Key Points
In the refractory cohort of the CLL2H trial PFS was significantly longer in patients with NOTCH1 mutation.
SF3B1 mutation had no impact on response rates or survival times in fludarabine-refractory patients.
Abstract
We studied the incidences, associations, and prognostic roles of NOTCH1 and SF3B1 mutations (NOTCH1mut, SF3B1mut) as compared with TP53mut in fludarabine-refractory chronic lymphocytic leukemia (CLL) patients treated with alemtuzumab in the CLL2H trial. We found NOTCH1mut, SF3B1mut, and TP53mut in 13.4%, 17.5%, and 37.4% of patients, respectively. NOTCH1mut and SF3B1mut were mutually exclusive, whereas TP53mut were evenly distributed within both subgroups. Apart from correlation of SF3B1mut with 11q deletion (P = .029), there were no other significant associations of the mutations with any baseline characteristics or response rates. However, NOTCH1mut cases had a significantly longer progression-free survival (PFS) compared with wild-type cases (15.47 vs 6.74 months; P = .025), although there was no significant difference with overall survival (OS). SF3B1mut had no significant impact on PFS and OS. In multivariable analyses, NOTCH1mut was identified as an independent favorable marker for PFS. This clinical trial is registered at www.clinicaltrials.gov as #NCT00274976.
Introduction
Genomic features are important pathogenic and prognostic factors in chronic lymphocytic leukemia (CLL).1,2 Recently, mutations in NOTCH1 and SF3B1 have been identified in CLL by next-generation sequencing.3-6 NOTCH1mut occurred in 5% to 15% of patients and resulted in a truncated, yet constitutively active protein.7 SF3B1mut have been identified in 10% to 20% of patients and have also notably been reported in myelodysplastic syndrome, pointing to the pathogenic role of pre-mRNA splicing in hematologic malignancies.5,6,8 Correlations have been described in CLL for NOTCH1mut with trisomy 12 and for SF3B1mut with deletion 11q. Both mutations have been associated with unmutated immunoglobulin heavy chain variable gene (IGHV) status, rapid progression, refractory disease, and short survival in initial studies on heterogeneous cohorts outside clinical trials.3-5,8-10 Refractory CLL can be explained only in about half of all patients by 17p deletion and/or TP53 mutation, but initial data have pointed to NOTCH1mut and SF3B1mut as alternative causes of resistance.3-5,8-11 Here, we evaluated NOTCH1mut and SF3B1mut compared with TP53mut in the CLL2H trial, a multicenter phase 2 trial of subcutaneous alemtuzumab in fludarabine-refractory CLL.12,13
Study design
Clinical results of the CLL2H trial of the German CLL Study Group (GCLLSG) have been reported.13 Genomic aberrations, IGHV status, TP53mut, β2-microglobulin, and thymidine kinase were analyzed in the central reference laboratories of the GCLLSG.14-17 NOTCH1 was studied by Sanger sequencing of 2 polymerase chain reaction fragments from the PEST domain (exon 34, chr9:139 390,619-139 391,290), whereas SF3B1 (exons 13-16) was analyzed by DHPLC (WAVE 3500HT; Transgenomic Inc.), with a limit of detection of 5% to 10% mutant allele, followed by sequencing (Big Dye Terminator Kit, ABI 3100 sequencer; Applied Biosystems, Germany). Details are available on request. Tumor load was at least 80%. Somatic origin of mutations was confirmed in CD19 negative fractions wherever available. Statistical analyses were performed by Fisher’s exact test, Kaplan-Meier estimates, and Cox proportional hazards regression with significance as P < .05 (two-sided). The study was approved by the ethics committee of Ulm University, and was conducted in accordance with the Declaration of Helsinki.
Results and discussion
The incidences of NOTCH1mut, SF3B1mut, and TP53mut were 13.4% (13 of 97), 17.5% (17 of 97), and 39.0% (39 of 100), respectively. The mutation frequencies of NOTCH1 and SF3B1, therefore, were not markedly increased in the CLL2H cohort of F-refractory CLL, as compared with recent data from large trials of patients in need of first treatment.18,19 This is in contrast to the much higher incidence of TP53mut in CLL2H as compared with the first-line trials, pointing to a greater role of p53 as a cause of refractoriness to F-based therapy.
