Key Points
The crystal structure of pro-pseutarin C reveals how the prothrombinase complex assembles and suggests a mechanism of prothrombin processing.
Abstract
The prothrombinase complex, composed of the protease factor (f)Xa and cofactor fVa, efficiently converts prothrombin to thrombin by specific sequential cleavage at 2 sites. How the complex assembles and its mechanism of prothrombin processing are of central importance to human health and disease, because insufficient thrombin generation is the root cause of hemophilia, and excessive thrombin production results in thrombosis. Efforts to determine the crystal structure of the prothrombinase complex have been thwarted by the dependence of complex formation on phospholipid membrane association. Pseutarin C is an intrinsically stable prothrombinase complex preassembled in the venom gland of the Australian Eastern Brown Snake (Pseudonaja textilis). Here we report the crystal structures of the fX-fV complex and of activated fXa from P textilis venom and the derived model of active pseutarin C. Structural analysis supports a single substrate binding channel on fVa, to which prothrombin and the intermediate meizothrombin bind in 2 different orientations, providing insight into the architecture and mechanism of the prothrombinase complex—the molecular engine of blood coagulation.
Introduction
Blood coagulation (hemostasis) is a critical part of the innate immune response.1,2 It is initiated by tissue damage and results in the production of large amounts of thrombin, the effector hemostatic enzyme.3,4 Thrombin is a serine protease that cleaves multiple substrates, including the platelet receptor and fibrinogen, which polymerizes to form a fibrin clot.5 These activities are crucial for human health, with insufficient thrombin generation resulting in bleeding, including hemophilia, and excessive thrombin production causing thrombosis.6 Consequently, the complex that converts prothrombin to thrombin, the prothrombinase complex (fXa-fVa), lies at the heart of the hemostatic response. Factor Xa is a multidomain serine protease composed of a membrane-anchoring γ-carboxyglutamic acid (Gla) domain, 2 epidermal growth factor (EGF) domains, and a catalytic domain (Figure 1A). The cofactor, fVa, is composed of 3 A domains, an inhibitory B domain (removed on activation), and 2 membrane-anchoring C domains (A1-A2-B-A3-C1-C2, from N to C terminus; Figure 1A). In the blood, factors X and V circulate in zymogen and procofactor states until activated at the site of tissue damage. The 2 active proteins (fXa and fVa) associate with high affinity on activated cellular surfaces7 and convert prothrombin to thrombin by sequential cleavage of 2 sites8 (a schematic is given as supplemental Figure 1 on the Blood website). Prothrombin binds to the activated membrane surface and docks onto the assembled prothrombinase complex in a manner that promotes cleavage at the 320 site. This cleavage event and the concomitant conformational change (conversion from prothrombin to the partially active intermediate meizothrombin) results in the presentation of the second cleavage site (Arg271) and the subsequent release of soluble thrombin from its membrane-anchored profragment.9 Efficient processing of prothrombin by the prothrombinase complex is required to form a stable blood clot at the site of tissue damage. Thus, understanding how prothrombinase is assembled is of critical importance for determining how coagulation is regulated, understanding the basis of bleeding and thrombotic disorders, and to design novel pro- and anticoagulant therapies. Previous efforts to determine the crystal structure of human prothrombinase have been thwarted by the membrane dependence of stable complex formation. The Australian Eastern Brown Snake (Pseudonaja textilis) has “weaponized” its liver-expressed plasma versions of fV and fX for expression in the venom gland,10 where the activated cofactor and protease assemble in a tight complex (called pseutarin C) capable of efficient thrombin generation in the absence of membranes.11 Pseutarin C cleaves human prothrombin in a sequential manner, similar to human prothrombinase in the presence of phospholipid vesicles (supplemental Figure 2); however, P textilis fVa has evolved to bind to membranes with low affinity. We solved crystal structures of the complex formed between P textilis venom fV and fX and of fXa bound to a key fV peptide. These structures were used to assemble a model of full-length active pseutarin C. Analysis of the structural features of the complex and putative fVa glycosylation sites across all available sequences suggest a conserved prothrombin binding site and a mechanism by which sequential processing is conferred.
