL2; 293T cells transiently transfected with ER71-MYC and/or HA-OVOL2 were immunostained with antibodies against MYC or HA, and then analyzed by confocal microscopy. Red and green denote ER71 and OVOL2, respectively. DAPI was used for visualization of nucleus (blue), and all fluorescent images were overlaid with phase contrast images (Merge, last column of panels). All images are magnified views of the original images in supplemental Figure 2. Scale bars, 10 μm. (F) GST-ER71 pull-down with in vitro translated OVOL2. Purified GST-ER71 was incubated with each 35S-labeled WT or mutants OVOL2. Subsequently, pull-down products were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by analysis of radioactive signals by phosphor imager. The phosphor image of input amount of mutant OVOL2 given for binding (top); the phosphor image of mutant OVOL2 that was precipitated by GST-ER71 (middle), and Coomassie blue staining for GST-mER71 that was used for each binding (bottom). (G) Quantification of radioactive signals from in vitro pull-down assays. Binding affinity between GST-ER71 and 35S-labeled OVOL2 mutants were analyzed and presented in the graph. The density of 35S-labeled OVOL2 was normalized against that of GST-ER71, after which it was renormalized against OVOL2 input signal. Value from WT OVOL2 was given at 100%. Results are mean ± standard error of the mean (SEM) from 3 replications. (*P < .05 and ***P < .001 compared with WT). (H-I) Peptide competition assay. (H) The phosphor image of 35S-labeled OVOL2 precipitated with GST-ER71 in the absence or presence of ZF peptide. (I) Quantification of radioactive signals from peptide competition assays. Binding affinity between GST-ER71 and 35S-labeled OVOL2 were analyzed. The density of OVOL2 was normalized against that of GST-ER71. Results are mean ± SEM from 3 replications (***P < .001 compared with no peptide control).
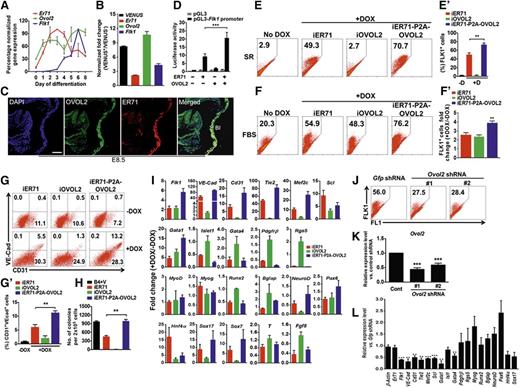
As reported previously,5 the expression of Er71 reached its peak at D3, followed by a sharp decrease in ESC differentiation, whereas that of Ovol2 increased steadily up to D6 (Figure 2A). The Flk1 message was detectable after the induction of Ovol2 or Er71. Further, Ovol2, as well as Flk1 message, was enriched in ER71-VENUS+ cells and ER71-VENUS+FLK1+ from E8.5 mouse embryos, in which VENUS expression was controlled by the endogenous Er71 locus (Figure 2B and supplemental Figure 4).6 Taken together, with the finding that ER71 and OVOL2 are coexpressed in the BIs at E8.5 (Figure 2C), these results suggest a functional significance of ER71-OVOL2 interaction in regulating FLK1+ cell generation and differentiation. To further test this, we performed a luciferase-based promoter assay and found that coexpression of OVOL2 and ER71 doubled the Flk1 promoter activity compared with ER71 alone (Figure 2D). OVOL2 itself did not increase the transcriptional activity of the Flk1 promoter used in this assay. Next, we generated doxycycline (DOX) inducible ESCs expressing: 1) FLAG-tagged ER71 (iER71), 2) HA-tagged OVOL2 (iOVOL2), and 3) both FLAG-tagged ER71 and HA-tagged OVOL2 (iER71-P2A-OVOL2) (supplemental Figure 5).18,19 We first confirmed the interaction between ER71 and OVOL2 in iER71-P2A-OVOL2 ESCs by coimmunoprecipitation (Figure 1D). Next, upon differentiation in a serum-free media,5 overexpression of ER71 significantly induced the generation of FLK1+ cells (Figure 2E). However, such de novo generation of FLK1+ cells was not observed in iOVOL2, iETS1, or iETS2 (Figure 2E and supplemental Figure 6). Consistent with the analysis of the Flk1 promoter (Figure 2D), the percentage of FLK1+ cells was higher in iER71-P2A-OVOL2 than in iER71 (73.4 ± 3.35% vs 50.2 ± 4.08%; Figure 2E-E’). We also found such a cooperative effect under differentiation conditions in the presence of serum (Figure 2F-F’). Interestingly, we found that the levels of ER71 were increased in cells overexpressing OVOL2-HA, which may also contribute to such a cooperative effect (supplemental