Of note, in none of our 97 patients concurrent NOTCH1mut and SF3B1mut were found (Figure 1). This is consistent with previous studies on nonrefractory patients18-20 and points to different driver mutations in CLL. In contrast, TP53mut was found together with NOTCH1mut in 3 of 13 (23.1%; P = .349) cases and with SF3B1mut in 7 of 17 (41%; P = 1.000) cases, whereas in some previous studies mutual exclusivity was reported.8,10
Incidences and distributions of genetic markers. (A) Incidences of genetic markers for all patients with mutation data of NOTCH1 and SF3B1 available (n = 97). Rows represent genetic markers and columns represent individual patients, color-coded based on the marker status (red: NOTCH1mut; orange: SF3B1mut; yellow: TP53mut; green: 17p deletion; blue: 11q deletion; purple: trisomy 12; dark blue: 13q deletion; gray: IGHV unmutated [UM]). Hatched gray indicates missing data. (B) Circos diagrams illustrating pairwise co-occurrence of gene mutations with genomic aberrations (left) and with IGHV status (right). Circle segments depict the relative frequency of mutations, aberrations, or IGHV status and their co-occurrence. The width of the ribbons corresponds to the proportion of pairwise co-occurring mutations and aberrations or IGHV status. Unoccupied parts of the segments represent cases with only a single mutation or aberration.
Incidences and distributions of genetic markers. (A) Incidences of genetic markers for all patients with mutation data of NOTCH1 and SF3B1 available (n = 97). Rows represent genetic markers and columns represent individual patients, color-coded based on the marker status (red: NOTCH1mut; orange: SF3B1mut; yellow: TP53mut; green: 17p deletion; blue: 11q deletion; purple: trisomy 12; dark blue: 13q deletion; gray: IGHV unmutated [UM]). Hatched gray indicates missing data. (B) Circos diagrams illustrating pairwise co-occurrence of gene mutations with genomic aberrations (left) and with IGHV status (right). Circle segments depict the relative frequency of mutations, aberrations, or IGHV status and their co-occurrence. The width of the ribbons corresponds to the proportion of pairwise co-occurring mutations and aberrations or IGHV status. Unoccupied parts of the segments represent cases with only a single mutation or aberration.
The most common NOTCH1mut was the published deletion of 2 base pairs in exon 34 (c.7541_7542delCT; n = 11).3 Furthermore, a single base pair deletion was identified (c.7444delC; n = 2). The most common SF3B1mut was p.K700E (c.A2098G; n = 7). In addition, 10 different point mutations were found in 1 case each (supplemental Table 1, available on the Blood Web site). All SF3B1mut were heterozygous missense mutations in exons 14, 15 and 16, resulting in 10 different amino acid changes. No patient harbored more than 1 mutation and the mutation profile observed was in-line with previous studies on nonrefractory cohorts (supplemental Table 1).3,5,6,10,18,20
Sequential samples, obtained prior to first therapy and at the time of inclusion into CLL2H (refractory disease), were available for 13 patients (supplemental Table 2). NOTCH1mut were detected in 3 of 13 patients prior to therapy and 1 of these patients harbored 2 mutations (c.7541_7542delCT and c.7444delC). Interestingly, this patient had lost the delCT mutation at the time of refractory disease, whereas the other 3 NOTCH1mut remained present. Of note, 1 patient had a SF3B1mut prior to first therapy that was not detected anymore at inclusion into CLL2H. TP53mut were present in 3 of 13 patients before treatment and remained unchanged.
IGHV was unmutated in 71 of 90 (79%) cases overall, and in 100% of the NOTCH1mut (P = .062) cases, but only in 60.0% of the SF3B1mut patients (P = .078) (supplemental Table 3). We observed the expected increased incidences of 17p deletion in TP53mut (63%; P < .0001), and 11q deletion in SF3B1mut (47%; P = .029), but only a weak association of trisomy 12 with NOTCH1mut, which was likely due to small numbers (23%; P = .201).6,9,18,21 Of note, no SF3B1mut case harbored trisomy 12 (see also Oscier et al18 ), and there was a high rate of sole TP53mut (without 17p deletion) in this group (Figure 1).
Comparing baseline characteristics between patients with or without mutations, no significant differences were found for age, sex, stage, “B”-symptoms, prior lines of treatment, blood cell counts, lactate dehydrogenase, β2-microglobulin, thymidine kinase levels, or lymphadenopathy (supplemental Table 3).