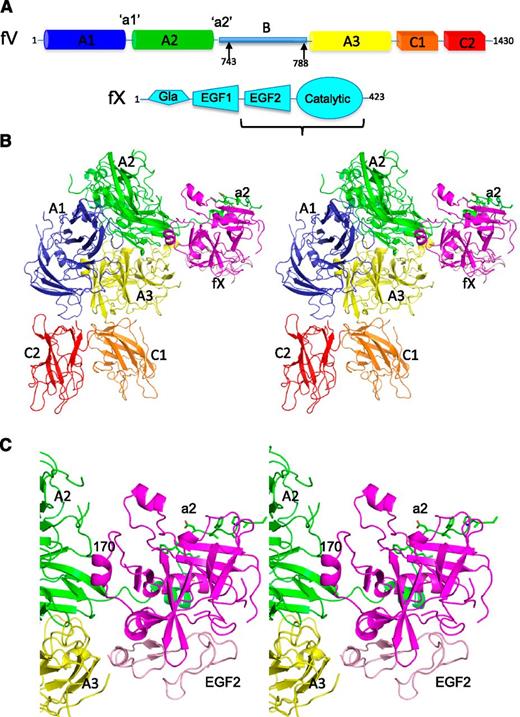
Domain organization and crystallographic structure of pro-pseutarin C. (A) Schematics (not to scale) are given depicting the domain organization of P textilis fV and fX. Factor V has a short B domain that can be released by thrombin cleavage at the 2 sites indicated by arrows; however, such processing is unnecessary for cofactor activity. The fX construct used in crystallization studies (EGF2-catalytic domain) is indicated by the bracket. (B) A stereo view of the crystal structure of the fV-fX complex is shown in cartoon representation, with fV colored according to domain (A1 domain in blue, A2 domain in green, A3 domain in yellow, C1 domain in orange, and C2 domain in red) and fX in magenta (EGF2 domain in light pink). The a2 region is shown as green sticks. The orientation shown here is referred to as the front throughout. (C) Close-up of the fV-fX interface.
Domain organization and crystallographic structure of pro-pseutarin C. (A) Schematics (not to scale) are given depicting the domain organization of P textilis fV and fX. Factor V has a short B domain that can be released by thrombin cleavage at the 2 sites indicated by arrows; however, such processing is unnecessary for cofactor activity. The fX construct used in crystallization studies (EGF2-catalytic domain) is indicated by the bracket. (B) A stereo view of the crystal structure of the fV-fX complex is shown in cartoon representation, with fV colored according to domain (A1 domain in blue, A2 domain in green, A3 domain in yellow, C1 domain in orange, and C2 domain in red) and fX in magenta (EGF2 domain in light pink). The a2 region is shown as green sticks. The orientation shown here is referred to as the front throughout. (C) Close-up of the fV-fX interface.
Methods
Proteins
Full-length venom P textilis fX DNA (accession number AY631238.1) was synthesized (GeneArt), and the region encoding EGF2-catalytic domains (residues 86-423) was amplified, digested using EcoRI/HindIII, and ligated into the pET23 vector (Novagen). Protein was expressed in BL21 Star (DE3) Escherichia coli (Invitrogen), which were grown in 2× TY and induced with 1 mM isopropyl β-d-1-thiogalactopyranoside for 4 hours at 37°C. The fX construct was refolded and purified using a method previously employed to refold prethrombin-2.12 Briefly, cells were lysed by sonication, and inclusion bodies were separated by centrifugation, washed 3 times, and solubilized in 10 mL (per liter of bacterial culture) of 0.01% (v/v) trifluoroacetic acid, 6.3 M GndHCl, and 30 mM l-cysteine for 90 minutes at room temperature. After centrifugation, solubilized protein was diluted with 40 mL of 6 M GndHCl in refolding buffer (0.6 M l-arginine, 50 mM Tris, 0.5 M NaCl, 1 mM EDTA, 10% glycerol, 0.2% [w/v] Brij58, and 1 mM l-cysteine, pH 8.3) and refolded by slow (8 mL/min) addition of 2 L of refolding buffer with stirring, followed by stationary overnight incubation at room temperature. Refolded protein was concentrated, dialyzed overnight against 5 L of 20 mM Tris, 0.25 M NaCl, 0.1% (w/v) polyethylene glycol 6000, pH 8.0, filtered, loaded onto a 5-mL HiTrap Heparin HP column (GE Healthcare) equilibrated in 20 mM Tris and 0.25 M NaCl, pH 8.0, and eluted with a 0.25-L M NaCl gradient. Contaminating proteins were removed with a HiTrap Q HP column (GE Healthcare) equilibrated in the same buffer. The gene coding the venom form of P textilis fV (AY168281.1) with the human signal sequence was purchased from GeneArt and cloned into the pED vector for expression in BHK M cells, as previously described.11 Arg residues 742 and 788, corresponding to activation sites, were mutated to glutamine to improve the homogeneity of the protein preparations (fV-QQ). A 2-step purification using Q FF and S FF ion exchange columns was used.