Figure 7). Intriguingly, OVOL2 induction alone did not stimulate the Flk1 promoter nor did it generate FLK1+ cells in a serum-free differentiation condition (Figure 2E-E’), whereas in a serum condition, OVOL2 generated FLK1+ cells to a level comparable to that induced by ER71 (Figure 2F-F’). This indirectly suggests that OVOL2 might depend on other transcriptional regulators (eg, ER71) present in a serum condition to generate FLK1+ cells. Consistent with this, Er71 message was approximately fivefold higher in serum, compared with serum-free differentiation conditions (supplemental Figure 8). In a subsequent study, we found that the generation of endothelial cells (CD31+VE-Cad+) was increased by ER71-OVOL2 coexpression (11.3 ± 1.35%), compared with iER71 (6.1 ± 1.90%) or iOVOL2 ESCs (2.0 ± 1.69%) (Figure 2G-G’). Also, coexpression of ER71 and OVOL2 led to a further increase in the number of erythroid colonies, compared with iER71 or iOVOL2 (Figure 2H). Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis (Figure 2I) showed that the expression level of the genes critical for endothelial cell development (Flk1, VE-Cad, Cd31, Mef2c, and Tie2)5,9 were significantly higher when ER71 and OVOL2 were co-induced, compared with cells induced with either ER71 or OVOL2. Gata1, a critical gene for erythropoiesis,23 was also upregulated in iER71-P2A-OVOL2, compared with iER71 or OVOL2. In contrast, both Islet1 and Gata4, critical genes for cardiac development,24 were downregulated in iER71-P2A-OVOL2. Pdgfrβ and Rgs5 (smooth muscle cell markers) were further reduced in iER71-P2A-OVOL2 cells. MyoD and Myogenin (skeletal muscle markers) and Runx2 and Bglap (osteteoblast markers) were not significantly changed in the 3 cell lines. While other germ layer markers such as NeuroD and Pax6 (neuroectoderm) were similar or slightly enhanced upon coexpression of ER71 and OVOL2, Hinf4, Sox17, and Sox7 (endoderm) were similar in iER71 and iER71-P2A-OVOL2 cells and were reduced in iOVOL2 cells. The expressions of posterior primitive streak genes (T,Fgf8) were also not significantly changed in any of the cell lines.
OVOL2 enhances ER71-mediated FLK1+ cell generation, as well as hematopoietic and endothelial cell lineage development. (A) Expression of Er71, Ovol2, and Flk1 in ESC differentiation. The expression in each day point was normalized against Gapdh and 100% denotes the level where its maximum expression was reached. (B) VENUS+ cells and VENUS− cells from E8.5 ER71-VENUS embryos were subjected to qRT-PCR analysis. (C) E8.5 embryos were sectioned and subjected to immunohistochemical analysis of OVOL2 (green) and ER71 (red). DAPI (blue) was used for nucleus staining. Scale bar, 100 µm. (D) Synergistic activation of Flk1 promoter by ER71 and OVOL2. Expression constructs were transfected into 293T cells with pGL3 or pGL3-Flk1 promoter-luciferase reporter plasmids. The relative luciferase activity was gained by normalizing Firefly luciferase activity to Renilla luciferase activity at 48 hours posttransfection (***P < .001). (E-F) iER71, iOVOL2, and iER71-OVOL2 ESCs were differentiated either in a serum-free medium for 4 days (E), or in a serum-containing medium for 3.5 days (F), and subjected to fluorescence-activated cell sorter (FACS) analysis. Numbers in the plots denote the percentages of FLK1+ cells. DOX was added at D2 and D1 in a serum-free or a serum-containing differentiation medium, respectively. (E’) The percentage of FLK1+ cells in iER71, iOVOL2, and iER71-OVOL2 differentiated in a serum-free condition in the absence (-D) or presence (+D) of DOX. Results are mean ± SEM from 3 replications (**P < .01). (F’) Fold change of the generation of FLK1+ cells in ESCs after DOX treatment (+DOX/-DOX) in a serum-containing media. Results are mean ± SEM from 3 replications (**P < .01). (G, G’) FACS analysis for CD31/PECAM1 and VE-Cad in D6.5 EBs. iER71, iOVOL2, and iER71-OVOL2 ESCs differentiated in a serum-containing medium were treated with DOX at D3.5, followed by