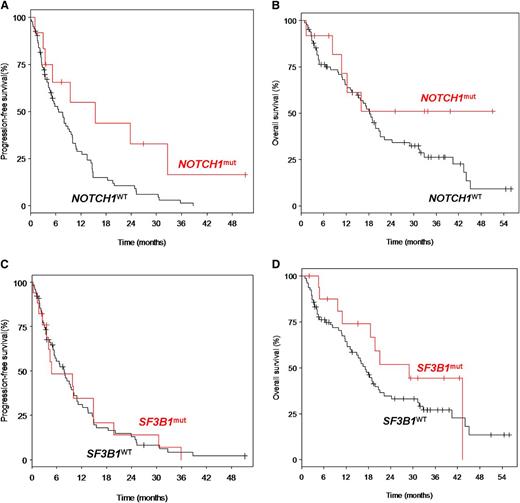
The overall response rate (ORR) among the 97 cases was 34% (3% complete remissions [CRs] and 31% partial remissions [PRs]). ORR was 50% (0% CR, 50% PR) and 41.2% (0% CR, 41.2% PR) for NOTCH1mut and SF3B1mut cases, respectively. After a median follow-up time of 33.9 months, the entire cohort had a median progression-free survival (PFS) of 7.7 months and a median overall survival (OS) of 18.3 months. NOTCH1mut cases showed a significantly longer PFS compared with wild-type cases (median 15.5 months vs 6.7 months; P = .025), but there was no significant difference in OS (median not reached vs 18.3 months; P = .181) (Figure 2). The PFS benefit may not translate into OS prolongation due to lack of efficacy of salvage chemotherapy after alemtuzumab failure, in particular in the NOTCH1mut subgroup. Moreover, the prognostic impact of NOTCH1mut seems to be dependent on the treatment context: other trials showed no impact on outcome after allo-stem cell transplantation22 and a differential impact on outcome after fludarabine/cyclophosphamide or fludarabine/cyclophosphamide/rituximab treatment.19 For SF3B1mut, there was no significant difference in PFS (median 4.8 vs 7.7; P = .974) and OS (median 29.0 vs 17.1; P = .243). There was no significant impact of TP53mut on ORR (P = .19), PFS (P = .80), or OS (P = .48). Confirmation of these findings is pending from other trials and the discrepancy between different series may be due to patient characteristics and treatments.
Estimated survival times according to NOTCH1mut and SF3B1mut. PFS was defined as the time from first drug administration to disease progression or death, and was censored at the initiation of subsequent treatment without progression. OS was defined as the time from first drug administration to death, and was censored at the time of allo-SCT. (A) Progression-free survival (PFS) of patients with NOTCH1mut (n = 12; median PFS: 15.47 months) was significantly longer as compared with patients with NOTCH1WT (n = 82: median: 6.74 months) (P = .025). (B) OS of patients with NOTCH1mut (n = 12; median OS: not reached) was not significantly different as compared with patients with NOTCH1WT (n = 82; median OS: 18.3 months) (P = .181). (C) PFS of patients with SF3B1mut (n = 17; median PFS: 4.76 months) was not significantly different as compared with patients with SF3B1WT (n = 77; median PFS: 7.72 months) (P = .974). (D) OS of patients with SF3B1mut (n = 17; median OS: 29.0 months) was not significantly different as compared with patients with SF3B1WT (n = 77; median OS: 17.1 months) (P = .243). WT, wild-type.
Estimated survival times according to NOTCH1mut and SF3B1mut. PFS was defined as the time from first drug administration to disease progression or death, and was censored at the initiation of subsequent treatment without progression. OS was defined as the time from first drug administration to death, and was censored at the time of allo-SCT. (A) Progression-free survival (PFS) of patients with NOTCH1mut (n = 12; median PFS: 15.47 months) was significantly longer as compared with patients with NOTCH1WT (n = 82: median: 6.74 months) (P = .025). (B) OS of patients with NOTCH1mut (n = 12; median OS: not reached) was not significantly different as compared with patients with NOTCH1WT (n = 82; median OS: 18.3 months) (P = .181). (C) PFS of patients with SF3B1mut (n = 17; median PFS: 4.76 months) was not significantly different as compared with patients with SF3B1WT (n = 77; median PFS: 7.72 months) (P = .974). (D) OS of patients with SF3B1mut (n = 17; median OS: 29.0 months) was not significantly different as compared with patients with SF3B1WT (n = 77; median OS: 17.1 months) (P = .243). WT, wild-type.