Structure of the fV-fX complex
The S195A mutation (catalytic Ser-to-Ala, chymotrypsin numbering) was made to fX using the QuikChange mutagenesis kit (Stratagene), and the protein was expressed and purified as detailed above. Purified P textilis venom fV-QQ and S195A fX were individually concentrated to 13 and 3.3 mg/mL, respectively, and dialyzed into crystallization buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 2 mM CaCl2, and 50 µM CuCl2). Factor X was then added to fV in a 1.2-fold molar excess and incubated for 30 minutes at room temperature to allow complex formation. The complex was then purified from free fX on a HiLoad 16/600 Superdex 200 column (GE Healthcare) equilibrated in crystallization buffer. Fractions containing the complex, as detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, were pooled and concentrated to 16.9 mg/mL. Crystallization trials were established using the sitting drop vapor-diffusion method in 96-well Medical Research Council (United Kingdom) plates with 200 nL of protein and 200 nL of precipitant. Initial crystals appeared after 3 days at 20°C in 0.1 M Morpheus buffer 3, 30% Morpheus P20K_P550MME, and 100 mM Morpheus carboxylic acids (Morpheus HT; Molecular Dimensions). A single diffraction quality crystal was obtained in 0.1 M Morpheus buffer 3, 20% Morpheus P20K_P550MME, and 50 mM Morpheus carboxylic acids using the hanging drop vapor-diffusion method with 2 µL protein and 1 µL reservoir solution after 24 days at 37°C. The crystal was harvested after 34 days and cryoprotected by replacing the 20% P20K_P550MME with 25% P550MME and a subsequent stepwise increase to 35% P550MME. The crystal was cryocooled in liquid nitrogen, and diffraction data were collected at Diamond Light Source (DLS) beamline I04 to ∼4 Å. The data were indexed with iMosflm in space group I43212 and scaled and merged using Scala and ctruncate from the CCP4 suite.13 Phases were obtained by molecular replacement with Phaser14 using the high-resolution structure of P textilis venom fV-QQ (unpublished) and bovine fXa (EGF2-catalytic domain construct, PDB ID code 1KIG15 ) as search models. Model building and refinement were conducted in Refmac16 and Coot.17
After 7 months at 20°C, a single large crystal appeared in a drop containing 3 µL protein and 1 µL reservoir solution (0.1 M Morpheus buffer 3, 20% Morpheus P20K_P550MME, and 50 mM Morpheus carboxylic acids). The crystal was cryoprotected as before, and a full dataset to 3.3 Å was collected at DLS beamline I04 (100 K; wavelength, 0.88560 Å). Data processing and model building were performed as described above. The previous Rfree set was extended to 3.3 Å, and the low-resolution structure was used as the initial model. The 2.7-Å structure of P textilis fXa (see below) was used in later rounds of refinement as a new starting model for fX. Refinement was conducted with Refmac, and statistics are given in supplemental Table 1. Ramachandran analysis of the final model (using Rampage) gave 95.4% residues in favored or allowed regions (81.4% favored, 14% in allowed, 4.6% outliers).