FACS analysis for CD31 and VE-Cad at D6.5 (**P < .01). (H) Hematopoietic replating assay. iER71, iOVOL2, and iER71-P2A-OVOL2 ESCs differentiated in a serum-free condition were treated with DOX at D3 and subjected to hematopoietic replating assay at D6. Colonies were counted 4 days later. B4, BMP4; V, VEGF-A. Results are mean ± SEM from 3 replications (**P < .01). (I) Gene expression profiling in D3.5 EBs overexpressing either ER71 (iER71) or OVOL2 (iOVOL2), or both ER71 and OVOL2 (iER71-P2A-OVOL2) in a serum-containing medium. DOX was added at D1 and RNA was prepared at D3.5. Expression of each gene was normalized against Gapdh, and the fold change of its expression level (+DOX/-DOX) was calculated. (J-L) iER71 ESCs infected with lentiviral shRNA particles against Ovol2 or Gfp control were differentiated in a serum-containing medium, and subjected to FACS analysis for FLK1 (J), and qRT-PCR for Ovol2 (K) (***P < .001). (L) Gene expression analysis in D3.5 EBs that had been infected with lentiviral particles of Ovol2 shRNAs (**P < .01; ***P < .001).
OVOL2 enhances ER71-mediated FLK1+ cell generation, as well as hematopoietic and endothelial cell lineage development. (A) Expression of Er71, Ovol2, and Flk1 in ESC differentiation. The expression in each day point was normalized against Gapdh and 100% denotes the level where its maximum expression was reached. (B) VENUS+ cells and VENUS− cells from E8.5 ER71-VENUS embryos were subjected to qRT-PCR analysis. (C) E8.5 embryos were sectioned and subjected to immunohistochemical analysis of OVOL2 (green) and ER71 (red). DAPI (blue) was used for nucleus staining. Scale bar, 100 µm. (D) Synergistic activation of Flk1 promoter by ER71 and OVOL2. Expression constructs were transfected into 293T cells with pGL3 or pGL3-Flk1 promoter-luciferase reporter plasmids. The relative luciferase activity was gained by normalizing Firefly luciferase activity to Renilla luciferase activity at 48 hours posttransfection (***P < .001). (E-F) iER71, iOVOL2, and iER71-OVOL2 ESCs were differentiated either in a serum-free medium for 4 days (E), or in a serum-containing medium for 3.5 days (F), and subjected to fluorescence-activated cell sorter (FACS) analysis. Numbers in the plots denote the percentages of FLK1+ cells. DOX was added at D2 and D1 in a serum-free or a serum-containing differentiation medium, respectively. (E’) The percentage of FLK1+ cells in iER71, iOVOL2, and iER71-OVOL2 differentiated in a serum-free condition in the absence (-D) or presence (+D) of DOX. Results are mean ± SEM from 3 replications (**P < .01). (F’) Fold change of the generation of FLK1+ cells in ESCs after DOX treatment (+DOX/-DOX) in a serum-containing media. Results are mean ± SEM from 3 replications (**P < .01). (G, G’) FACS analysis for CD31/PECAM1 and VE-Cad in D6.5 EBs. iER71, iOVOL2, and iER71-OVOL2 ESCs differentiated in a serum-containing medium were treated with DOX at D3.5, followed by FACS analysis for CD31 and VE-Cad at D6.5 (**P < .01). (H) Hematopoietic replating assay. iER71, iOVOL2, and iER71-P2A-OVOL2 ESCs differentiated in a serum-free condition were treated with DOX at D3 and subjected to hematopoietic replating assay at D6. Colonies were counted 4 days later. B4, BMP4; V, VEGF-A. Results are mean ± SEM from 3 replications (**P < .01). (I) Gene expression profiling in D3.5 EBs overexpressing either ER71 (iER71) or OVOL2 (iOVOL2), or both ER71 and OVOL2 (iER71-P2A-OVOL2) in a serum-containing medium. DOX was added at D1 and RNA was prepared at D3.5. Expression of each gene was normalized against Gapdh, and the fold change of its expression level (+DOX/-DOX) was calculated. (J-L) iER71 ESCs infected with lentiviral shRNA particles against Ovol2 or Gfp control were differentiated in a serum-containing medium, and subjected to FACS analysis for FLK1 (J), and qRT-PCR for Ovol2 (K) (***P < .001). (L) Gene expression analysis in D3.5 EBs that had been infected with lentiviral particles of Ovol2 shRNAs (**P < .01; ***P < .001).