To identify markers of independent prognostic impact, we performed multivariable analyses, including clinical and biological parameters (age, Eastern Cooperative Oncology Group [ECOG] performance status, white blood cell count, thymidine kinase, β2-microglobulin, IGHV status, 11q deletion, 17p deletion, NOTCH1mut, SF3B1mut, TP53mut). Regarding PFS, ECOG performance status > 1 (hazard ratio [HR] 2.080; P = .057), increased thymidine kinase (>26.5 U/L [median], HR 1.479; P = .007), and elevated β2-microglobulin (>4.41 mg/L [median], HR 1.188; P = .0003) were identified as independent adverse factors, whereas NOTCH1mut (HR 0.375; P = .049) was a favorable marker. Regarding OS, none of the gene mutations showed significant prognostic impact, whereas age (at increments of 10 years) (HR 1.035; P = .024), ECOG performance status > 1 (HR 2.575; P = .017), and increased thymidine kinase (HR 1.568, P = .002) were identified as independent unfavorable factors.
In summary, our analysis performed in a homogenous and uniformly treated cohort of fludarabine-refractory CLL patients derived from a multicenter prospective trial showed NOTCH1mut in 13.4%, SF3B1mut in 17.5%, and TP53mut in 39.0% of the cases, with NOTCH1mut and SF3B1mut being mutually exclusive. As compared with CLL cohorts in need of first treatment, the incidence of NOTCH1mut and SF3B1mut appeared not to be markedly increased in contrast with TP53mut. NOTCH1mut was found as a favorable prognostic marker for PFS, but none of the mutations impacted OS in this trial.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Else Kröner Fresenius Stiftung (Else Kröner Forschungskolleg Ulm), the CLL Global Research Foundation (Alliance), the Virtual Helmholtz Institute (VH-VI-404, TP2), and the DFG (SFB 1074 project B2).
Authorship
Contribution: A.S., M.C., K.D., D.M., R.B., S.M., M.H., H.D., and S.S. designed research; T.Z., H.D., and S.S. provided patients and samples; A.S., P.P., M.R., T.Z., A.B., D.W., M.C., K.D., J.E., S.K., D.M., R.B., S.M., H.D., M.H., and S.S. performed research and analyzed data; M.H., H.D., and S.S. gave administrative support; and S.S. wrote the paper.
Conflict-of-interest disclosure: S.S. received research support and honoraria from Genzyme and Sanofi. The remaining authors declare no competing financial interests.
Correspondence: Stephan Stilgenbauer, Department of Internal Medicine III, Ulm University, Albert Einstein Allee 23, Ulm, 89081 Germany; e-mail: stephan.stilgenbauer@uniklinik-ulm.de.

![Figure 1. Incidences and distributions of genetic markers. (A) Incidences of genetic markers for all patients with mutation data of NOTCH1 and SF3B1 available (n = 97). Rows represent genetic markers and columns represent individual patients, color-coded based on the marker status (red: NOTCH1mut; orange: SF3B1mut; yellow: TP53mut; green: 17p deletion; blue: 11q deletion; purple: trisomy 12; dark blue: 13q deletion; gray: IGHV unmutated [UM]). Hatched gray indicates missing data. (B) Circos diagrams illustrating pairwise co-occurrence of gene mutations with genomic aberrations (left) and with IGHV status (right). Circle segments depict the relative frequency of mutations, aberrations, or IGHV status and their co-occurrence. The width of the ribbons corresponds to the proportion of pairwise co-occurring mutations and aberrations or IGHV status. Unoccupied parts of the segments represent cases with only a single mutation or aberration.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/7/10.1182_blood-2013-03-488197/4/m_1266f1.jpeg?Expires=1764368998&Signature=aHWaTPoM9QHLyvt9mqr383tmH7AyMfpkdhrgy9ZCzSexoxOXneuDhOCiQJjho6GOhPC1DurSoyk9Aio9GrAh7-J3wVmoIaOyyjYGjytTSSTVQebzrygWqOyyhdSo4JaeIbQ9Xz0W7V2-B~e-fiorZfFqjN-SsLDG2DJkH5qH7SHxi-kK1EFEsC1KUdxte-ifVj0xfVQ4ONyJKNilzkFGAQMh4K2BAqGYeGBKhMQ8tQ8KDZi1iuL5u44c3YhRVTyX4hgFItDlpT6rE9szejcOk9XeWJLpfRenD-NIo2W~h9VooIFyqt88vbeS8CgUPRkEEOrN8-Yru5ak2ZUvWJUzfQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)