Structure of the fXa-Glu-Gly-Arg-chloromethylketone-a2 peptide complex
Activation of our fX construct in sufficient amounts for crystallography required the replacement of the native P textilis activation site (PDIR) with the human sequence (NLTR) by site-directed mutagenesis. For activation, the protein was dialyzed into 20 mM Tris, pH 7.4, 100 mM NaCl, 1 mM CaCl2, 0.002% NaN3, and 0.5% Triton X100, and furin (5 U/mg; New England Biolabs) was added to cleave between the EGF2 domain and the activation peptide. After full cleavage, the protein was repurified using a HiTrap Heparin HP column, as before, and subsequently activated with RVV-X (1:50 [w/w]; Haematologic Technologies) in the presence of 10 mM benzamidine HCl and a twofold excess of Glu-Gly-Arg-chloromethylketone (EGRCK; Haematologic Technologies). Activation was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions. After full activation, a fresh fivefold excess of EGRCK was added to the protein to ensure complete inhibition. The complex was repurified by HiTrap Heparin HP chromatography. The a2 peptide (663GNEEEEEDDGDIFADIFI680) was synthesized by Peptide Synthetics (Fareham, United Kingdom) and was resuspended in water/NaOH (final pH 5.0), aliquoted, and dehydrated in a vacuum concentrator. The EGRCK-inhibited fXa was then concentrated to 5.75 mg/mL, dialyzed against 20 mM Tris, pH 8, and 100 mM NaCl, and added to a fivefold molar excess of the dehydrated peptide. This complex was crystallized in 96-well sitting drop plates as above, yielding small crystals in 100 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.5, and 12% polyvinylpyrrolidone K12 (PVP) (MIDAS-HT screen; Molecular Dimensions). After optimization, a diffraction quality crystal grew in 45 mM MES, pH 5.5, and 5.6% PVP K12, with initial incubation at 10°C and subsequent transfer to 20°C, and was cryoprotected in 25% glycerol, 12% PVP K12, and 0.1 M MES, pH 5.5, and then cooled in liquid nitrogen. Diffraction data were collected at DLS beamline I04 (100 K; wavelength, 0.97950 Å). Data from 1 crystal diffracting to 2.7 Å were indexed in iMosflm in space group I213. Data suffered from twinning with an apparent space group of I4132 and were scaled and merged in Aimless from the CCP4 suite. Molecular replacement was conducted with Phaser using a trimmed homology search model (prepared in MODELER18 ) of P textilis fXa based on human fXa (PDB ID code 1HCG19 ). Refinement and model building were performed in Refmac (twinned refinement) and Coot, respectively. Ramachandran analysis of the final model (Rampage) gave 93.3% in favored, 6.5% in allowed, and 0.2% in outlier regions. Statistics are given in supplemental Table 1.
Modeling
All figures were made using PyMol.20 Modeling was conduced using Modeler18 and Coot. All 37 known and putative fV sequences were aligned to identify potential N-linked glycosylation sites. Only when found in ≥2 sequences was a site selected. These included residues 20, 23, 28, 104, 126, 150, 174, 194, 198, 212, 270, 355, 374, 376, 433, 441, 527, 561, 640, 874, 914, 971, 1062, 1150, 1166, 1217, 1367, 1389, and 1415 (in P textilis fV numbering, ignoring the B domains).
Results
Structure of pro-pseutarin C
Full-length P textilis fV was expressed in BHK-M cells (nonactivatable QQ variant11 ), and a truncated version of fX (EGF2-catalytic domain, S195A variant) was produced in E coli. Unprocessed P textilis venom fV is fully active in the absence of proteolytic processing11,21 and was used for crystallization and functional studies. We found that the complex of fV with our truncated fXa was able to convert prothrombin to thrombin in the absence and presence of membranes at rates comparable to the full-length construct (supplemental Figure 3). In addition, the unactivated fX bound to fV with comparable affinity to fXa (supplemental Figure 4), and this “zymogen” complex (pro-pseutarin C) was able to convert prothrombin to thrombin, although at a very low rate (see “Methods”; supplemental Figure 5). Early difficulties in activating venom fX (“Methods”) made it necessary to established crystal trials of the pro-pseutarin C. However, because the unactivated components fV and fX assembled with high affinity and formed a partially active complex, we concluded that it was likely to resemble the activated complex. A single crystal of pro-pseutarin C provided useful X-ray diffraction data to 3.3-Å resolution, and the structure was solved by molecular replacement using a high-resolution structure of P textilis fV (unpublished data; S. Krishnaswamy) and bovine EGF2-catalytic domain fXa15 (Figure 1B; supplemental Table 1). Despite the moderate resolution, electron density was of high quality and revealed several features not present in the molecular replacement models, including branched N-linked carbohydrate chains (supplemental Figure 6A), a coordinated copper ion (supplemental Figure 6B), and a 30° rotation of the C2 domain (supplemental Figure 6C). In fV, the 3 A domains are organized into an equilateral triangle, similar to previous structures of ceruloplasmin and fVIII (supplemental Figure 7A-B), with the membrane-anchoring C domains protruding from the triangle, like short legs. The C domains have moved relative to the positions observed in fVIII and the apo-structure of P textilis fV, perhaps reflecting a degree of independent C domain mobility (supplemental Figure 7B-C). A single copper ion was found at the interface of the A1 and A3 domains, coordinating side chain atoms from each domain (supplemental Figure 6B), and calcium ions were found coordinated to homologous positions of the A1 and A3 domains. Overall, 1262 of 1430 residues of fV were modeled into electron density, with 130 of 168 missing residues contributed by the short B domain. For fX, 248 of 339 residues were modeled into electron density, with missing regions corresponding to the zymogen activation domain of chymotrypsin family serine proteases (including the 140, 180, and 220 loops in chymotrypsin numbering; supplemental Figure 8).22 The 90 loop of P textilis fX contains an 8-residue insert relative to mammalian fX orthologs and is fully resolved in electron density.