To further determine whether ER71-OVOL2 interaction is functional in FLK1+ cell generation, we infected Ovol2 lentiviral short hairpin RNA (shRNA) particles into iER71 cells, and found that the FLK1+ cells induced by ER71 were significantly decreased in Ovol2 shRNAs-infected EBs compared with the Gfp shRNA-infected control (Figure 2J-K and supplemental Figure 9). Also, qRT-PCR analysis showed that Ovol2 knockdown led to a greater reduction of expression of the genes known to be induced by ER71 (Figure 2L). Other markers representing non-hemo/endo mesoderm derivatives (Isl1, Gata4, Pdgfrb, Rsg5, MyoD, Myogenin, Runx2, and Bglap) were slightly upregulated, or not affected in Ovol2 shRNA-infected EBs. While the expression of neuroectoderm markers (NeuroD and Pax6) appeared augmented in Ovol2 shRNA-infected EBs, messages of endodermal markers (Hinf4a and Sox17) were decreased. Thus, these results suggest that ER71-mediated FLK1+ cell generation is partly dependent upon OVOL2.
In summary, we demonstrate that ER71 interacts with OVOL2, and that such interaction cooperatively enhances the generation of FLK1+ cells, as well as endothelial and hematopoietic cells. A recent study reported that OVOL2 acting downstream of bone morphogenetic protein signaling can expand mesendoderm at the expense of neuroectoderm.25 However, our analysis clearly showed that OVOL2 in a serum-free condition failed to induce FLK1+ cell generation, suggesting that Ovol2 is not sufficient for FLK1+ mesoderm formation. Rather, OVOL2, in collaboration with ER71, directly activated the FLK1 promoter activity and was required for optimal endothelial and hematopoietic cell generation without altering the formation of the mesoderm.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Novina for providing lentiviral packaging and envelope plasmids, and Emory+Children's Pediatric Research Flow Cytometry Core.
This study was supported by grants from the March of Dimes Foundation (5-FY12-44) (C.P.), the American Heart Association (11SDG7390074) (C.P.), National Institutes of Health, National Heart, Lung, and Blood Institute (HL119291) (C.P.) and (HL105732) (D.M.O), and the National Research Foundation Stem Cell Program, Republic of Korea (NRF-2014M3A9B4043056) (J.-I.C.).
Authorship
Contribution: J.Y.K., R.H.L., and T.M.K., performed experiments and analyzed data; D.-W.K., Y.-J.J., S.-H.H., and S.-Y.O. performed experiments; M.K., H.K., K.C., and D.M.O. provided experimental materials; J.-I.C. conceived, designed research, analyzed data, and wrote the manuscript; C.P. conceived, designed, and performed experiments, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Current address for D.-W.K. is Department of Microbiology, Immunology, and Cancer Biology, School of Medicine, University of Virginia, Charlottesville, VA.
Correspondence: Jung-Il Chae, 664, 1Ga DeokJin, Jeonju 561-756, Republic of Korea; e-mail: jichae@jbnu.ac.kr; and Changwon Park, 2015 Uppergate Dr, #208B, Atlanta, GA 30322; e-mail: cpark23@emory.edu.
References
Author notes
J.Y.K., R.H.L., and T.M.K. contributed equally to this study.