It is known that the A2 domain of fVa is critical for fXa binding,23 because its release on cleavage by activated protein C dissociates the complex.24,25 The A3 domain has also been implicated in direct binding to fXa.26 In our structure, fX is bound to the interface between the A2 and A3 domains of fV (Figure 1C). The 170 loop of the protease domain binds to the A2 domain, and the EGF2 domain interacts with the A3 domain. The unstructured region following the A2 domain (the highly acidic a2 region preceding the B domain) wraps around fX in the vicinity of its heparin-binding site (analogous to exosite II of thrombin), making electrostatic and hydrophobic interactions, consistent with previous biochemical data.27,28 A total of 2366 Å2 is buried at the interface between the 2 proteins, with contacting residues listed in supplemental Table 2.
Structure of the fXa-a2 peptide complex
Although clear density for a portion of the a2 region was observed in the fV-fX structure, the low resolution and ionic nature of the interface made it difficult to determine its precise position and register. To improve the structure of fX and to independently establish the mode of interaction between the a2 region and fX in pro-pseutarin C, we determined the crystal structure of P textilis fXa (inhibited by the covalent active-site inhibitor EGRCK) with the peptide GNEEEEEDDGDIFADIFI, corresponding to residues 663 to 680 of the a2 region of fV (supplemental Table 1; supplemental Figure 9). Conversion of P textilis fX to fXa was made possible by replacing the sequence of the activation loop with that of the human ortholog (“Methods”). The Glu-rich region of the a2 peptide could not be resolved in electron density; however, it is in position to make several ionic interactions with the highly basic heparin binding site of fXa. The lack of fixed binding mode for ionic interactions is consistent with the nondirectional nature of salt bridges and the sandwiching of the Glu-rich region between 2 heparin binding sites in the asymmetric unit.29 Interestingly, the resolved hydrophobic portion of the peptide corresponds well to what was originally observed in the fV-fX structure (supplemental Figure 10A). Previous studies have demonstrated the importance of the heparin binding region of fXa,30 and the a2 regions of known fV sequences all contain a highly acidic region followed by a hydrophobic stretch (supplemental Figure 10B), suggesting that the a2-fXa interaction is an important and conserved part of prothrombinase assembly.
Model of pseutarin C
A model of activated, full-length pseutarin C was created based on the structures described above. Missing loops and side chains were built and minimized for fVa, and our fXa structure was superimposed on fX, and the missing Gla and EGF1 domains were modeled based on the structure of full-length porcine fIXa.31 In the resulting model of pseutarin C, the membrane-binding domains of fVa (C1 and C2) and fXa (Gla) are coplanar, allowing the complex to dock on a membrane surface without any conformational rearrangement (Figure 2A). A perpendicular arrangement of the complex on a membrane results in a fXa active site-to-membrane distance of ∼60 Å and a fVa Cys540-to-membrane distance of ∼90 Å, consistent with previous fluorescence resonance energy transfer measurements.32,33 The total buried surface area for the complex is 4543 Å2, with large contributions from the A2, a2, and A3 portions of fVa and the catalytic and EGF domains of fXa (Figure 2B-C; supplemental Table 2). The interface is largely electrostatic, with fXa presenting a highly basic face to an acidic surface of fVa, including the a2 peptide. To help interpret previous biochemical work and to guide future studies, the numbering for human fVa and fXa is also provided in supplemental Table 2.
Model of full-length membrane-docked pseutarin C. (A) A model of the full-length pseutarin C complex (fXa in cyan and fVa colored as in Fig. 1), including missing loops and domains (the a1 loop in magenta), is shown bound to a phospholipid membrane surface (a lipid bilayer is shown as sticks). The Gla domain of fXa (calcium ions depicted as green spheres) and the membrane-binding C domains of fVa are coplanar, suggesting a similar extent of membrane penetration. A ∼10-Å penetration into the membrane is consistent with previous fluorescence resonance energy transfer measurements from the membrane to residue Cys540 of the A2 domain and to the active site of fXa in human prothrombinase (measurements indicated in right panel; EGRCK inhibitor shown as spheres). (B) The nature of the fVa-fXa interface is illustrated by coloring the surface of fVa according to electrostatic potential (red, negative; blue, positive) and showing fXa as a semitransparent cartoon. (C) The surface of fXa is shown colored according to electrostatics (highly basic region indicated by oval), with fVa (semitransparent, colored as in panel A).
Model of full-length membrane-docked pseutarin C. (A) A model of the full-length pseutarin C complex (fXa in cyan and fVa colored as in Fig. 1), including missing loops and domains (the a1 loop in magenta), is shown bound to a phospholipid membrane surface (a lipid bilayer is shown as sticks). The Gla domain of fXa (calcium ions depicted as green spheres) and the membrane-binding C domains of fVa are coplanar, suggesting a similar extent of membrane penetration. A ∼10-Å penetration into the membrane is consistent with previous fluorescence resonance energy transfer measurements from the membrane to residue Cys540 of the A2 domain and to the active site of fXa in human prothrombinase (measurements indicated in right panel; EGRCK inhibitor shown as spheres). (B) The nature of the fVa-fXa interface is illustrated by coloring the surface of fVa according to electrostatic potential (red, negative; blue, positive) and showing fXa as a semitransparent cartoon. (C) The surface of fXa is shown colored according to electrostatics (highly basic region indicated by oval), with fVa (semitransparent, colored as in panel A).
Location of the prothrombin binding site
How prothrombinase recognizes its substrate is of critical importance for understanding the cofactor effect of fVa and the efficient and sequential processing of prothrombin. Although our structures and the resulting model do not include prothrombin or the intermediate meizothrombin, they do provide clues as to how and where prothrombin might bind. Prothrombin consists of 2 rigid domains separated by a long unstructured linker34 (supplemental Figure 11). The crystal structure of the N-terminal portion, composed of the Gla and kringle 1 domains (together known as F1), revealed a tight association between the 2 domains into a single rigid unit.35 The C-terminal portion of prothrombin is composed of the second kringle domain (K2) and the catalytic domain, which also associate into a rigid unit due to the tight interaction of K2 with the catalytic domain (together known as prethrombin-1 [Pre1]36 ). F1 and Pre1 are connected by a 26-residue unstructured linker, suggesting that each rigid domain binds independently to prothrombinase. The F1 will anchor Pre1 (the domain containing the 2 cleavage sites) a certain distance from the membrane surface, and it is likely that direct interactions between Pre1 and fVa will orient the 2 cleavage sites (320 and 271) with respect to the active site of fXa.
Biochemical and mutagenesis studies have attempted to define the prothrombin binding site on fVa, but with mixed and often conflicting results.37 We are now able to address the issue of prothrombin docking and sequential cleavage in a structurally directed manner. An important and interesting observation from our structure is that the active site of fXa is pointing away from the membrane toward the A1-A2 domain interface (up and to the left in the orientation shown in Figure 2A, left), suggesting that Pre1 docks on the ledge formed by the A1 and A2 domains and the connecting a1 loop. This hypothesis is strengthened by an analysis of potential N-linked glycosylation sites in all available fV sequences (supplemental Figure 12), which reveals a unique glycosylation-free area available for protein-protein interaction on the A1-a1-A2 ledge. This approach is supported by the complete lack of potential glycosylation sites in the large region of fVa that interacts with fXa in our model of pseutarin C (supplemental Figure 13), as well as previous site-directed glycosylation studies on human prothrombinase.26 Where F1 binds is less clear, but analysis of the glycosylation pattern revealed a continuous unglycosylated channel running from the C2 domain on the membrane to the active site of fXa, including the A1-a1-A2 ledge.
Substrate allostery confers sequential processing
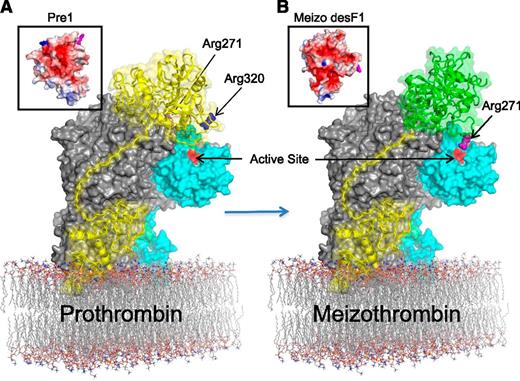
On the basis of our structures, the derived model, and analysis, we propose a hypothesis for the initial docking of prothrombin and the mechanism of the sequential cleavage of the 320 and 271 sites (Figure 3). F1 docks on the membrane in the vicinity of the C2 domain, perhaps making favorable contacts,38 and the linker region will thread along the unglycosylated channel to place Pre1 on the A1-a1-A2 ledge. The proposed Pre1 binding site on fVa is highly basic in nature (supplemental Figure 12E) and would therefore predictably bind to an acidic surface of Pre1. Indeed, an analysis of the electrostatic properties of Pre1 reveals a single acidic face spanning the catalytic domain and the kringle 2 domain (Figure 3A, left inset). The positioning of this acidic face of Pre1 on the basic A1-a1-A2 ledge places the 320 loop of prothrombin close to the active site of fXa. We predict that when prothrombin is initially docked, the loop containing the 271 site makes contacts with fVa in a position where it is not accessible to the active site of fXa.
Proposal for the mechanism of sequential prothrombin cleavage by prothrombinase. (A) The surface of pseutarin C is shown, with fVa in gray and fXa in cyan and the active site colored red. Prothrombin (yellow cartoon with semitransparent surface) is docked onto the side of fVa, with F1 (Gla-EGF1) binding to the C2 domain. The Pre1 portion of prothrombin (Pre1 surface electrostatic representation as inset) docks onto fVa in a manner that feeds Arg320 (blue) into the active site of fXa while keeping the 271 site (magenta) remote. (B) Cleavage of the 320 site causes the zymogen-to-protease conformational change in the catalytic domain of prothrombin (yielding the active intermediate meizothrombin; right inset) and an altered interaction with the K2 domain (indicated by change from yellow to green). The change in surface properties of meizothrombin (Meizo des F1; inset) result in an adjusted interaction with fVa, and the presentation of Arg271 to the active site of fXa.
Proposal for the mechanism of sequential prothrombin cleavage by prothrombinase. (A) The surface of pseutarin C is shown, with fVa in gray and fXa in cyan and the active site colored red. Prothrombin (yellow cartoon with semitransparent surface) is docked onto the side of fVa, with F1 (Gla-EGF1) binding to the C2 domain. The Pre1 portion of prothrombin (Pre1 surface electrostatic representation as inset) docks onto fVa in a manner that feeds Arg320 (blue) into the active site of fXa while keeping the 271 site (magenta) remote. (B) Cleavage of the 320 site causes the zymogen-to-protease conformational change in the catalytic domain of prothrombin (yielding the active intermediate meizothrombin; right inset) and an altered interaction with the K2 domain (indicated by change from yellow to green). The change in surface properties of meizothrombin (Meizo des F1; inset) result in an adjusted interaction with fVa, and the presentation of Arg271 to the active site of fXa.
The high affinity and intimacy of the pseutarin C complex supports the 2-substrate model of sequential cleavage of prothrombin,39 where prothrombin docks in 1 orientation and conversion to the intermediate meizothrombin results in a different substrate-binding mode (a ratcheting 2-substrate40 model, rather than a ping-pong 2-enzyme model41 ). It has been demonstrated that the zymogen-to-protease conformational change after cleavage of the 320 site drives the reorientation of meizothrombin on fVa.42 The structural features of Pre1 and meizothrombin, coupled with the identification of the putative substrate binding site on prothrombinase help to rationalize the ratcheting mechanism. The structures of Pre1 and meizothrombin36,43 show that Pre1 undergoes a large conformational rearrangement when converted to meizothrombin. The conformational change involves the ordering of the catalytic domain and a repositioning of the K2 domain. The result is a large alteration to the surface electrostatics (Figure 3, insets) that would drive the reorientation of meizothrombin to present the 271-loop to the active site of fXa (Figure 3B). Determination of the precise details of the interactions between fVa and its 2 substrates (Pre1 and meizothrombin) will ultimately require crystal structures; however, our model suggests a plausible mechanism for the previously described allosteric ratcheting of prothrombin.
Plasma vs venom prothrombinase
P textilis adapted its regulated plasma prothrombinase into a weapons-grade enzyme (pseutarin C) with 116 mutations: 49 in fV (21 of which are in the B domain) and 67 in fX (supplemental Figure 14). The paucity of mutations in fV (only 3.4%) suggests that the brunt of the selective pressure was borne by fX (19.8%). Five mutations in the C domains of fV replace basic residues with neutral or acidic residues, predictably reducing the affinity of pseutarin C for negatively charged membranes. Surprisingly, none of the mutations in fV is in position to affect binding to fXa; however, several lie within the predicted prothrombin binding site and might result in improved processing of the substrate in the absence of membranes. The fX mutations include the surface loops that contact fV in our structure and loops that line the active site cleft. The effect of the fX mutations might be to improve affinity for fVa, to improve substrate recognition and processing, and to slow inhibition of the complex by the principal endogenous fXa inhibitor in mammalian blood: antithrombin.44 Indeed, we found that P textilis fXa was inhibited by human AT with a second-order rate constant of ∼5 M−1s−1, in the absence or presence of fV, giving pseutarin C a half-life of ∼17 hours in human blood (supplemental Figure 15). Together, these mutations ensure that the pseutarin C injected into prey or victim will persist and circulate throughout the vasculature converting prothrombin to thrombin, resulting in the systemic consumption of fibrinogen and ultimately in hemorrhagic death.
Discussion
In this study, we exploited the high affinity and membrane independence of the P textilis venom fV-fX complex to determine its crystal structure. Although not in its activated form, the fV-fX complex was found to be capable of thrombin formation. This is perhaps not surprising considering the full activity of the unprocessed P textilis fV11 and the ability of cofactors, such as fVa, to allosterically activate zymogen or zymogen-like forms of proteases. Examples include the allosteric activation of fVII/fVIIa by tissue factor at the initiation stage of blood coagulation45 and the activation of dead mutants of thrombin46 and fXa47 by their cofactors thrombomodulin and fVa, respectively. There are several examples of unprocessed zymogens with nearly full catalytic activity, including the fibrinolytic factor single-chain tissue plasminogen actvator. In addition, a recent report has demonstrated the formation of a human antithrombin-fX complex, suggesting that human fX is also catalytically competent to some extent.48 The similar affinities between the components of the activated and zymogen forms of pseutarin C also support a single conserved mode of binding. We therefore consider it reasonable to conclude that the interface between fV and fX revealed in our crystal structure is the same as that formed with the active components of pseutarin C. Due to the low resolution of the structure, some details of the interaction are not observed, and the flexibility inherent in certain regions of the zymogen fX also influenced what could be observed in the structure. However, the positions of the 2 proteins and the interfaces used for binding are clear. In addition, we solved a medium-resolution structure of P textilis fXa bound to an active site inhibitor and the a2 peptide that provided independent validation of the a2-fX interaction and allowed a simple superposition to arrive at a model of fully active pseutarin C. Structurally directed mutagenesis studies on orthologs will be needed to determine whether the observed assembly of pseutarin C is general to prothrombinase across species. However, the evolutionary heritage (gene duplication to derive the venom from the plasma form), the sequence identity/similarity to all known fV and fX sequences, and the preserved order of prothrombin cleavage argue strongly for a single conserved architecture of the prothrombinase complex. The structures and derived model also provide important clues as to how prothrombin binds and is processed, supporting a mechanism where an allosteric change in the substrate, after the first cleavage event, alters the binding mode to present the second cleavage site. In conclusion, the structure and analysis presented here provide insight into the formation and function of the prothrombinase complex—the molecular engine of physiological and pathological blood coagulation.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the British Heart Foundation (to J.A.H.) and National Institutes of Health, National Heart, Lung, and Blood Institute grants R01-HL88010 and P01 HL74124 (Project 2) (to R.M.C.). This work was carried out with the support of the Diamond Light Source.
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 4BXS and 4BXW.
Authorship
Contribution: B.C.L. crystallized pseutarin C and the fXa-a2 complex, collected diffraction data, and solved and refined the structures; T.A.M.-R. expressed and purified fV and determined binding affinities and activities; D.J.D.J. determined rates of fXa inhibition by antithrombin; T.E.A. assessed the activities of prothrombinase complexes under various conditions; S.K. provided the coordinates of P textilis fV; R.M.C. helped with protein expression; and J.A.H. conceived the experiments, helped in solving and refining the structures, analyzed the structures, conducted molecular modeling, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for B.C.L. is Program in Apoptosis and Cell Death Research, Sanford-Burnham Medical Research Institute, La Jolla, CA.
Correspondence: James A. Huntington, University of Cambridge, Department of Haematology, Cambridge Institute for Medical Research, Wellcome Trust/MRC Building, Hills Rd, Cambridge, CB2 0XY, United Kingdom; e-mail: jah52@cam.ac.uk.
References
Author notes
B.C.L. and T.A.M.-R. contributed equally to this work.